CesH Represses Cereulide Synthesis as an Alpha/Beta Fold Hydrolase in Bacillus cereus
Abstract
1. Introduction
2. Results
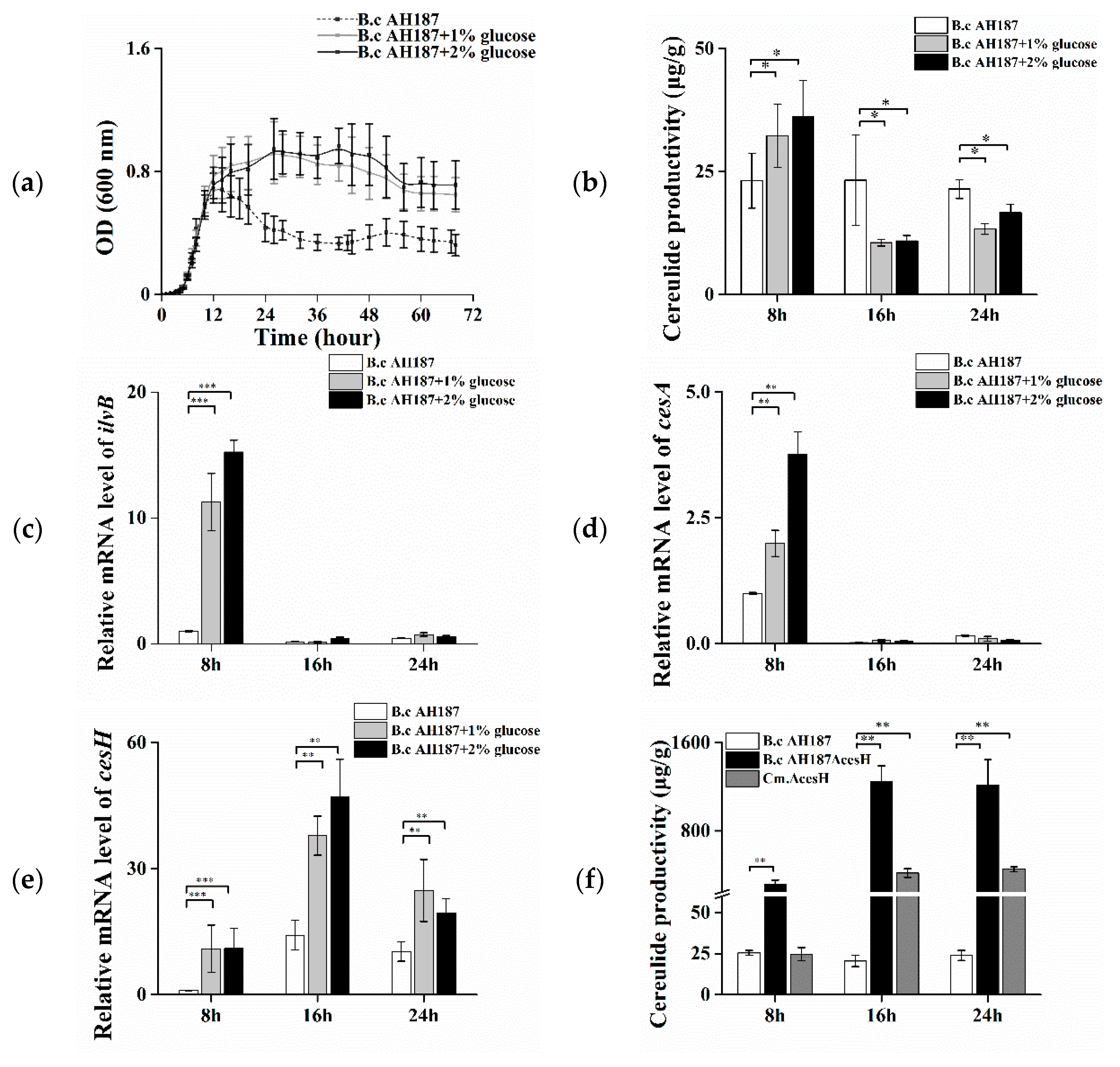
2.1. The Negative Effect of cesH on Cereulide Production
2.2. Prediction and Characterization of CesH as a Membrane Associated Esterase
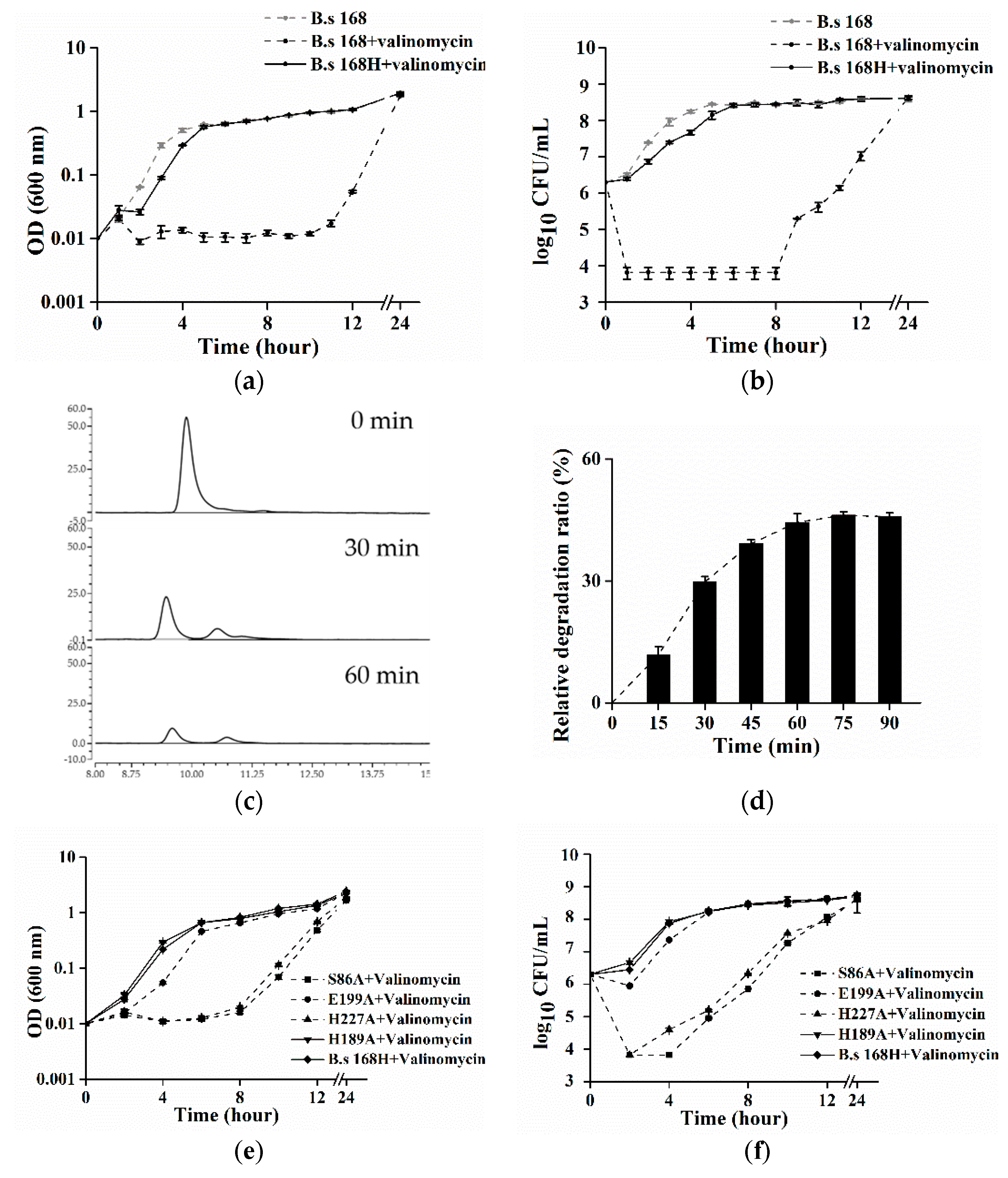
2.3. The Effect of CesH on Cereulide Analogue–Valinomycin In Vitro and In Vivo
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, Primers, and Growth Condition
4.2. Extraction and Quantification of Cereulide via High Performance Liquid Chromatography (HPLC)
4.3. Bioinformatic Analysis of CesH
4.4. RT-PCR and DNA Manipulations
4.5. Growth Measurements
4.6. Activity Assays of CesH
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Makarasen, A.; Yoza, K.; Isobe, M. Higher structure of cereulide, an emetic toxin from Bacillus cereus, and special comparison with valinomycin, an antibiotic from Streptomyces fulvissimus. Chem. Asian J. 2009, 4, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Dommel, M.K.; Frenzel, E.; Strasser, B.; Blöchinger, C.; Scherer, S.; Ehling-Schulz, M. Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods. Appl. Environ. Microbiol. 2010, 76, 1232–1240. [Google Scholar] [CrossRef]
- Suwan, S.; Isobe, M.; Ohtani, I.; Agata, N.; Mori, M.; Ohta, M. Structure of cereulide, a cyclic dodecadepsipeptide toxin from Bacillus cereus and studies on NMR characteristics of its alkali metal complexes including a conformational structure of the K+ complex. J. Chem. Soc. Perkin Trans. 1 1995, 1, 765–775. [Google Scholar] [CrossRef]
- Daniele, R.P.; Holian, S.K. A potassium ionophore (valinomycin) inhibits lymphocyte proliferation by its effects on the cell membrane. Proc. Natl. Acad. Sci. USA 1976, 73, 3599–3602. [Google Scholar] [CrossRef] [PubMed]
- Hoornstra, D.; Andersson, M.A.; Teplova, V.V.; Mikkola, R.; Uotila, L.M.; Andersson, L.C.; Roivainen, M.; Gahmberg, C.G.; Salkinoja-Salonen, M.S. Potato crop as a source of emetic Bacillus cereus and cereulide induced mammalian cell toxicity. Appl. Environ. Microbiol. 2013, 79, 3534–3543. [Google Scholar] [CrossRef] [PubMed]
