Traditional and Non-Traditional Clustering Techniques for Identifying Chrononutrition Patterns in University Students
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Data Collection and Variables
2.2.1. Data Collection
2.2.2. Sociodemographic Characteristics
2.2.3. Anthropometric Measurements
2.2.4. Food Intake Quality Assessment
2.2.5. Chronotype Assessment
2.2.6. Chrononutrition Assessment
2.3. Statistical Analysis
2.3.1. Descriptive Analyses
2.3.2. Clustering Methods
2.3.3. Clustering Performance and Methods Concordance
2.3.4. Software and Statistical Significance
3. Results
3.1. Sample Characteristics
3.2. Meal Timing Patterns by Traditional and Non-Traditional Clustering Methods
3.2.1. K-Means Clustering
3.2.2. Hierarchical Clustering
3.2.3. Gaussian Mixture Models
3.2.4. Spectral Clustering
3.3. Sociodemographic Characteristics by Meal Timing Patterns
3.4. Chrononutrition Variables
3.5. Anthropometric and Food Intake Quality by Meal Timing Patterns
3.6. Clustering Performance and Method Concordance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ARI | Adjusted Rand Index |
| BIC | Bayesian Information Criterion |
| BMI | Body Mass Index |
| CIOMS | Council for International Organizations of Medical Sciences |
| Gamma | Hubert’s Gamma statistic |
| GMM | Gaussian Mixture Models |
| HOMA-IR | Homeostatic Model Assessment for Insulin Resistance |
| IQR | Interquartile Range |
| MEQ | Morningness-Eveningness Questionnaire |
| Mini-ECCA | Mini-Survey to Evaluate Food Intake Quality |
| NHANES | National Health and Nutrition Examination Survey |
| PCA | Principal Component Analysis |
| WHO | World Health Organization |
References
- Dashti, H.S.; St-Onge, M.P.; Engeda, J.C.; Guo, J.; Gundersen, E.P.; Haas, M.C.; Hall, K.D.; Jager, L.R.; Laferrère, B.; Migueles, J.H.; et al. Advancing Chrononutrition for Cardiometabolic Health: A 2023 NHLBI Workshop Report. J. Am. Heart Assoc. 2025, 14, e039373. [Google Scholar] [CrossRef] [PubMed]
- Raji, O.E.; Kyeremah, E.B.; Sears, D.D.; St-Onge, M.P.; Makarem, N. Chrononutrition and cardiometabolic health: An overview of epidemiological evidence and key future research directions. Nutrients 2024, 16, 2332. [Google Scholar] [CrossRef] [PubMed]
- Paoli, A.; Tinsley, G.; Bianco, A.; Moro, T. The influence of meal frequency and timing on health in humans: The role of fasting. Nutrients 2019, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Minguez, J.; Gómez-Abellán, P.; Garaulet, M. Timing of breakfast, lunch, and dinner. Effects on obesity and metabolic risk. Nutrients 2019, 11, 2624. [Google Scholar] [CrossRef]
- Verde, L.; Di Lorenzo, T.; Savastano, S.; Colao, A.; Barrea, L.; Muscogiuri, G. Chrononutrition in type 2 diabetes mellitus and obesity: A narrative review. Diabetes. Metab. Res. Rev. 2024, 40, e3778. [Google Scholar] [CrossRef]
- Papakonstantinou, E.; Oikonomou, C.; Nychas, G.; Dimitriadis, G.D. Effects of diet, lifestyle, chrononutrition and alternative dietary interventions on postprandial glycemia and insulin resistance. Nutrients 2022, 14, 823. [Google Scholar] [CrossRef]
- McHill, A.W.; Phillips, A.J.; Czeisler, C.A.; Keating, L.; Yee, K.; Barger, L.K.; Garaulet, M.; Scheer, F.A.; Klerman, E.B. Later circadian timing of food intake is associated with increased body fat. Am. J. Clin. Nutr. 2017, 106, 1213–1219. [Google Scholar] [CrossRef]
- Bernardes da Cunha, N.; Teixeira, G.P.; Madalena Rinaldi, A.E.; Azeredo, C.M.; Crispim, C.A. Late meal intake is associated with abdominal obesity and metabolic disorders related to metabolic syndrome: A chrononutrition approach using data from NHANES 2015–2018. Clin. Nutr. 2023, 42, 1798–1805. [Google Scholar] [CrossRef]
- Zerón-Rugerio, M.F.; Hernáez, Á.; Porras-Loaiza, A.P.; Cambras, T.; Izquierdo-Pulido, M. Eating jet lag: A marker of the variability in meal timing and its association with body mass index. Nutrients 2019, 11, 2980. [Google Scholar] [CrossRef]
- Kim, N.; Conlon, R.K.; Farsijani, S.; Hawkins, M.S. Association between chrononutrition patterns and multidimensional sleep health. Nutrients 2024, 16, 3724. [Google Scholar] [CrossRef]
- Intemann, T.; Bogl, L.H.; Hunsberger, M.; Lauria, F.; De Henauw, S.; Molnár, D.; Moreno, L.A.; Tornaritis, M.; Veidebaum, T.; Ahrens, W.; et al. A late meal timing pattern is associated with insulin resistance in European children and adolescents. Pediatr. Diabetes 2024, 2024, 6623357. [Google Scholar] [CrossRef] [PubMed]
