The Cytotoxicity Effect of Resveratrol: Cell Cycle Arrest and Induced Apoptosis of Breast Cancer 4T1 Cells
Abstract
1. Introduction
2. Results
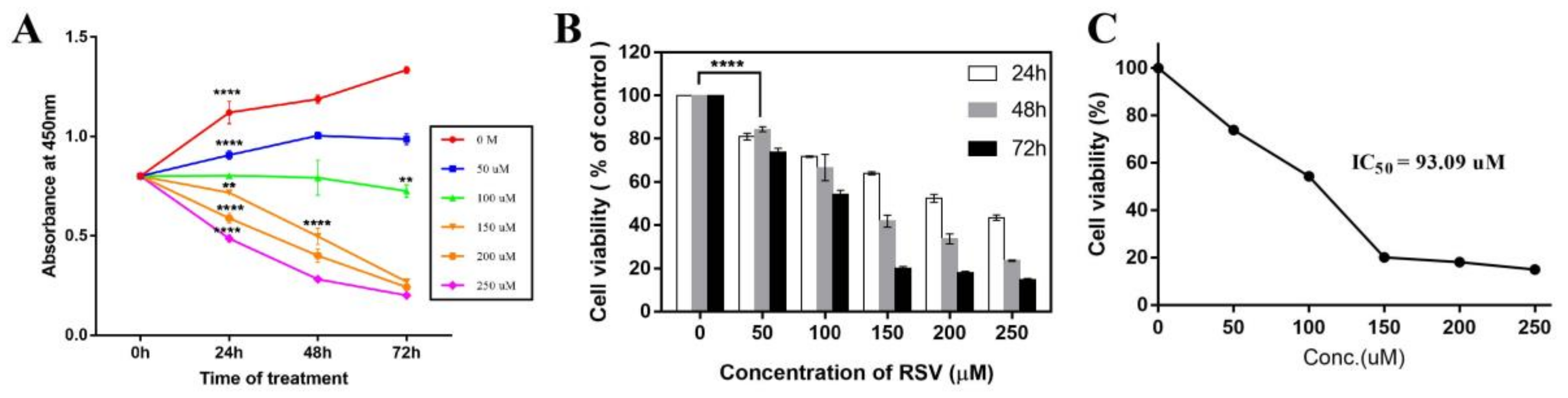
2.1. Resveratrol Significantly Inhibits the Proliferation of Cancer Cells
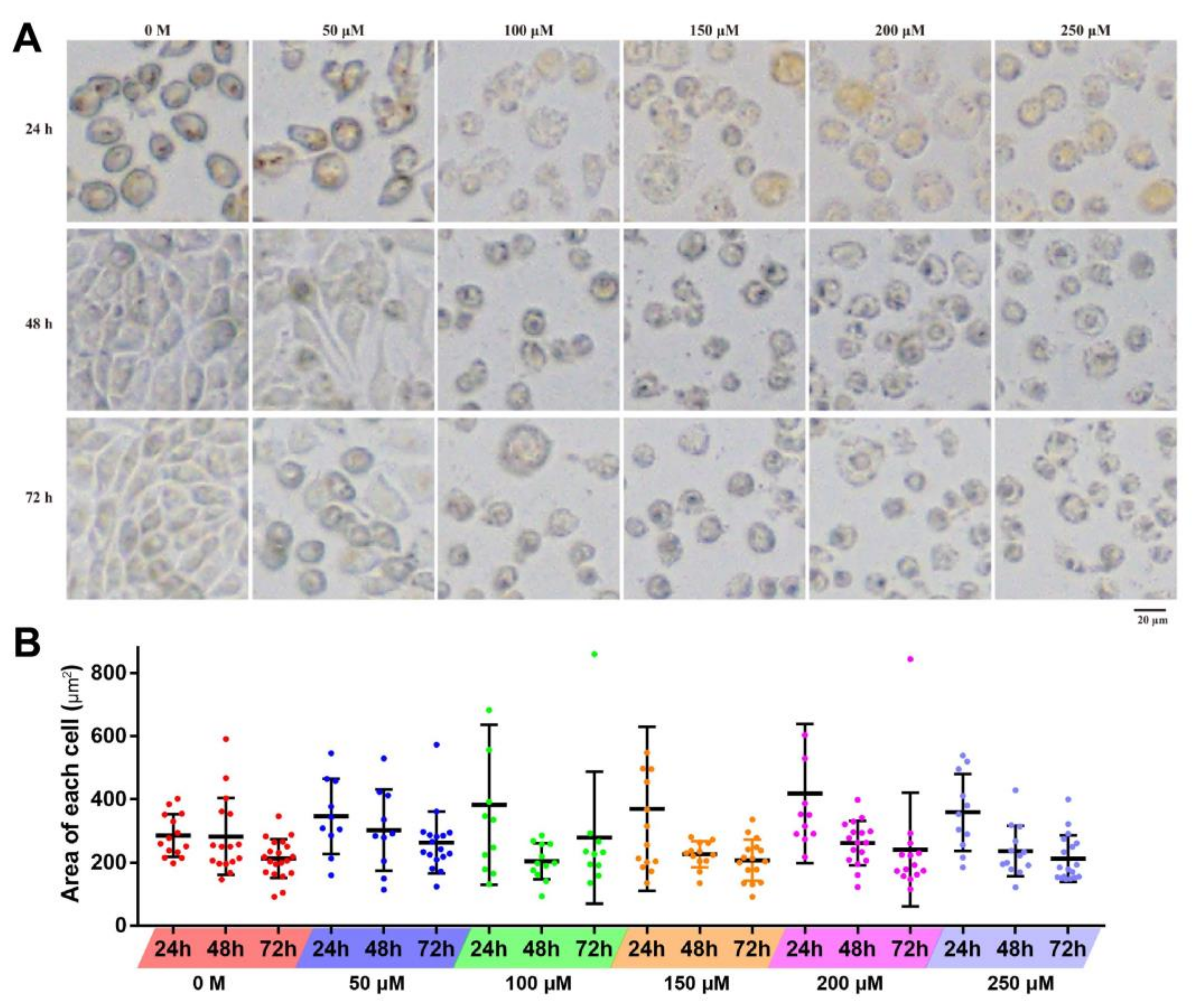
2.2. Effects of Resveratrol on the Cell Morphology
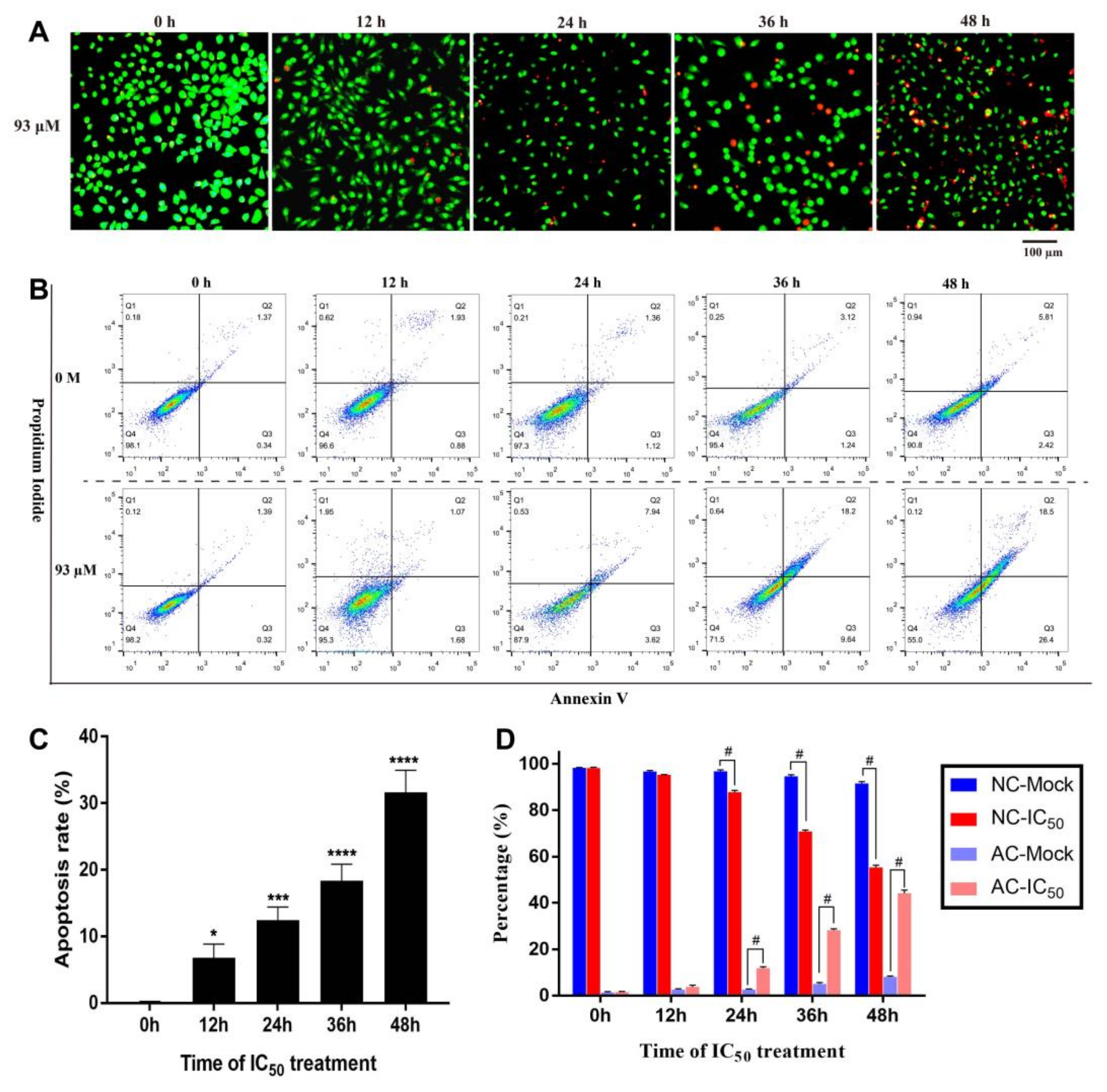
2.3. Resveratrol Induced Apoptosis of 4T1 Cells
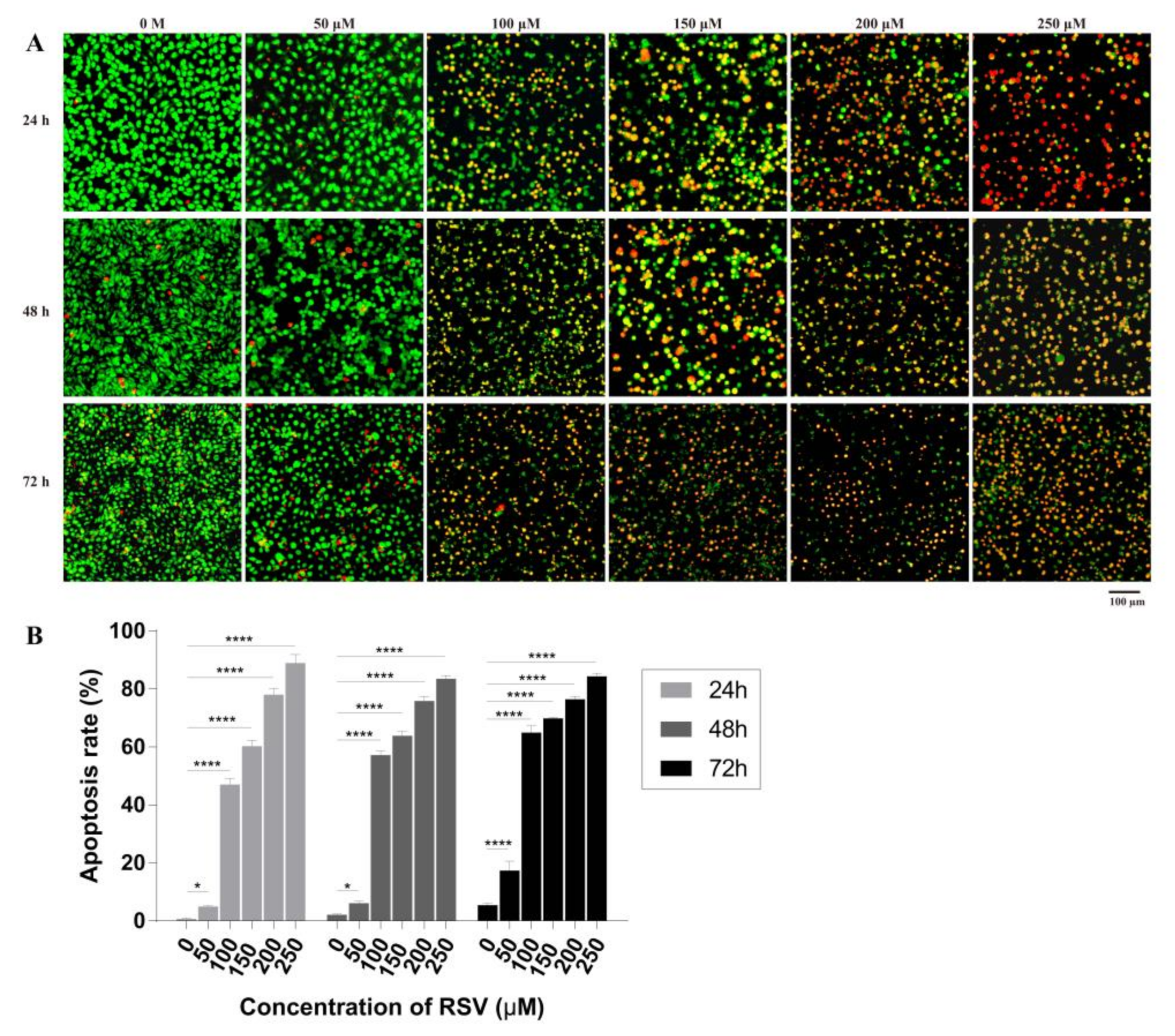
2.3.1. Detection of Cell Apoptosis
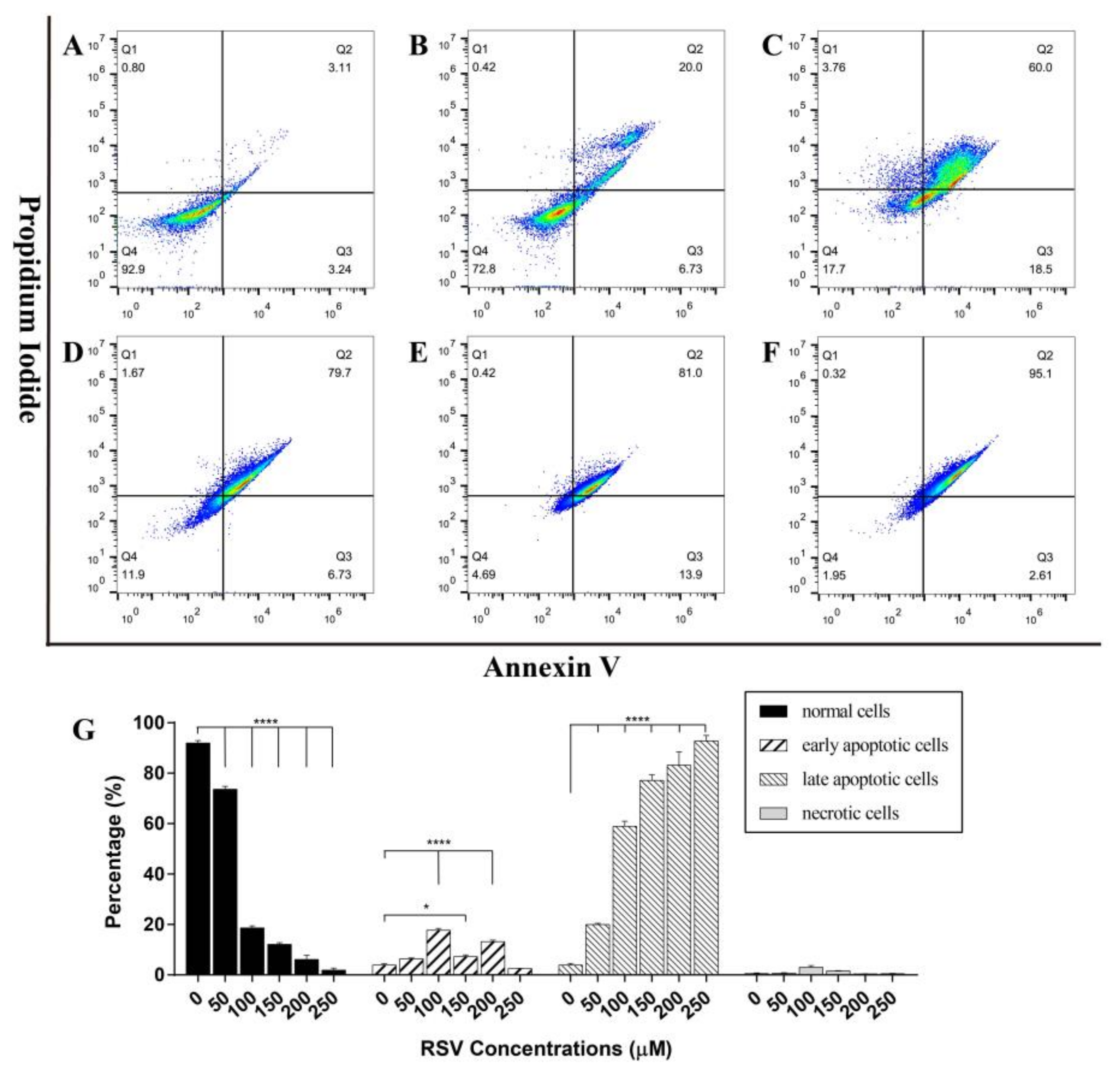
2.3.2. Concentration Dependent Apoptosis Assay
2.3.3. Time Dependent Apoptosis Assay
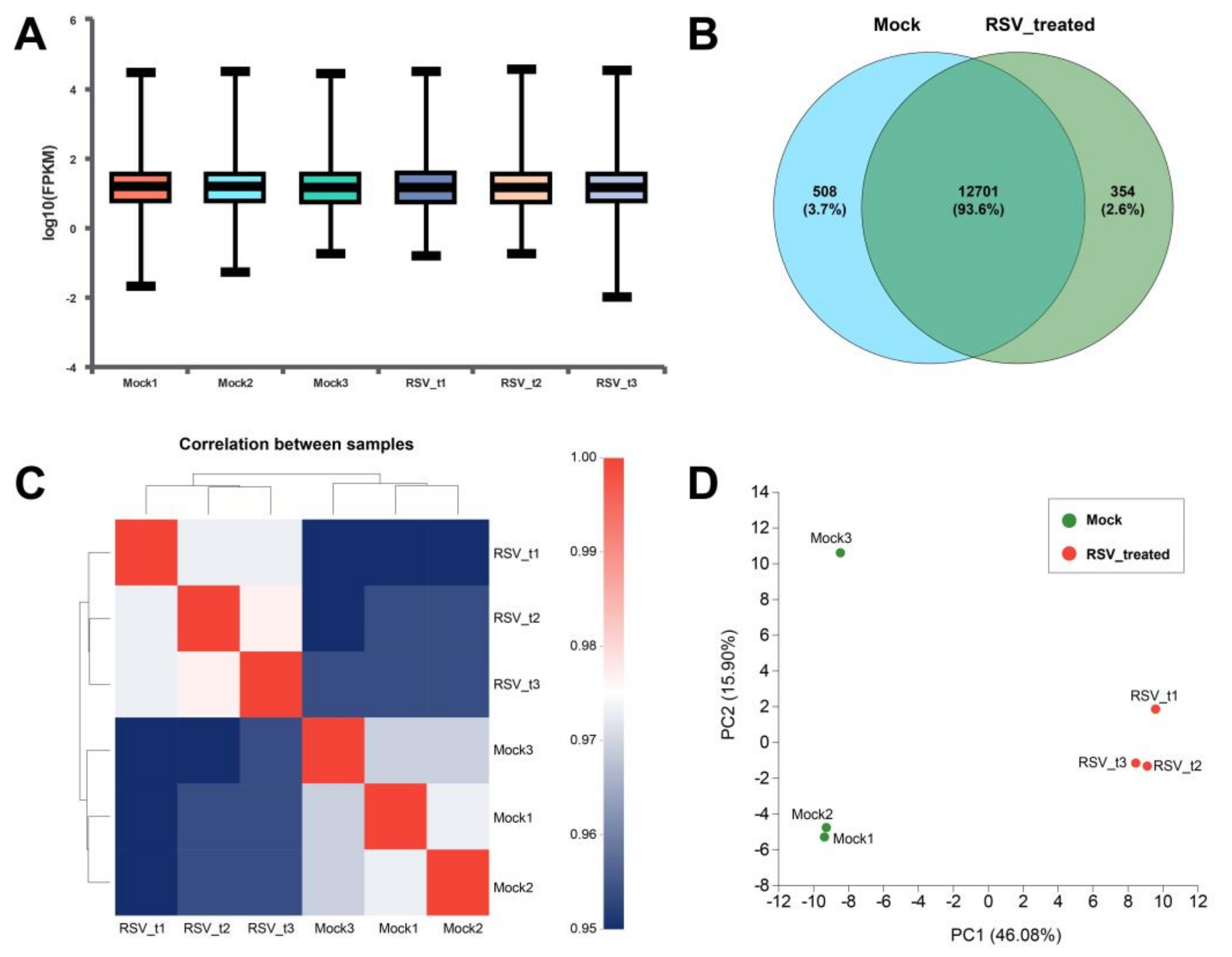
2.4. Transcriptome Analysis of 4T1 Cells
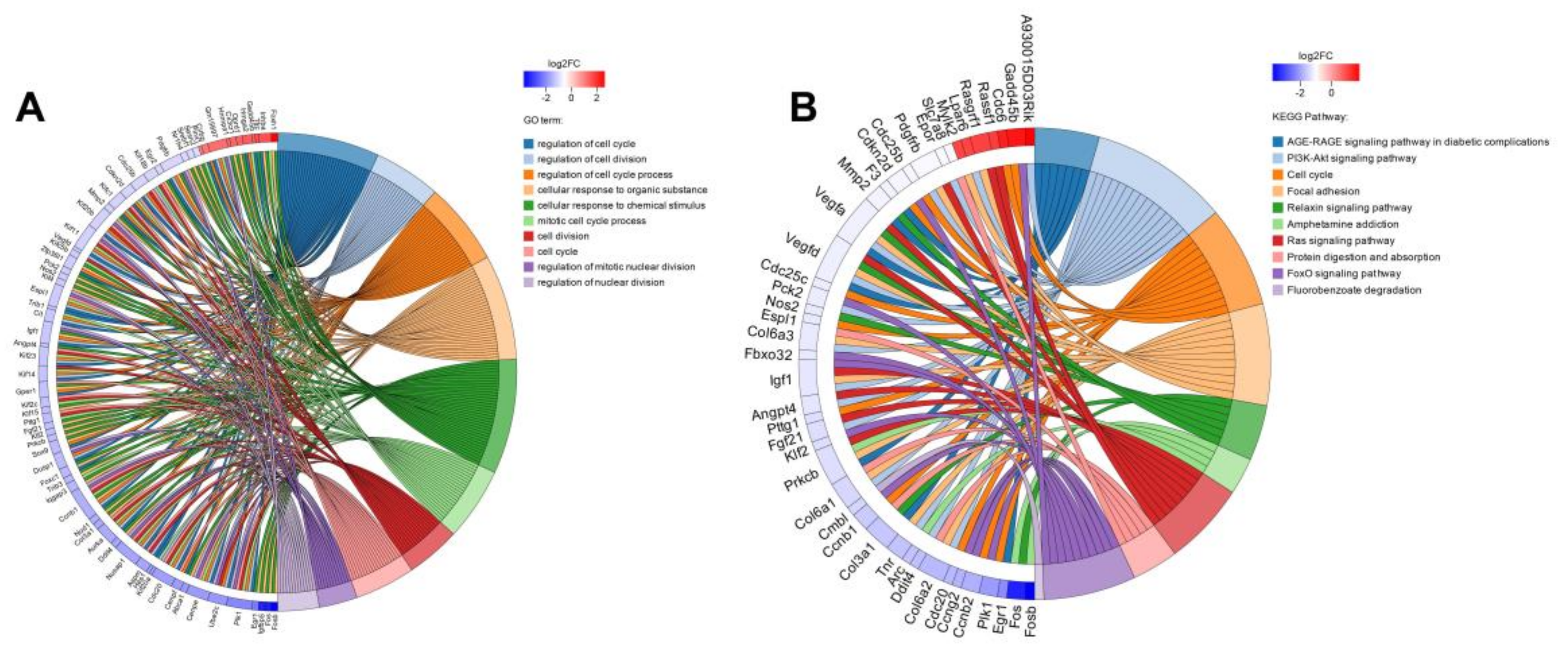
2.4.1. Screening and Functional Enrichment Analysis of Differential Genes
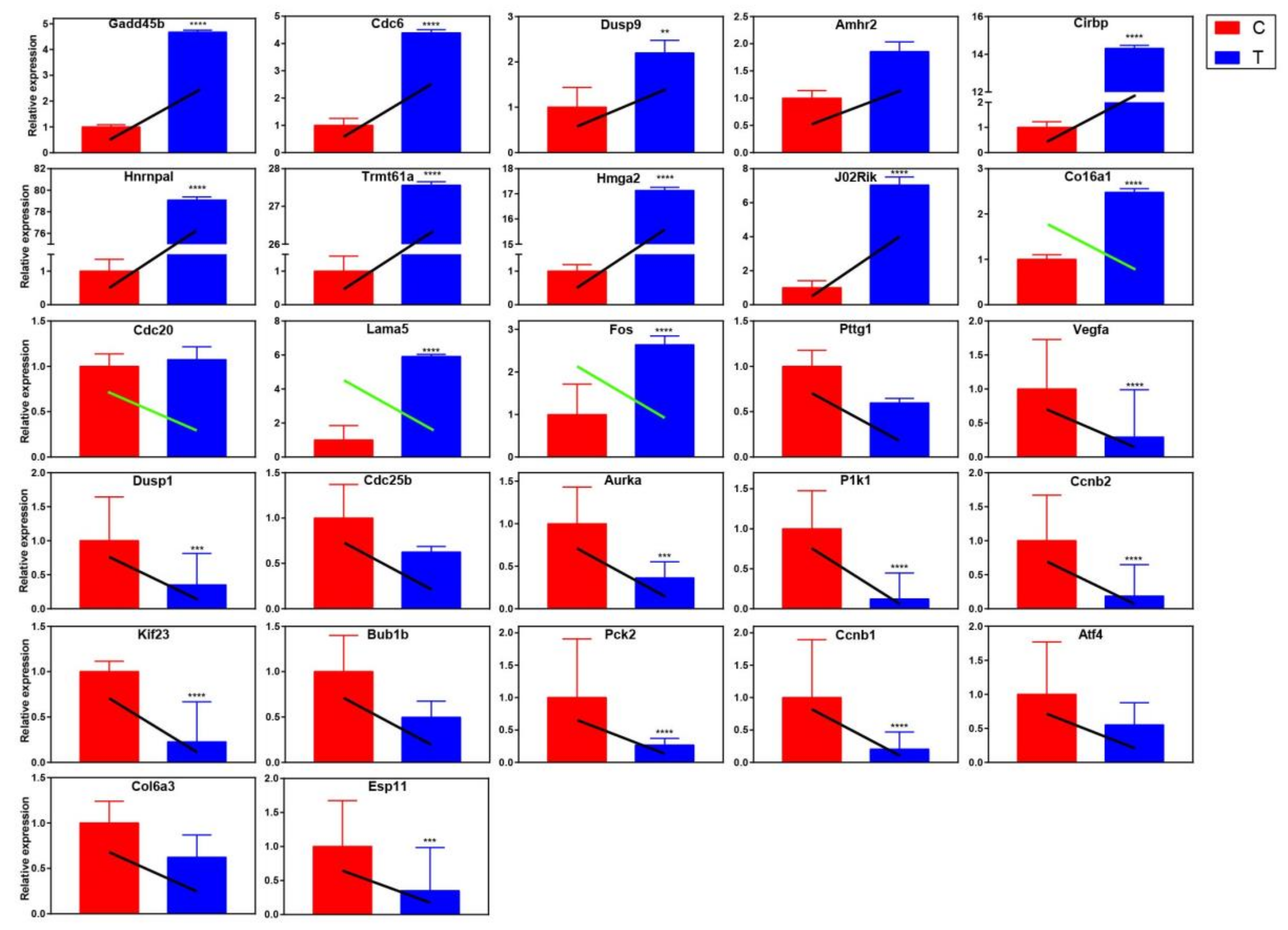
2.4.2. qRT-PCR Validation of Differentially Expressed Genes
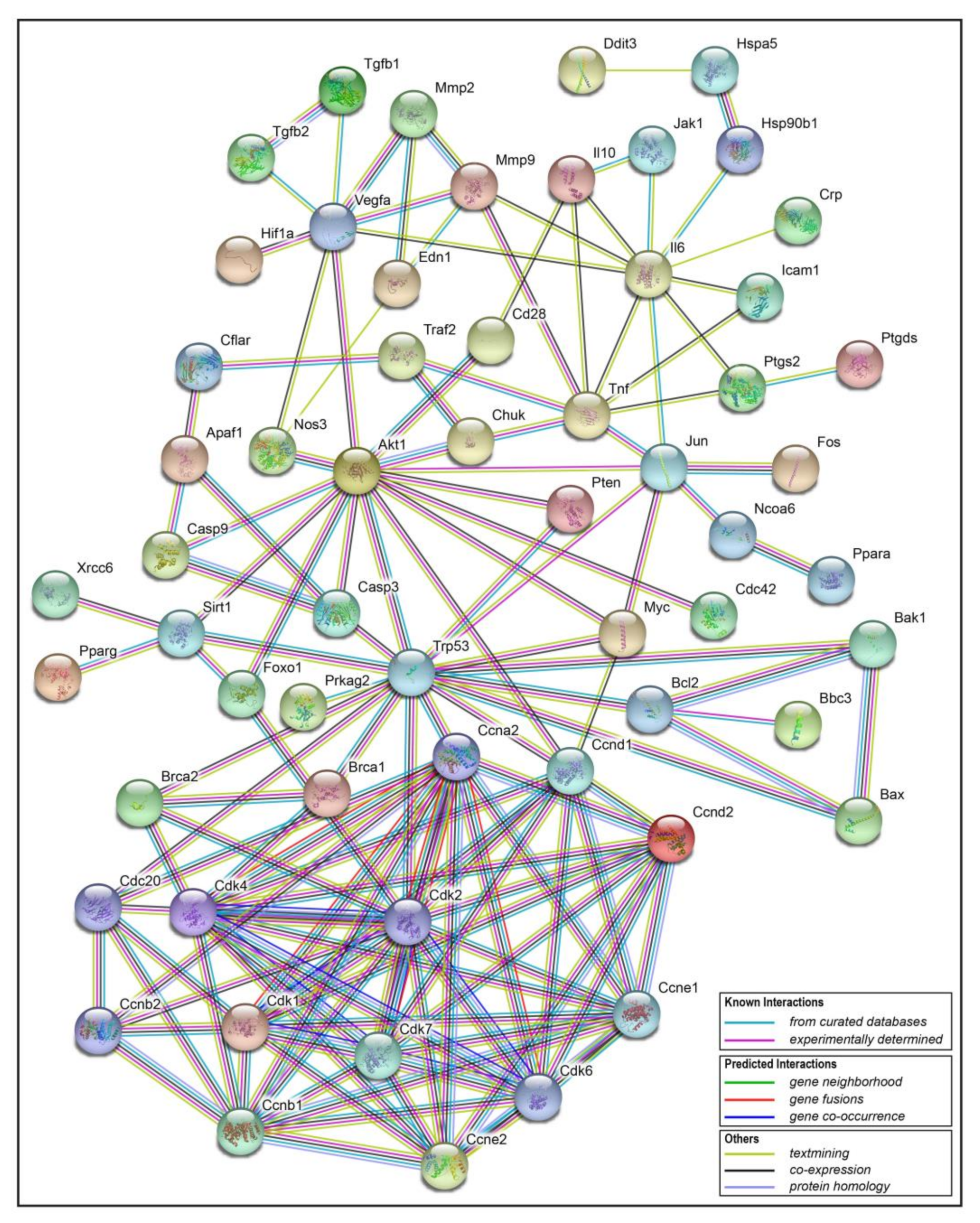
2.5. Interaction Network Analysis of Resveratrol’s Protein Targets
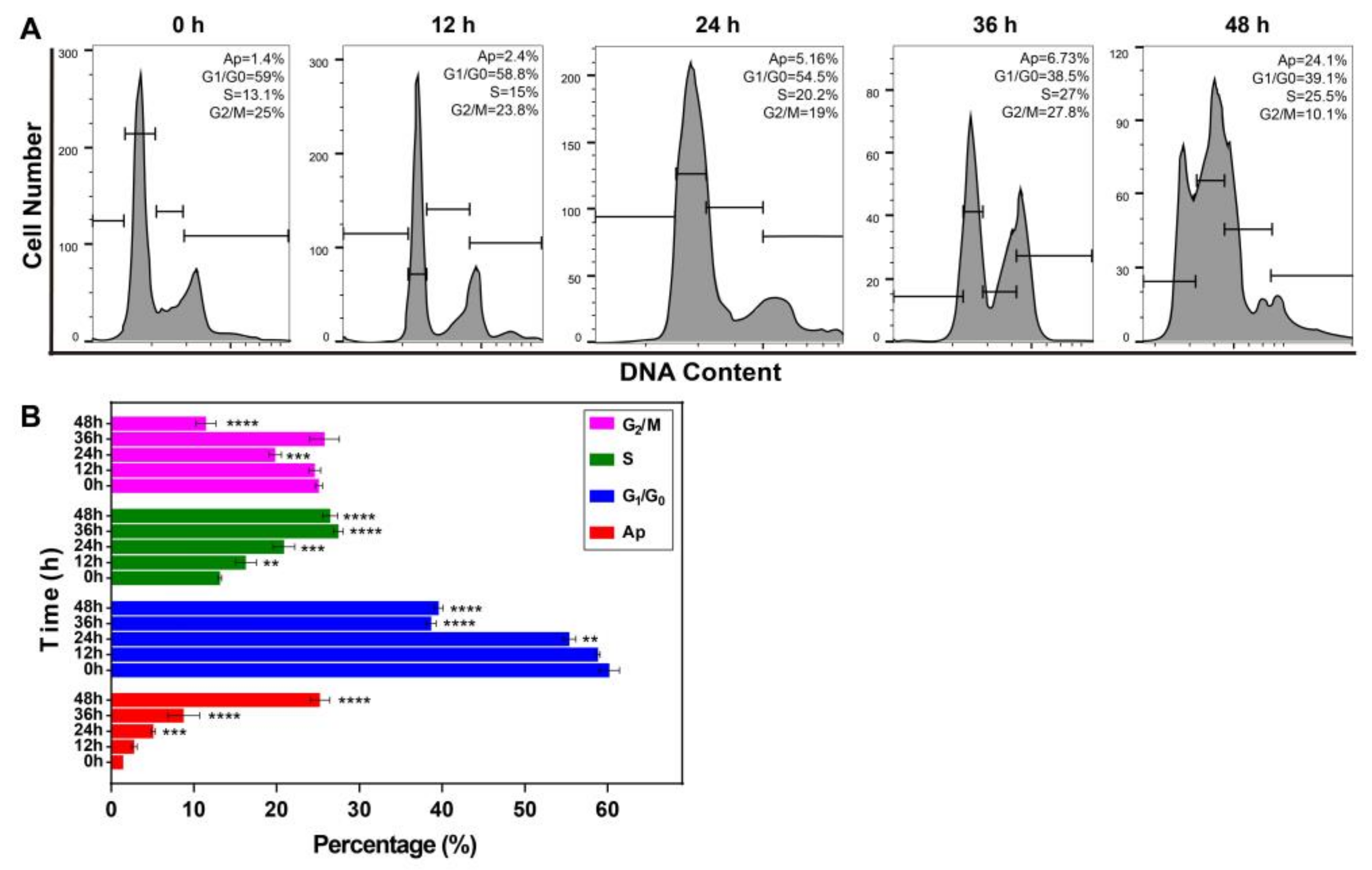
2.6. Resveratrol Changed the Phase Distribution of the Cell Cycle
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Preparation of Resveratrol
5.2. Cell Culture
5.3. Morphological Analysis of Cells
5.4. Cell Viability Measurement
5.5. AO/EB Staining
5.6. Cell Apoptosis and Cell Cycle Detection
5.7. RNA-Sequencing and Transcriptome Analysis
5.8. qRT-PCR Verification
5.9. Statistical Analysis
5.10. Image Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Gene. | ENSEMBL# | Primer Sequence (5′ - 3′) |
|---|---|---|
| Ppia | ENSMUSG00000071866 | F: GCTGGACCAAACACAAACGG |
| R: CAAAACGCTCCATGGCTTCC | ||
| Gadd45b | ENSMUSG00000015312 | F: CAGCGTGGTCTTGTGCCT |
| R: CGGTTGTGCCCAATGTCT | ||
| Cdc6 | ENSMUSG00000017499 | F: TGCAACACTCCCCGGTTATC |
| R: GCGGAGGAGGAACGTATCTG | ||
| Dusp9 | ENSMUSG00000031383 | F: TCTGTCCCAGTTCTTTCCG |
| R: TCGTAGGCATCGTTGAGTG | ||
| Amhr2 | ENSMUSG00000023047 | F: GGCTCCTTATGCCACTACTT |
| R: GGGCGATACCTGGTTTGT | ||
| Cirbp | ENSMUSG00000045193 | F: AGACTACTATGCCAGCCGGA |
| R: CTCGTTGTGTGTAGCATAACTGT | ||