- Hensley, J.R.; Hanson, J.B. The action of valinomycin in uncoupling corn mitochondria. Plant Physiol. 1975, 56, 13–18. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Furlong, I.J.; Mediavilla, C.L.; Ascaso, R.; Rivas, A.L.; Collins, M.K. Induction of apoptosis by valinomycin: Mitochondrial permeability transition causes intracellular acidification. Cell Death Differ. 1998, 5, 214. [Google Scholar] [CrossRef]
- Saiz-Jimenez, C. Molecular Biology and Cultural Heritage, 1st ed.; Routledge: London, UK, 2017; pp. 93–105. [Google Scholar]
- Cui, Y.; Liu, Y.; Liu, X.; Xia, X.; Ding, S.; Zhu, K. Evaluation of the toxicity and toxicokinetics of cereulide from an emetic Bacillus cereus strain of milk origin. Toxins 2016, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Granum, P.E.; Lund, T. Bacillus cereus and its food poisoning toxins. FEMS. Microbiol. Lett. 1997, 157, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Häggblom, M.M.; Apetroaie, C.; Andersson, M.A.; Salkinoja-Salonen, M.S. Quantitative analysis of cereulide, the emetic toxin of Bacillus cereus, produced under various conditions. Appl. Environ. Microbiol. 2002, 68, 2479–2483. [Google Scholar] [CrossRef] [PubMed]
- Fricker, M.; Messelhäußer, U.; Busch, U.; Scherer, S.; Ehling-Schulz, M. Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl. Environ. Microbiol. 2007, 73, 1892–1898. [Google Scholar] [CrossRef]
- Naranjo, M.; Denayer, S.; Botteldoorn, N.; Delbrassinne, L.; Veys, J.; Waegenaere, J.; Sirtaine, N.; Driesen, R.B.; Sipido, K.R.; Mahillon, J. Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 2011, 49, 4379–4381. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.; Al, M.N.; Doloy, A.; Jacqmin, S.; Ghiglione, S.; Verroust, N.; Poyart, C.; Ozier, Y. Bacillus cereus, an unusual cause of fulminant liver failure: Diagnosis may prevent liver transplantation. J. Med. Microbiol. 2012, 61, 743–745. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altayar, M.; Sutherland, A. Bacillus cereus is common in the environment but emetic toxin producing isolates are rare. J. Appl. Microbiol. 2006, 100, 7–14. [Google Scholar] [CrossRef]
- Hoton, F.M.; Fornelos, N.; N’guessan, E.; Hu, X.; Swiecicka, I.; Dierick, K.; Jääskeläinen, E.; Salkinoja-Salonen, M.; Mahillon, J. Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ. Microbiol. Rep. 2009, 1, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, E.; Kranzler, M.; Stark, T.D.; Hofmann, T.; Ehling-Schulz, M. The endospore-forming pathogen Bacillus cereus exploits a small colony variant-based diversification strategy in response to aminoglycoside exposure. mBio 2015, 6, e01172-15. [Google Scholar] [CrossRef]