- Santonja, I.; Bogl, L.H.; Degenfellner, J.; Klösch, G.; Seidel, S.; Schernhammer, E.; Papantoniou, K. Meal-timing patterns and chronic disease prevalence in two representative Austrian studies. Eur. J. Nutr. 2023, 62, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Pons-Muzzo, L.; de Cid, R.; Obón-Santacana, M.; Straif, K.; Papantoniou, K.; Santonja, I.; Kogevinas, M.; Palomar-Cros, A.; Lassale, C. Sex-specific chrono-nutritional patterns and association with body weight in a general population in Spain (GCAT study). Int. J. Behav. Nutr. Phys. Act. 2024, 21, 102. [Google Scholar] [CrossRef]
- Lesani, A.; Mojani-Qomi, M.S. Cross-sectional associations between meal timing patterns and diet quality indices in Iranian women. Sci. Rep. 2025, 15, 21135. [Google Scholar] [CrossRef]
- Horn, C.; Laupsa-Borge, J.; Andersen, A.I.O.; Dyer, L.; Revheim, I.; Leikanger, T.; Næsheim, N.T.; Storås, I.; Johannessen, K.K.; Mellgren, G.; et al. Meal patterns associated with energy intake in people with obesity. Br. J. Nutr. 2022, 128, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Chau, C.A.; Pan, W.H.; Chen, H.J. Employment status and temporal patterns of energy intake: Nutrition and Health Survey in Taiwan, 2005–2008. Public Health Nutr. 2017, 20, 3295–3303. [Google Scholar] [CrossRef]
- O’Hara, C.; Gibney, E.R. Meal Pattern Analysis in Nutritional Science: Recent Methods and Findings. Adv. Nutr. 2021, 12, 1365–1378. [Google Scholar] [CrossRef]
- Lo Siou, G.; Yasui, Y.; Csizmadi, I.; McGregor, S.E.; Robson, P.J. Exploring statistical approaches to diminish subjectivity of cluster analysis to derive dietary patterns: The Tomorrow Project. Am. J. Epidemiol. 2011, 173, 956–967. [Google Scholar] [CrossRef]
- Greve, B.; Pigeot, I.; Huybrechts, I.; Pala, V.; Börnhorst, C. A comparison of heuristic and model-based clustering methods for dietary pattern analysis. Public Health Nutr. 2016, 19, 255–264. [Google Scholar] [CrossRef]
- van Houwelingen, M.L.; Zhu, Y. Identifying and predicting dietary patterns in the Dutch population using machine learning. Eur. J. Nutr. 2025, 64, 305. [Google Scholar] [CrossRef]
- Lo Siou, G.; Akawung, A.K.; Solbak, N.M.; McDonald, K.L.; Al Rajabi, A.; Whelan, H.K.; Kirkpatrick, S.I. The effect of different methods to identify, and scenarios used to address energy intake misestimation on dietary patterns derived by cluster analysis. Nutr. J. 2021, 20, 42. [Google Scholar] [CrossRef]
- Sauvageot, N.; Schritz, A.; Leite, S.; Alkerwi, A.; Stranges, S.; Zannad, F.; Streel, S.; Hoge, A.; Donneau, A.F.; Albert, A.; et al. Stability-based validation of dietary patterns obtained by cluster analysis. Nutr. J. 2017, 16, 4. [Google Scholar] [CrossRef] [PubMed]
- Nava-Amante, P.A.; Betancourt-Núñez, A.; Díaz-López, A.; Bernal-Orozco, M.F.; De la Cruz-Mosso, U.; Márquez-Sandoval, F.; Vizmanos, B. Clusters of Sociodemographic Characteristics and Their Association with Food Insecurity in Mexican University Students. Foods 2024, 13, 2507. [Google Scholar] [CrossRef]
- Etikan, I.; Musa, S.A.; Alkassim, R.S. Comparison of Convenience Sampling and Purposive Sampling. Am. J. Theor. Appl. Stat. 2016, 5, 1–4. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- Council for International Organizations of Medical Sciences (CIOMS). International Ethical Guidelines for Health-Related Research Involving Humans, 4th ed.; CIOMS: Geneva, Switzerland, 2016. [Google Scholar]
- International Society for the Advancement of Kinanthropometry. International Standards for Anthropometric Assessment; International Society for the Advancement of Kinanthropometry: Underdale, SA, Australia, 2001. [Google Scholar]
- WHO. WHO Consultation on Obesity. In Obesity: Preventing and Managing the Global Epidemic; WHO Technical Report Series 894; World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Bernal-Orozco, M.F.; Salmeron-Curiel, P.B.; Prado-Arriaga, R.J.; Orozco-Gutiérrez, J.F.; Badillo-Camacho, N.; Márquez-Sandoval, F.; Altamirano-Martínez, M.B.; González-Gómez, M.; Gutiérrez-González, P.; Vizmanos, B.; et al. Second Version of a Mini-Survey to Evaluate Food Intake Quality (Mini-ECCA v.2): Reproducibility and Ability to Identify Dietary Patterns in University Students. Nutrients 2020, 12, 809. [Google Scholar] [CrossRef]
- Terman, M.; Rifkin, J.B.; Jacobs, J.; White, T.M. Cuestionario de Matutinidad-Vespertinidad (MEQ-SA-ESP); Center for Environmental Therapeutics: Worcester, MA, USA, 2018; Available online: https://cet.org/wp-content/uploads/2018/01/MEQ-SA-ESP.pdf (accessed on 2 December 2025).