| Hnrnpa1 | ENSMUSG00000046434 | F: TTGTGGAACCTAAGAGAGCCATC |
| R: CCTCTTTTTCCCACTGCCTCT | ||
| Trmt61a | ENSMUSG00000060950 | F: CCGGTTCTGTGGTCTGTGAA |
| R: CCGTTGCTGGTGGAACTCTA | ||
| Hmga2 | ENSMUSG00000056758 | F: TGAGTTTGGAGAACGCACCA |
| R: GTCTCCCATGAGAGTGGAAGC | ||
| Dlgap1 | ENSMUSG00000003279 | F: GGAAGGACTGACCAGGTTCC |
| R: TGTAGCTACCACTGCGCATC | ||
| J02Rik | ENSMUSG00000033688 | F: CCCTGTGCGGATGGAAAGAT |
| R: GCATGGCCTCAAACACTGTC | ||
| Col6a1 | ENSMUSG00000001119 | F: AAAGGCACCTACACCGACTG |
| R: GCATGGTTCCTTGTAGCCCT | ||
| Cdc20 | ENSMUSG00000006398 | F: GTGACCGCTTTATCCCCCAA |
| R: ATTCTGAGGTTTGCCGCTGA | ||
| Lama5 | ENSMUSG00000015647 | F: AGAGAGCCAGTTCTTGTGCC |
| R: AAACACTTGGATCGCCTTGC | ||
| Pttg1 | ENSMUSG00000020415 | F: CTATGAAGACTGGCAAACC |
| R: CATCAGGAGCAGGAACAG | ||
| Fos | ENSMUSG00000021250 | F: AGACCGTGTCAGGAGGCA |
| R: CCATCTTATTCCGTTCCCT | ||
| Vegfa | ENSMUSG00000023951 | F: CCCACGTCAGAGAGCAACAT |
| R: TGCGCTTTCGTTTTTGACCC | ||
| Dusp1 | ENSMUSG00000024190 | F: TGCCTATCACGCTTCTCG |
| R: CCTCCACAGGGATGCTCTT | ||
| Cdc25b | ENSMUSG00000027330 | F: ATTCAGGCAGCCAGTCGG |
| R: GCTCGGGAGTTGGTGATG | ||
| Aurka | ENSMUSG00000027496 | F: GGAGCCAGGGACCTCATTTC |
| R: CTAGTGTGGCCAGTTGGAGG | ||
| Plk1 | ENSMUSG00000030867 | F: AGTTTTGGAGCTCTGTCGCA |
| R: TACTGGCAGCCCAGGACTAT | ||
| Ccnb2 | ENSMUSG00000032218 | F: GCTAGCTCCCAAGGATCGTC |
| R: CTGCAGAGCTGAGGGTTCTC | ||
| Kif23 | ENSMUSG00000032254 | F: CCATAAAGCCCAAACTCC |
| R: AACACGCTATGTGAACGA | ||
| Bub1b | ENSMUSG00000040084 | F: GATCTATGCCGGAGTTGGGG |
| R: CCCAGCTCCTTCTTGCTGAA | ||
| Pck2 | ENSMUSG00000040618 | F: CAACTCCGGGCCATCAAC |
| R: CCGCCATCACTGGTCTCA | ||
| Ccnb1 | ENSMUSG00000041431 | F: GAAACATCTGGATGTGCGCC |
| R: GTTTGGGTCAGCCCCATCAT | ||
| Atf4 | ENSMUSG00000042406 | F: GCCTGACTCTGCTGCTTACA |
| R: AAGGCAGATTGTCTGGTGGG | ||
| Col6a3 | ENSMUSG00000048126 | F: ATGGCACCTCTCAGGACTCT |
| R: TGTTTTTCCCAACGTTGTCGG | ||
| Espl1 | ENSMUSG00000058290 | F: TGGGCTCTATGCTCACCTCT |
| R: GCTCTGCCAGTTCAGGTGAT |
References