- Valero, M.; Salmeron, M. Antibacterial activity of 11 essential oils against Bacillus cereus in tyndallized carrot broth. Int. J. Food Microbiol. 2003, 85, 73–81. [Google Scholar] [CrossRef]
- Morente, E.O.; Abriouel, H.; López, R.L.; Omar, N.B.; Gálvez, A. Antibacterial activity of carvacrol and 2-nitro-1-propanol against single and mixed populations of foodborne pathogenic bacteria in corn flour dough. Food Microbiol. 2010, 27, 274–279. [Google Scholar] [CrossRef]
- Kranzler, M.; Stollewerk, K.; Rouzeau-Szynalski, K.; Blayo, L.; Sulyok, M.; Ehling-Schulz, M. Temperature exerts control of Bacillus cereus emetic toxin production on post-transcriptional levels. Front. Microbiol. 2016, 7, 1640. [Google Scholar] [CrossRef]
- Jääskeläinen, E.; Häggblom, M.; Andersson, M.; Salkinoja-Salonen, M. Atmospheric oxygen and other conditions affecting the production of cereulide by Bacillus cereus in food. Int. J. Food Microbiol. 2004, 96, 75–83. [Google Scholar] [CrossRef]
- Biesta-Peters, E.G.; Reij, M.W.; Blaauw, R.H.; Rajkovic, A.; Ehling-Schulz, M.; Abee, T. Quantification of the emetic toxin cereulide in food products by liquid chromatography-mass spectrometry using synthetic cereulide as a standard. Appl. Environ. Microbiol. 2010, 76, 7466–7472. [Google Scholar] [CrossRef]
- Toh, M.; Moffitt, M.; Henrichsen, L.; Raftery, M.; Barrow, K.; Cox, J.; Marquis, C.; Neilan, B. Cereulide, the emetic toxin of Bacillus cereus, is putatively a product of nonribosomal peptide synthesis. J. Appl. Microbiol. 2004, 97, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Ehling-Schulz, M.; Svensson, B.; Guinebretiere, M.-H.; Lindbäck, T.; Andersson, M.; Schulz, A.; Fricker, M.; Christiansson, A.; Granum, P.E.; Märtlbauer, E. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 2005, 151, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Shivers, R.P.; Sonenshein, A.L. Bacillus subtilisilvB operon: An intersection of global regulons. Mol. Microbiol. 2005, 56, 1549–1559. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vidal, M.; Jayasinghe, S.; Ladokhin, A.S.; White, S.H. Folding Amphipathic Helices Into Membranes: Amphiphilicity Trumps Hydrophobicity. J. Mol. Biol. 2007, 370, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Szeto, T.H.; Rowland, S.L.; Rothfield, L.I.; King, G.F. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl. Acad. Sci. USA 2002, 99, 15693–15698. [Google Scholar] [CrossRef] [PubMed]