- Horne, J.A.; Östberg, O. A Self-Assessment Questionnaire to Determine Morningness-Eveningness in Human Circadian Rhythms. Int. J. Chronobiol. 1976, 4, 97–110. [Google Scholar]
- Lloyd, S. Least Squares Quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Ward, J., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Dempster, A.; Laird, N.; Rubin, D. Maximum Likelihood from Incomplete Data via the EM Algorithm. J. R. Stat. Soc. Ser. B 1977, 39, 1–22. [Google Scholar] [CrossRef]
- von Luxburg, U. A Tutorial on Spectral Clustering. Stat. Comput. 2007, 17, 395–416. [Google Scholar] [CrossRef]
- Rousseeuw, P. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the Number of Clusters in a Data Set via the Gap Statistic. J. R. Stat. Soc. Ser. B 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Ikotun, A.M.; Habyarimana, F.; Ezugwu, A.E. Cluster Validity Indices for Automatic Clustering: A Comprehensive Review. Heliyon 2025, 11, e41953. [Google Scholar] [CrossRef] [PubMed]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Dunn, J.C. Well-Separated Clusters and Optimal Fuzzy Partitions. J. Cybern. 1974, 4, 95–104. [Google Scholar] [CrossRef]
- Caliński, T.; Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Hubert, L.J. A General Statistical Framework for Assessing Categorical Clustering in Free Recall. Psychol. Bull. 1976, 83, 1072–1080. [Google Scholar] [CrossRef]
- Davies, D.L.; Bouldin, D.W. A Cluster Separation Measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, PAMI-1, 224–227. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing Partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Wickham, H.; Miller, E.; Smith, D. R Package, Version 2.5.5; Haven: Import and Export ’SPSS’, ’Stata’ and ’SAS’ Files. 2025. Available online: https://CRAN.R-project.org/package=haven (accessed on 2 December 2025).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. R Package, Version 1.1.4; Dplyr: A Grammar of Data Manipulation. 2023. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 2 December 2025).
- Wickham, H.; Vaughan, D.; Girlich, M. R Package, Version 1.3.1; Tidyr: Tidy Messy Data. 2024. Available online: https://CRAN.R-project.org/package=tidyr (accessed on 2 December 2025).
- Müller, K. R Package, Version 1.1.4; Hms: Pretty Time of Day. 2025. Available online: https://CRAN.R-project.org/package=hms (accessed on 2 December 2025).
- Ogle, D.H.; Doll, J.C.; Wheeler, A.P.; Dinno, A. R Package, Version 0.10.0; FSA: Simple Fisheries Stock Assessment Methods. 2025. Available online: https://CRAN.R-project.org/package=FSA (accessed on 2 December 2025).
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. R Package, Version 2.1.6; Cluster: Cluster Analysis Basics and Extensions. 2023. Available online: https://CRAN.R-project.org/package=cluster (accessed on 2 December 2025).
- Scrucca, L.; Fraley, C.; Murphy, T.B.; Raftery, A.E. Model-Based Clustering, Classification, and Density Estimation Using mclust in R; Chapman and Hall/CRC: Boca Raton, FL, USA, 2023. [Google Scholar] [CrossRef]
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. kernlab—An S4 Package for Kernel Methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. R Package, Version 1.0.7; Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 2 December 2025).
- Hennig, C. R Package, Version 2.2-13; Fpc: Flexible Procedures for Clustering. 2024. Available online: https://CRAN.R-project.org/package=fpc (accessed on 2 December 2025).