- DeSantis, C.E.; Fedewa, S.A.; Goding, S.A.; Kramer, J.L.; Smith, R.A.; Jemal, A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J. Clin. 2016, 66, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, J.; Kinghorn, A.D. Development of Anticancer Agents from Plant-Derived Sesquiterpene Lactones. Curr. Med. Chem. 2016, 23, 2397–2420. [Google Scholar] [CrossRef] [PubMed]
- Iwuchukwu, O.F.; Nagar, S. Resveratrol (trans-resveratrol, 3,5,4′-trihydroxy-trans-stilbene) glucuronidation exhibits atypical enzyme kinetics in various protein sources. Drug Metab. Dispos. 2008, 36, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Das, D.K. Resveratrol: A therapeutic promise for cardiovascular diseases. Recent Pat. Cardiovasc. Drug Discov. 2007, 2, 133–138. [Google Scholar] [PubMed]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 5, 493–506. [Google Scholar] [CrossRef]
- Shakibaei, M.; Harikumar, K.B.; Aggarwal, B.B. Resveratrol addiction: To die or not to die. Mol. Nutr. Food Res. 2009, 53, 115–128. [Google Scholar] [CrossRef]
- Yang, X.; Li, X.; Ren, J. From French Paradox to cancer treatment: Anti-cancer activities and mechanisms of resveratrol. Anticancer Agents Med. Chem. 2014, 14, 806–825. [Google Scholar] [CrossRef]
- Tomé-Carneiro, J.; Larrosa, M.; González-Sarrías, A.; Tomás-Barberán, F.A.; García-Conesa, M.T.; Espín, J.C. Resveratrol and clinical trials: The crossroad from in vitro studies to human evidence. Curr. Pharm. Des. 2013, 19, 6064–6093. [Google Scholar] [CrossRef]
- Fulda, S. Resveratrol and derivatives for the prevention and treatment of cancer. Drug Discov. 2010, 15, 757–765. [Google Scholar] [CrossRef]
- Athar, M.; Back, J.H.; Tang, X.; Kim, K.H.; Kopelovich, L.; Bickers, D.R.; Kim, A.L. Resveratrol: A review of preclinical studies for human cancer prevention. Toxicol. Appl. Pharmacol. 2007, 224, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Lim, Y.H. Resveratrol antibacterial activity against Escherichia coli is mediated by Z-ring formation inhibitionvia suppression of FtsZ expression. Sci. Rep. 2015, 5, 10029. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, M.; Chen, K.; Chen, B.; Zhao, Y.; Gong, H.; Zhao, X.; Qi, R. Suppression of TLR4 activation by resveratrol is associated with STAT3 and Akt inhibition in oxidized low-density lipoprotein-activated platelets. Eur. J. Pharmacol. 