- Lücking, G. Molecular Characterization of Cereulide Synthesis in Emetic Bacillus cereus. Doctoral Dissertation, Technische Universität München, München, Germany, 2009. [Google Scholar]
- Park, H.I.; Lee, S.; Ullah, A.; Cao, Q.; Sang, Q.-X.A. Effects of detergents on catalytic activity of human endometase/matrilysin 2, a putative cancer biomarker. Anal. Biochem. 2010, 396, 262–268. [Google Scholar] [CrossRef]
- Tempelaars, M.H.; Rodrigues, S.; Abee, T. Comparative analysis of antimicrobial activity of valinomycin and cereulide, the Bacillus cereus emetic toxin. Appl. Environ. Microbiol. 2011, 77, 2755–2762. [Google Scholar] [CrossRef] [PubMed]
- Lücking, G.; Frenzel, E.; Rütschle, A.; Marxen, S.; Stark, T.D.; Hofmann, T.; Scherer, S.; Ehling-Schulz, M. Ces locus embedded proteins control the non-ribosomal synthesis of the cereulide toxin in emetic Bacillus cereus on multiple levels. Front. Microbiol. 2015, 6, 1101. [Google Scholar] [CrossRef]
- Hotelier, T.; Renault, L.; Cousin, X.; Negre, V.; Marchot, P.; Chatonnet, A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res. 2004, 32, D145–D147. [Google Scholar] [CrossRef]
- Mindrebo, J.T.; Nartey, C.M.; Seto, Y.; Burkart, M.D.; Noel, J.P. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016, 41, 233–246. [Google Scholar] [CrossRef]
- Suplatov, D.; Besenmatter, W.; Švedas, V.; Svendsen, A. Bioinformatic analysis of alpha/beta-hydrolase fold enzymes reveals subfamily-specific positions responsible for discrimination of amidase and lipase activities. Protein Eng. Des. Sel. 2012, 25, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Holmquist, M. Alpha beta-hydrolase fold enzymes structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef] [PubMed]
- Ladeuze, S.; Lentz, N.; Delbrassinne, L.; Hu, X.; Mahillon, J. Antifungal activity displayed by cereulide, the emetic toxin produced by Bacillus cereus. Appl. Environ. Microbiol. 2011, 77, 2555–2558. [Google Scholar] [CrossRef]
- Agata, N.; Ohta, M.; Mori, M.; Shibayama, K. Growth conditions of and emetic toxin production by Bacillus cereus in a defined medium with amino acids. Microbiol. Immunol. 1999, 43, 15–18. [Google Scholar] [CrossRef]