- Walesiak, M.; Dudek, A. R Package, Version 0.51-5; ClusterSim: Searching for Optimal Clustering Procedure for a Data Set. 2024. Available online: https://CRAN.R-project.org/package=clusterSim (accessed on 2 December 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pedersen, T.L. R Package, Version 1.3.2; Patchwork: The Composer of Plots. 2025. Available online: https://CRAN.R-project.org/package=patchwork (accessed on 2 December 2025).
- Fransen, H.P.; May, A.M.; Stricker, M.D.; Boer, J.M.A.; Hennig, C.; Rosseel, Y.; Ocké, M.C.; Peeters, P.H.M.; Beulens, J.W.J. A posteriori dietary patterns: How many patterns to retain? J. Nutr. 2014, 144, 1274–1282. [Google Scholar] [CrossRef]
- Phoi, Y.Y.; Rogers, M.; Bonham, M.P.; Dorrian, J.; Coates, A.M. A scoping review of chronotype and temporal patterns of eating of adults: Tools used, findings, and future directions. Nutr. Res. Rev. 2022, 35, 112–135. [Google Scholar] [CrossRef]
- van der Merwe, C.; Münch, M.; Kruger, R. Chronotype Differences in Body Composition, Dietary Intake and Eating Behavior Outcomes: A Scoping Systematic Review. Adv. Nutr. 2022, 13, 2357–2405. [Google Scholar] [CrossRef]
- Bonaccio, M.; Ruggiero, E.; Di Castelnuovo, A.; Martínez, C.F.; Esposito, S.; Costanzo, S.; Cerletti, C.; Donati, M.B.; de Gaetano, G.; Iacoviello, L. Association between Late-Eating Pattern and Higher Consumption of Ultra-Processed Food among Italian Adults: Findings from the INHES Study. Nutrients 2023, 15, 1497. [Google Scholar] [CrossRef]
- Fiore, G.; Scapaticci, S.; Neri, C.R.; Azaryah, H.; Escudero-Marín, M.; Pascuzzi, M.C.; Mendola, A.L.; Mameli, C.; Chiarelli, F.; Campoy, C.; et al. Chrononutrition and metabolic health in children and adolescents: A systematic review and meta-analysis. Nutr. Rev. 2024, 82, 1309–1354. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, G.P.; Barreto, A.C.F.; Mota, M.C.; Crispim, C.A. Caloric midpoint is associated with total calorie and macronutrient intake and body mass index in undergraduate students. Chronobiol. Int. 2019, 36, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Baron, K.G.; Reid, K.J.; Kern, A.S.; Zee, P.C. Role of sleep timing in caloric intake and BMI. Obesity 2011, 19, 1374–1381. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Total (n = 459) | No Meal Skipping (n = 388) | ≥1 Meal Skipped (n = 71) | p-Value |
|---|---|---|---|---|
| Demographics | ||||
| Age (years) | 20.0 [19.0, 22.0] | 20.0 [19.0, 22.0] | 20.0 [19.0, 21.5] | 0.173 |
| Sex n (%) | ||||