2018, 836, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dembic, M.; Andersen, H.S.; Bastin, J.; Doktor, T.K.; Corydon, T.J.; Sass, J.O.; Lopes, C.A.; Djouadi, F.; Andresen, B.S. Next generation sequencing of RNA reveals novel targets of resveratrol with possible implications for Canavan disease. Mol. Genet. Metab. 2019, 126, 64–76. [Google Scholar] [CrossRef]
- Juškaitė, V.; Ramanauskienė, K.; Briedis, V. Testing of resveratrol microemulsion photostability and protective effect against UV induced oxidative stress. Acta. Pharm. 2017, 67, 247–256. [Google Scholar] [CrossRef][Green Version]
- Shao, A.-W.; Wu, H.-J.; Chen, S.; Ammar, A.-B.; Zhang, J.-M.; Hong, Y. Resveratrol attenuates early brain injury after subarachnoid hemorrhage through inhibition of NF-κB-dependent inflammatory/MMP-9 pathway. CNS Neurosci. Ther. 2014, 20, 182–185. [Google Scholar] [CrossRef]
- Charles-Henry, C.; Nivet-Antoine, V.; Jean-Louis, B. Review of recent data on the metabolism, biological effects, and toxicity of resveratrol in humans. Mol. Nutr. Food Res. 2014, 58, 7–21. [Google Scholar]
- Hausenblas, H.A.; Schoulda, J.A.; Smoliga, J.M. Resveratrol treatment as an adjunct to pharmacological management in type 2 diabetes mellitus--systematic review and meta-analysis. Mol. Nutr. Food Res. 2015, 59, 147–159. [Google Scholar] [CrossRef]
- Ozog, S.; Timberlake, N.D.; Hermann, K.; Garijo, O.; Haworth, K.G.; Shi, G.L.; Glinkerman, C.M.; Schefter, L.E.; D’Souza, S.; Simpson, E.; et al. Resveratrol trimer enhances gene delivery to hematopoietic stem cells by reducing antiviral restriction at endosomes. Blood 2019, 134, 1298–1311. [Google Scholar] [CrossRef]
- TCMSP. Available online: http://tcmspw.com/tcmsp.php (accessed on 12 December 2019).
- STRING Software. Available online: http://string-db.org (accessed on 12 December 2019).
- Chin, Y.-T.; Hsieh, M.-T.; Yang, S.-H.; Tsai, P.-W.; Wang, S.-H.; Wang, C.-C.; Lee, Y.-S.; Cheng, G.-Y.; HuangFu, W.-C.; London, D.; et al. Anti-proliferative and gene expression actions of resveratrol in breast cancer cells in vitro. Oncotarget 2014, 5, 12891–12907. [Google Scholar] [CrossRef]