- UniProt Consortium, T. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2018, 46, 2699. [Google Scholar] [CrossRef]
- Velluet, E.; Lenfant, N.; Marchot, P.; Hotelier, T.; Bourne, Y.; Chatonnet, A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: Tools to explore diversity of functions. Nucleic Acids Res. 2012, 41, D423–D429. [Google Scholar]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, 372–379. [Google Scholar] [CrossRef]
- Ge, Y.; Hu, X.; Zhao, N.; Shi, T.; Cai, Q.; Yuan, Z. A new tubRZ operon involved in the maintenance of the Bacillus sphaericus mosquitocidal plasmid pBsph. Microbiology 2014, 160, 1112–1124. [Google Scholar] [CrossRef]
- Zhao, N.; Ge, Y.; Shi, T.; Hu, X.; Yuan, Z. Collagen-like glycoprotein BclS is involved in the formation of filamentous structures of the Lysinibacillus sphaericus exosporium. Appl. Environ. Microbiol. 2014, 80, 6656–6663. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmeitzl, C.; Varga, E.; Warth, B.; Kugler, K.G.; Malachová, A.; Michlmayr, H.; Wiesenberger, G.; Mayer, K.F.X.; Mewes, H.-W.; Krska, R.; et al. Identification and Characterization of Carboxylesterases from Brachypodium distachyon Deacetylating Trichothecene Mycotoxins. Toxins 2016, 8, 6. [Google Scholar] [CrossRef] [PubMed]

| Strains and Plasmids | Characteristics | Reference |
|---|---|---|
| B. cereus AH187 | Wild cereulide-producing isolate | Ehling-Schulz, 2005 |
| B. cereus AH187 ΔcesH | a kanamycin gene fragment was taken the place of cesH in AH187; KanaR | This study |
| B. cereus AH187 Cm.ΔcesH | AH187ΔcesH complemented with pHT315-cesH; KanaR EryR | This study |
| B. cereus AH187 gfp | AH187 containing pHT315-gfp | This study |
| B. cereus AH187 cesH-gfp | AH187 containing pHT315-cesH-gfp | pHT315-cesH-gfp |
| B. subtilis 168 | Wild isolate | Spizizen, 1958 |
| B. subtilis 168H | B. subtilis 168 containing pHT315-cesH for expression; EryR | This study |
| B. subtilis S86A | B. subtilis 168H containing an alanine substitution at serine 86 of CesH; EryR | This study |
| B. subtilis H189A | B. subtilis 168H containing an alanine substitution at histidine 189 of CesH; EryR | This study |
| B. subtilis E197A | B. subtilis 168H containing an alanine substitution at glutamate 197 of CesH; EryR | This study |
| B. subtilis E199A | B. subtilis 168H containing an alanine substitution at glutamate 199 of CesH; EryR | This study |