| Female | 334 (72.8) | 283 (72.9) | 51 (71.8) | 0.962 |
| Male | 125 (27.2) | 105 (27.1) | 20 (28.2) | |
| Marital status | ||||
| Single | 432 (94.1) | 364 (93.8) | 68 (95.8) | 0.711 |
| In relationship | 27 (5.9) | 24 (6.2) | 3 (4.2) | |
| Academic program | ||||
| Medicine | 89 (19.4) | 78 (20.1) | 11 (15.5) | 0.319 |
| Nutrition | 98 (21.4) | 87 (22.4) | 11 (15.5) | |
| Psychology | 61 (13.3) | 51 (13.1) | 10 (14.1) | |
| Other programs | 211 (45.9) | 172 (44.3) | 39 (54.9) | |
| Semester level | ||||
| 1–2 years | 272 (59.9) | 224 (58.5) | 48 (67.6) | 0.191 |
| 3–5 years | 182 (40.1) | 159 (41.5) | 23 (32.4) | |
| Employment status | ||||
| Employed | 150 (32.7) | 128 (33.0) | 22 (31.0) | 0.847 |
| Unemployed | 309 (67.3) | 260 (67.0) | 49 (69.0) | |
| Anthropometric measures | ||||
| BMI (kg/m2) | 22.8 [20.7, 26.0] | 22.7 [20.7, 25.8] | 23.1 [20.7, 26.4] | 0.318 |
| Waist circumference (cm) | 72.0 [66.5, 79.4] | 71.8 [66.3, 79.0] | 72.7 [67.2, 81.1] | 0.241 |
| Food intake quality, n (%) | ||||
| Healthy food intake | 126 (27.8) | 115 (29.9) | 11 (15.9) | <0.001 |
| Habits in need of improvement | 129 (28.4) | 116 (30.1) | 13 (18.8) | |
| Unhealthy food intake | 199 (43.8) | 154 (40.0) | 45 (65.2) | |
| Chronotype, n (%) | (n = 454) | (n = 385) | (n = 69) | |
| Morning type | 100 (22.0) | 90 (23.4) | 10 (14.5) | 0.261 |
| Intermediate | 318 (70.0) | 265 (68.8) | 53 (76.8) | |
| Evening type | 36 (7.9) | 30 (7.8) | 6 (8.7) | |
| Chrononutrition variables | ||||
| Breakfast time | 09:00 [08:00, 10:00] (n = 417) | 09:00 [07:54, 10:00] | 09:30 [09:00, 10:30] (n = 29) | 0.015 † |
| Lunch time | 15:30 [15:00, 16:00] (n = 449) | 15:30 [15:00, 16:00] | 16:00 [15:00, 17:00] (n = 61) | 0.025 † |
| Dinner time | 21:30 [21:00, 22:00] (n = 430) | 21:30 [21:00, 22:00] | 22:00 [21:00, 22:30] (n = 42) | 0.007 † |
| Eating midpoint | 15:15 [14:30, 16:00] (n = 459) | 15:15 [14:30, 15:45] | 17:00 [13:45, 19:00] (n = 61) | 0.002 † |
| Eating window (h) | 12.0 [11.0, 13.2] (n = 459) | 12.5 [11.5, 13.5] | 6.8 [6.0, 8.0] (n = 61) | <0.001 † |
| Overnight fasting (h) | 12.0 [10.8, 13.0] (n = 459) | 11.5 [10.5, 12.5] | 17.2 [16.0, 18.0] (n = 61) | <0.001 † |
| Breakfast-Lunch interval (h) | 6.5 [5.5, 7.5] (n = 410) | 6.5 [5.5, 7.5] | 6.5 [6.0, 7.5] (n = 22) | 0.722 † |
| Lunch-Dinner interval (h) | 6.0 [5.0, 6.5] (n = 420) | 6.0 [5.0, 6.5] | 6.4 [6.0, 7.5] (n = 32) | 0.004 † |
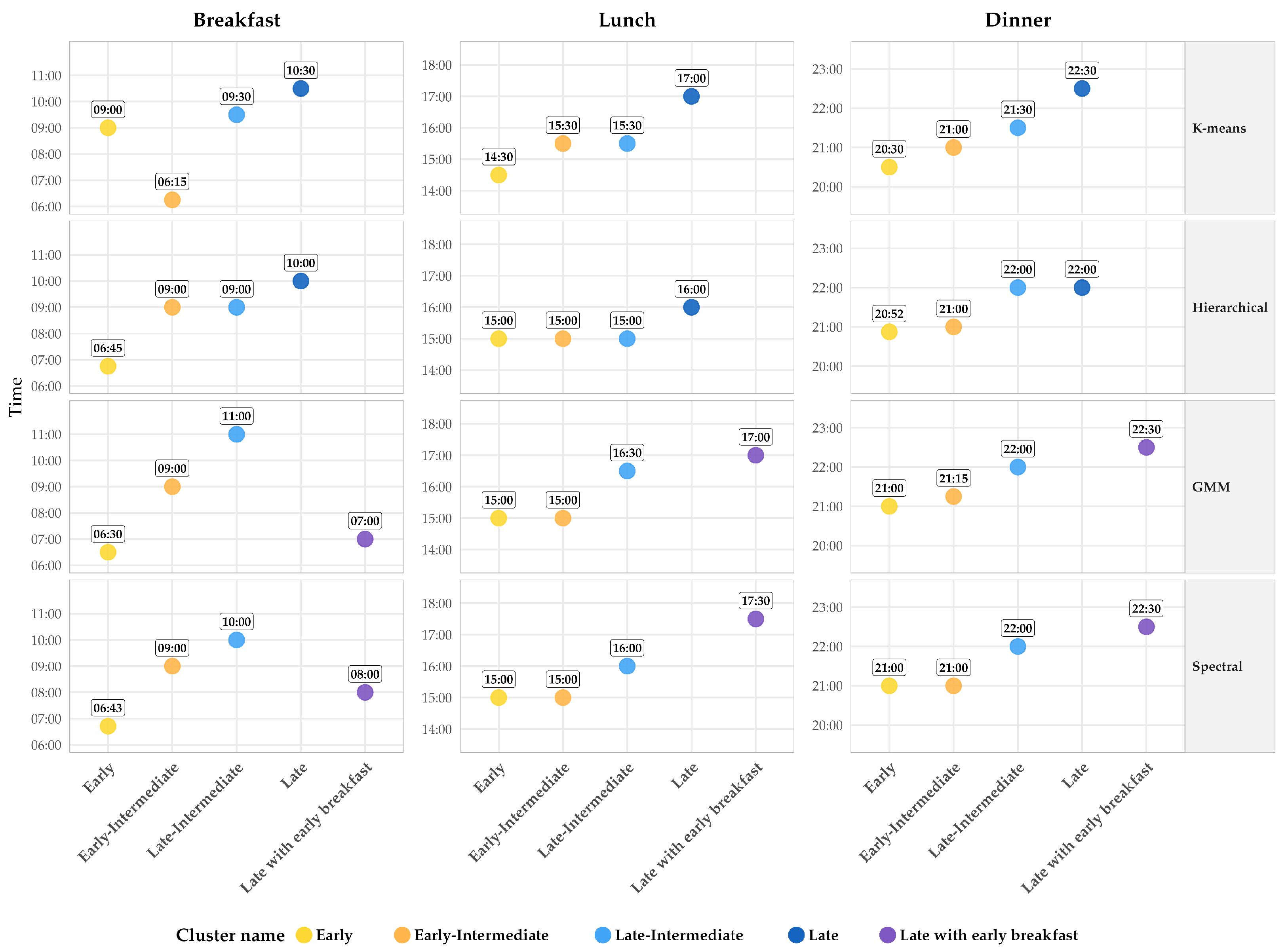
| K-means | |||||
| Mealtime | C1 Early n = 94 (24.2%) | C2 Early-Intermediate n = 68 (17.5%) | C3 Late n = 59 (15.2%) | C4 Late-Intermediate n = 167 (43.0%) | p-Value |