- Meng, J.; Guo, F.Q.; Xu, H.Y.; Liang, W.; Wang, C.; Yang, X.-D. Combination therapy using co-encapsulated resveratrol and paclitaxel in liposomes for drug resistance reversal in breast cancer cells in vivo. Sci. Rep. 2016, 6, 22390. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Sull, J.W.; Sung, H.J. Suppressing effect of resveratrol on the migration and invasion of human metastatic lung and cervical cancer cells. Mol. Biol. Rep. 2012, 39, 8709–8716. [Google Scholar] [CrossRef] [PubMed]
- Alobaedi, O.H.; Talib, W.H.; Basheti, I.A. Antitumor effect of thymoquinone combined with resveratrol on mice transplanted with breast cancer. Asian Pac. J. Trop. Med. 2017, 10, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Poschner, S.; Maier-Salamon, A.; Thalhammer, T.; Jäger, W. Resveratrol and other dietary polyphenols are inhibitors of estrogen metabolism in human breast cancer cells. Steroid Biochem. Mol. Biol. 2019, 190, 11–18. [Google Scholar] [CrossRef]
- Bove, K.; Lincoln, D.W.; Tsan, M.-F. Effect of resveratrol on growth of 4T1 breast cancer cells in vitro and in vivo. Biochem. Biophys. Res. Commun. 2002, 291, 1001–1005. [Google Scholar] [CrossRef]
- Lee, H.S.; Ha, A.W.; Kim, W.K. Effect of resveratrol on the metastasis of 4T1 mouse breast cancer cells in vitro and in vivo. Nutr. Res. Pract. 2012, 6, 294–300. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Tang, H.-Y.; Davis, F.B.; Davis, P.J. Resveratrol and apoptosis. Ann. N. Y. Acad. Sci. 2011, 1215, 79–88. [Google Scholar] [CrossRef]
- Huang, C.; Huang, Y.-L.; Wang, C.-C.; Pan, Y.-L.; Lai, Y.-H.; Huang, H.-C. Ampelopsins A and C Induce Apoptosis and Metastasis through Downregulating AxL, TYRO3, and FYN Expressions in MDA-MB-231 Breast Cancer Cells. J. Agric. Food Chem. 2019, 67, 2818–2830. [Google Scholar] [CrossRef]
- Roshan, M.M.; Young, A.; Reinheimer, K.; Rayat, J.; Dai, L.-J.; Warnock, G.L. Dynamic assessment of cell viability, proliferation and migration using real time cell analyzer system (RTCA). Cytotechnology 2015, 67, 379–386. [Google Scholar] [CrossRef]