| B. subtilis H227A | B. subtilis 168H containing an alanine substitution at histidine 227 of CesH; EryR | This study |
| E. coli BL21 (DE3) pET28a-cesH | E. coli BL21 (DE3) containing pET28a-cesH for protein expression | This study |
| pMD18T-Kana-gfp | pMD18T plasmid containing a kanamycin resistance cassette and a gfp gene | This study |
| pHT315ts-cesH::kana | Shutter vector containing the flank region of cesH and kanamycin resistance fragment, and a temperature sensitive replicon | This study |
| pHT315-cesH | Shutter plasmid containing the intact cesH fragment and its promoter | This study |
| pHT315-gfp | Shutter plasmid containing the intact gfp fragment and the promoter of cesH | This study |
| pHT315-cesH-gfp | Shutter plasmid containing the intact cesH with a gfp fragment fused in the c-terminal | This study |
| pET28a-cesH | Expression vector containing the complete sequence of cesH | This study |
| Order | Sequence | Features |
|---|---|---|
| 1 | GCGGGATCCGCAATCCCCCCTAGCTATG | Forward primer for cloning up stream of cesH (KpnI) |
| 2 | GCGGGTACCTCTAACACATTCATATAGTA | Reverse primer for cloning up stream of cesH (BamHI) |
| 3 | GCGAAGCTTTATTTCAATTTCATACGGGTA | Forward primer for cloning down stream of cesH (SalI) |
| 4 | GCGGTCGACAATTTTAGCTCTTTAGTTCC | Reverse primer for cloning down stream of cesH (HindIII) |
| 5 | CGGGGATCCAGCGAACCATTTGAGGTGATAG | Forward primer for cloning kanamycin resistance cassette (BamHI) |
| 6 | CGGGTCGACCTAGGTACTAAAACAATTCATCCAG | Reverse primer for cloning kanamycin resistance cassette (SalI) |
| 7 | GCGGGTACCCAAACAAATTAGATAAGTGGATAGAGAGACA | Forward primer for cloning promoter (KpnI) |
| 8 | GCGGTTCTTCTCCTTTACTCATGCAATCCCCCCTAGCTATG | Reverse primer for gfp element overlapping pcr |
| 9 | GCGGTGGCAATAGGTTTCGCTTTAGGATCCCAAGTA | Forward primer for generating mutation at residue 86 |
| 10 | GCGTACTTGGGATCCTAAAGCGAAACCTATTGCCAC | Reverse primer for generating mutation at residue 86 |
| 11 | GCGCATTCAGGGAATACTCAGCTAATATATTAGTTACTGT | Forward primer for generating mutation at residue 189 |
| 12 | GCGACAGTAACTAATATATTAGCTGAGTATTCCCTGAATG | Reverse primer for generating mutation at residue 189 |
| 13 | GCGATATTAGTTACTGTTGGTGCTAAAGAGAAAAAAATAATG | Forward primer for generating mutation at residue 197 |
| 14 | GCGCATTATTTTTTTCTCTTTAGCACCAACAGTAACTAATAT | Reverse primer for generating mutation at residue 197 |