| Breakfast | 09:00 [08:00, 09:30] b | 06:15 [05:50, 07:00] a | 10:30 [09:00, 11:00] d | 09:30 [09:00, 10:00] c | <0.001 |
| Lunch | 14:30 [14:00, 15:00] a | 15:30 [14:45, 16:00] b | 17:00 [16:38, 17:30] d | 15:30 [15:00, 16:00] c | <0.001 |
| Dinner | 20:30 [20:00, 21:00] a | 21:00 [20:45, 21:30] b | 22:30 [22:00, 23:00] d | 21:30 [21:00, 22:00] c | <0.001 |
| Hierarchical | |||||
| Mealtime | C1 Early n = 90 (23.2%) | C2 Late-Intermediate n = 60 (15.5%) | C3 Late n = 142 (36.6%) | C4 Early-Intermediate n = 96 (24.7%) | p-Value |
| Breakfast | 06:45 [06:00, 07:26] a | 09:00 [08:49, 09:30] b | 10:00 [09:00, 10:30] c | 09:00 [09:00, 09:30] b | <0.001 |
| Lunch | 15:00 [14:00, 15:30] a | 15:00 [14:30, 15:00] a | 16:00 [16:00, 17:00] c | 15:00 [15:00, 15:33] b | <0.001 |
| Dinner | 20:52 [20:00, 21:30] a | 22:00 [21:30, 22:30] b | 22:00 [21:30, 22:30] b | 21:00 [20:30, 21:00] a | <0.001 |
| Gaussian Mixture Models | |||||
| Mealtime | C1 Early n = 79 (20.4%) | C2 Late with Early Breakfast n = 17 (4.4%) | C3 Late-Intermediate n = 59 (15.2%) | C4 Early-Intermediate n = 233 (60.1%) | p-Value |
| Breakfast | 06:30 [06:00, 07:00] a | 07:00 [06:15, 07:30] a | 11:00 [10:30, 11:30] c | 09:00 [09:00, 10:00] b | <0.001 |
| Lunch | 15:00 [14:30, 15:30] a | 17:00 [17:00, 17:30] d | 16:30 [16:00, 17:15] c | 15:00 [15:00, 16:00] b | <0.001 |
| Dinner | 21:00 [20:22, 21:30] a | 22:30 [22:00, 22:45] c | 22:00 [21:30, 22:52] c | 21:15 [21:00, 22:00] b | <0.001 |
| Spectral | |||||
| Mealtime | C1 Early n = 86 (22.2%) | C2 Late-Intermediate n = 138 (35.6%) | C3 Early-Intermediate n = 125 (32.2%) | C4 Late with Early Breakfast n = 39 (10.1%) | p-Value |
| Breakfast | 06:43 [06:00, 07:26] a | 10:00 [09:00, 10:30] c | 09:00 [08:45, 09:30] b | 08:00 [07:00, 10:30] b | <0.001 |
| Lunch | 15:00 [14:04, 15:30] a | 16:00 [15:30, 16:30] b | 15:00 [14:30, 15:00] a | 17:30 [17:00, 17:52] c | <0.001 |
| Dinner | 21:00 [20:15, 21:30] a | 22:00 [21:00, 22:00] b | 21:00 [20:30, 21:30] a | 22:30 [22:00, 23:00] c | <0.001 |
| K-means | |||||
| Variable | C1 Early n = 94 (24.2%) | C2 Early–Intermediate n = 68 (17.5%) | C3 Late n = 59 (15.2%) | C4 Late–Intermediate n = 167 (43.0%) | p-Value |
| Age (years) | 21.0 [20.0, 23.0] | 20.0 [19.0, 22.0] | 20.0 [19.0, 22.0] | 20.0 [19.0, 22.0] | 0.129 |
| Sex, Female | 69 (73.4) a | 39 (57.4) a | 47 (79.7) b | 128 (76.6) b | 0.012 |
| Academic program | |||||
| Nutrition | 26 (27.7) a | 15 (22.1) a | 6 (10.2) a | 40 (24.0) a | 0.050 |