- Grimaldi, M.; Santin, G.; Insolia, V.; Bo, V.D.; Piccolini, V.M.; Veneroni, P.; Barni, S.; Verri, M.; Pascali, S.A.D.; Fanizzi, F.P.; et al. [Pt(O,O’-acac)(γ-acac)(DMS)] versus cisplatin: Apoptotic effects in B50 neuroblastoma cells. Histochem. Cell Biol. 2016, 145, 587–601. [Google Scholar] [CrossRef]
- He, Y.; Ruganzu, J.B.; Lin, C.; Ding, B.; Zheng, Q.; Wu, X.; Ma, R.; Liu, Q.; Wang, Y.; Jin, H.; et al. Tanshinone IIA ameliorates cognitive deficits by inhibiting endoplasmic reticulum stress-induced apoptosis in APP/PS1 transgenic mice. Neurochem. Int. 2019, 133, 104610. [Google Scholar] [CrossRef] [PubMed]
- Guzińska-Ustymowicz, K.; Pryczynicz, A.; Kemona, A.; Czyzewska, J. Correlation between proliferation markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer. Anticancer Res. 2009, 29, 3049–3052. [Google Scholar]
- Wojnar, A.; Pula, B.; Piotrowska, A.; Jethon, A.; Kujawa, K.; Kobierzycki, C.; Rys, J.; Podhorska-Okolow, M.; Dziegiel, P. Correlation of intensity of MT-I/II expression with Ki-67 and MCM-2 proteins in invasive ductal breast carcinoma. Anticancer Res. 2011, 31, 3027. [Google Scholar] [PubMed]
- Cramer, L.P.; Mitchison, T.J. Investigation of the mechanism of retraction of the cell margin and rearward flow of nodules during mitotic cell rounding. Mol. Biol. Cell. 1997, 8, 109–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yamakita, Y.; Totsukawa, G.; Yamashiro, S.; Fry, D.; Zhang, X.; Hanks, S.K.; Matsumura, F. Dissociation of FAK/p130(CAS)/c-Src complex during mitosis: Role of mitosis-specific serine phosphorylation of FAK. Cell Biol. 1999, 144, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Maddox, A.S.; Burridge, K. RhoA is required for cortical retraction and rigidity during mitotic cell rounding. Cell Biol. 2003, 160, 255–265. [Google Scholar] [CrossRef]
- Dao, V.T.; Dupuy, A.G.; Gavet, O.; Caron, E.; de Gunzburg, J. Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J. Cell. Sci. 2009, 122, 2996–3004. [Google Scholar] [CrossRef]