| 15 | GCGACTGTTGGTGAAAAAGCTAAAAAAATAATGAAGGAT | Forward primer for generating mutation at residue 199 |
| 16 | GCGATCCTTCATTATTTTTTTAGCTTTTTCACCAACAGT | Reverse primer for generating mutation at residue 199 |
| 17 | GCGATTCCTAAAATTGGTGCTGGGATACCTTTAGCA | Forward primer for generating mutation at residue 277 |
| 18 | GCGTGCTAAAGGTATCCCAGCACCAATTTTAGGAAT | Reverse primer for generating mutation at residue 277 |
| 19 | GCGCCCGCGCTGCCACTATTAGATACCAATTTCACTTCATC | Reverse primer for gfpligation |
| 20 | GCGGAATTCATGTATTATACAGAATTTGGAACGGATC | Forward primer for cloning cesH (EcoRI) |
| 21 | GCGAAGCTTTCAACTATTAGATACCAATTTCACTTC | Reverse primer for cloning cesH (HindIII) |
| 22 | GCGCATAGCTAGGGGGGATTGCATGAGTAAAGGAGAAGAAC | Forward primer for gfp element overlapping pcr |
| 23 | GCGGTACCTTATTTGTAGAGCTCATCCATGC | Reverse primer for cloning gfp element (KpnI) |
| 24 | AAGCCTGATGAATTAGTTATTG | Forward primer for ilvB in RT-PCR assay |
| 25 | CTGGTTGACACGATAGTAA | Reverse primer for ilvB in RT-PCR assay |
| 26 | GATTACGTTCGATTATTTGAAG | Forward primer for cesA in RT-PCR assay |
| 27 | CGTAGTGGCAATTTCGCAT | Reverse primer for cesA in RT-PCR assay |
| 28 | TGCTTAGTTCTTGACCTA | Forward primer for cesH in RT-PCR assay |
| 29 | CACAACAGACTTACCTTC | Reverse primer for cesH in RT-PCR assay |
| 30 | GGAGGAAGGTGGGGATGACG | Forward primer for 16s rrn in RT-PCR assay |
| 31 | ATGGTGTGACGGGCGGTGTG | Reverse primer for 16s rrn in RT-PCR assay |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Xiong, H.; Geng, P.; Yuan, Z.; Hu, X. CesH Represses Cereulide Synthesis as an Alpha/Beta Fold Hydrolase in Bacillus cereus. Toxins 2019, 11, 231. https://doi.org/10.3390/toxins11040231
Tian S, Xiong H, Geng P, Yuan Z, Hu X. CesH Represses Cereulide Synthesis as an Alpha/Beta Fold Hydrolase in Bacillus cereus. Toxins. 2019; 11(4):231. https://doi.org/10.3390/toxins11040231
Chicago/Turabian StyleTian, Shen, Hairong Xiong, Peiling Geng, Zhiming Yuan, and Xiaomin Hu. 2019. "CesH Represses Cereulide Synthesis as an Alpha/Beta Fold Hydrolase in Bacillus cereus" Toxins 11, no. 4: 231. https://doi.org/10.3390/toxins11040231
APA StyleTian, S., Xiong, H., Geng, P., Yuan, Z., & Hu, X. (2019). CesH Represses Cereulide Synthesis as an Alpha/Beta Fold Hydrolase in Bacillus cereus. Toxins, 11(4), 231. https://doi.org/10.3390/toxins11040231