| Medicine | 9 (9.6) a | 17 (25.0) b | 16 (27.1) b | 36 (21.6) ab | |
| Psychology | 13 (13.8) a | 10 (14.7) a | 5 (8.5) a | 23 (13.8) a | |
| Other programs | 46 (48.9) a | 26 (38.2) a | 32 (54.2) a | 68 (40.7) a | |
| Employment, Yes | 33 (35.1) | 17 (25.0) | 19 (32.2) | 59 (35.3) | 0.460 |
| Hierarchical | |||||
| Variable | C1 Earlyn = 90 (23.2%) | C2 Late–Intermediate n = 60 (15.5%) | C3 Late n = 142 (36.6%) | C4 Early–Intermediate n = 96 (24.7%) | p-Value |
| Age (years) | 20.0 [19.0, 22.0] | 21.0 [19.0, 22.0] | 20.0 [19.0, 22.0] | 21.0 [19.0, 23.0] | 0.794 |
| Sex, Female | 59 (65.6) | 45 (75.0) | 108 (76.1) | 71 (74.0) | 0.339 |
| Academic program | |||||
| Nutrition | 19 (21.1) a | 17 (28.3) a | 23 (16.2) a | 28 (29.2) a | 0.008 |
| Medicine | 17 (18.9) a | 16 (26.7) a | 33 (23.2) a | 12 (12.5) a | |
| Psychology | 13 (14.4) a | 13 (21.7) a | 14 (9.9) a | 11 (11.5) a | |
| Other programs | 41 (45.6) a | 14 (23.3) b | 72 (50.7) a | 45 (46.9) a | |
| Employment, Yes | 24 (26.7) | 20 (33.3) | 50 (35.2) | 34 (35.4) | 0.531 |
| Gaussian Mixture Models | |||||
| Variable | C1 Early n = 79 (20.4%) | C2 Late with Early Breakfast n = 17 (4.4%) | C3 Late–Intermediate n = 59 (15.2%) | C4 Early–Intermediate n = 233 (60.1%) | p-Value |
| Age (years) | 20.0 [19.0, 22.0] | 19.0 [19.0, 20.0] | 21.0 [19.0, 22.5] | 21.0 [19.0, 22.0] | 0.197 |
| Sex, Female | 52 (65.8) a | 8 (47.1) a | 46 (78.0) a | 177 (76.0) a | 0.022 |
| Academic program | |||||
| Nutrition | 18 (22.8) a | 0 (0.0) a | 12 (20.3) a | 57 (24.5) a | 0.044 |
| Medicine | 14 (17.7) a | 9 (52.9) b | 12 (20.3) ab | 43 (18.5) a | |
| Psychology | 10 (12.7) a | 1 (5.9) a | 5 (8.5) a | 35 (15.0) a | |
| Other programs | 37 (46.8) a | 7 (41.2) a | 30 (50.8) a | 98 (42.1) a | |
| Employment, Yes | 23 (29.1) | 5 (29.4) | 23 (39.0) | 77 (33.0) | 0.661 |
| Spectral | |||||
| Variable | C1 Early n = 86 (22.2%) | C2 Late-Intermediate n = 138 (35.6%) | C3 Early-Intermediate n = 125 (32.2%) | C4 Late with Early Breakfast n = 39 (10.1%) | p-Value |
| Age (years) | 20.0 [19.0, 22.0] | 20.0 [19.0, 22.0] | 21.0 [19.0, 22.0] | 20.0 [19.0, 22.0] | 0.401 |
| Sex, Female | 56 (65.1) | 110 (79.7) | 92 (73.6) | 25 (64.1) | 0.059 |
| Academic program | |||||
| Nutrition | 18 (20.9) | 30 (21.7) | 35 (28.0) | 4 (10.3) | 0.118 |
| Medicine | 16 (18.6) | 30 (21.7) | 19 (15.2) | 13 (33.3) | |
| Psychology | 12 (14.0) | 16 (11.6) | 21 (16.8) | 2 (5.1) | |
| Other programs | 40 (46.5) | 62 (44.9) | 50 (40.0) | 20 (51.3) | |