- Stewart, M.P.; Helenius, J.; Toyoda, Y.; Ramanathan, S.P.; Muller, D.J.; Hyman, A.A. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature 2011, 469, 226–230. [Google Scholar] [CrossRef]
- Ramanathan, S.P.; Helenius, J.; Stewart, M.P.; Cattin, C.J.; Hyman, A.A.; Muller, D.J. Cdk1-dependent mitotic enrichment of cortical myosin II promotes cell rounding against confinement. Nat. Cell Biol. 2015, 17, 148–159. [Google Scholar] [CrossRef]
- Kunda, P.; Baum, B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009, 19, 174–179. [Google Scholar] [CrossRef]
- Luxenburg, C.; Pasolli, H.A.; Williams, S.E.; Fuchs, E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat. Cell. Biol. 2011, 13, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, O.M.; Le, B.M.; Dimitracopoulos, A.; Bonazzi, D.; Zlotek-Zlotkiewicz, E.; Picone, R.; Duke, T.; Piel, M.; Baum, B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev. Cell. 2013, 25, 270–283. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Jacquemet, G.; Byron, A.; Jones, M.C.; Warwood, S.; Selley, J.N.; Knight, D.; Humphries, J.D.; Humphries, M.J. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat. Commun. 2015, 6, 6265. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.C.; Askari, J.A.; Humphries, J.D.; Humphries, M.J. Cell adhesion is regulated by CDK1 during the cell cycle. Cell. Biol. 2018, 217, 3203–3218. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.-S.; Zhou, J.; Ong, C.-N.; Shen, H.-M. Luteolin induces G1 arrest in human nasopharyngeal carcinoma cells via the Akt-GSK-3β-Cyclin D1 pathway. Cancer Lett. 2010, 298, 167–175. [Google Scholar] [CrossRef]
- Gong, D.; Ferrell, J.E. The roles of cyclin A2, B1, and B2 in early and late mitotic events. Mol. Biol. Cell. 2010, 21, 3149–3161. [Google Scholar] [CrossRef]
- Gould, K.L.; Nurse, P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 1989, 342, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Parker, L.L.; Piwnica-Worms, H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science 1992, 257, 1955–1957. [Google Scholar] [CrossRef]
- Jackman, M.; Lindon, C.; Nigg, E.A.; Pines, J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003, 5, 143–148. [Google Scholar] [CrossRef]
- Krämer, A.; Mailand, N.; Lukas, C.; Syljuåsen, R.G.; Wilkinson, C.J.; Nigg, E.A.; Bartek, J.; Lukas, J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 2004, 6, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.V.; Sramkoski, R.M.; Lam, M.; Stefan, T.; Jacobberger, J.W. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle 2008, 7, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Gavet, O.; Pines, J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev. Cell 2010, 18, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Gavet, O.; Pines, J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. Cell Biol. 2010, 189, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Majorbio Cloud Platform. Available online: http://www.majorbio.com (accessed on 12 December 2019).
- Grimaldi, M.; Bo, V.D.; Ferrari, B.; Roda, E.; De Luca, F.; Veneroni, P.; Barni, S.; Verri, M.; De Pascalic, S.A.; Fanizzi, F.P.; et al. Long-term effects after treatment with platinum compounds, cisplatin and [Pt(O,O’-acac)(γ-acac)(DMS)]: Autophagy activation in rat B50 neuroblastoma cells. Toxicol. Appl. Pharmacol. 2019, 364, 1–11. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Chen, L.; Zhu, F.; Han, X.; Sun, L.; Chen, K. The Cytotoxicity Effect of Resveratrol: Cell Cycle Arrest and Induced Apoptosis of Breast Cancer 4T1 Cells. Toxins 2019, 11, 731. https://doi.org/10.3390/toxins11120731
Wu H, Chen L, Zhu F, Han X, Sun L, Chen K. The Cytotoxicity Effect of Resveratrol: Cell Cycle Arrest and Induced Apoptosis of Breast Cancer 4T1 Cells. Toxins. 2019; 11(12):731. https://doi.org/10.3390/toxins11120731
Chicago/Turabian StyleWu, Hong, Liang Chen, Feifei Zhu, Xu Han, Lindan Sun, and Keping Chen. 2019. "The Cytotoxicity Effect of Resveratrol: Cell Cycle Arrest and Induced Apoptosis of Breast Cancer 4T1 Cells" Toxins 11, no. 12: 731. https://doi.org/10.3390/toxins11120731
APA StyleWu, H., Chen, L., Zhu, F., Han, X., Sun, L., & Chen, K. (2019). The Cytotoxicity Effect of Resveratrol: Cell Cycle Arrest and Induced Apoptosis of Breast Cancer 4T1 Cells. Toxins, 11(12), 731. https://doi.org/10.3390/toxins11120731