| Employment, Yes | 23 (26.7) | 51 (37.0) | 41 (32.8) | 13 (33.3) | 0.475 |
| K-means | C1 Early n = 94 (24.2%) | C2 Early–Intermediate n = 68 (17.5%) | C3 Late n = 59 (15.2%) | C4 Late–Intermediate n = 167 (43.0%) | p-Value |
| Eating Window (h) | 11.8 [11.0, 12.5] a | 14.9 [14.0, 15.5] b | 12.2 [11.5, 13.0] c | 12.0 [11.0, 13.0] c | <0.001 |
| Overnight Fasting (h) | 12.2 [11.5, 13.0] a | 9.1 [8.5, 10.0] b | 11.8 [11.0, 12.5] c | 12.0 [11.0, 13.0] c | <0.001 |
| Eating Midpoint | 14:36 [14:00, 15:00] a | 13:45 [13:22, 14:02] b | 16:15 [15:30, 16:45] c | 15:30 [15:15, 16:00] d | <0.001 |
| Breakfast-Lunch (h) | 6.0 [5.0, 6.2] a | 9.0 [8.0, 9.8] b | 7.0 [6.0, 8.0] c | 6.0 [5.5, 6.9] a | <0.001 |
| Lunch-Dinner (h) | 6.0 [5.0, 6.5] a | 6.0 [5.0, 6.5] a | 5.0 [4.8, 6.0] b | 6.0 [5.5, 7.0] a | <0.001 |
| Chronotype, n (%) | |||||
| Morning | 34 (36.6) a | 25 (37.3) a | 4 (6.8) b | 27 (16.3) b | <0.001 |
| Intermediate | 51 (54.8) a | 41 (61.2) ab | 49 (83.1) b | 124 (74.7) b | |
| Evening | 8 (8.6) a | 1 (1.5) a | 6 (10.2) a | 15 (9.0) a | |
| Hierarchical | C1 Early n = 90 (23.2%) | C2 Late–Intermediate n = 60 (15.5%) | C3 Late n = 142 (36.6%) | C4 Early–Intermediate n = 96 (24.7%) | p-Value |
| Eating Window (h) | 14.0 [13.0, 15.0] a | 13.0 [12.0, 13.3] b | 12.0 [11.0, 13.0] c | 11.5 [11.0, 12.0] d | <0.001 |
| Overnight Fasting (h) | 10.0 [9.0, 11.0] a | 11.0 [10.7, 12.0] b | 12.0 [11.0, 13.0] c | 12.5 [12.0, 13.0] d | <0.001 |
| Eating Midpoint | 13:45 [13:30, 14:00] a | 15:30 [15:15, 16:00] b | 15:45 [15:30, 16:28] c | 15:00 [14:45, 15:15] d | <0.001 |
| Breakfast–Lunch (h) | 8.0 [7.5, 9.0] a | 6.0 [5.0, 6.0] b | 6.5 [5.5, 7.5] c | 6.0 [5.0, 7.0] b | <0.001 |
| Lunch–Dinner (h) | 6.0 [5.5, 6.5] a | 7.0 [7.0, 7.8] b | 5.5 [5.0, 6.0] c | 5.5 [5.0, 6.0] c | <0.001 |
| Chronotype, n (%) | |||||
| Morning | 33 (36.7) a | 9 (15.0) b | 17 (12.1) b | 31 (33.0) a | <0.001 |
| Intermediate | 54 (60.0) a | 43 (71.7) ab | 111 (78.7) b | 57 (60.6) a | |
| Evening | 3 (3.3) a | 8 (13.3) a | 13 (9.2) a | 6 (6.4) a | |
| GMM | C1 Early n = 79 (20.4%) | C2 Late with Early Breakfast n = 17 (4.4%) | C3 Late-Intermediate n = 59 (15.2%) | C4 Early-Intermediate n = 233 (60.1%) | p-Value |
| Eating Window (h) | 14.3 [13.6, 15.1] a | 15.5 [14.8, 15.8] b | 11.5 [10.4, 12.0] c | 12.0 [11.0, 13.0] d | <0.001 |
| Overnight Fasting (h) | 9.7 [8.9, 10.4] a | 8.5 [8.2, 9.2] b | 12.5 [12.0, 13.6] c | 12.0 [11.0, 13.0] d | <0.001 |
| Eating Midpoint | 13:45 [13:21, 14:00] a | 14:45 [14:22, 15:00] b | 16:30 [16:04, 17:00] c | 15:15 [15:00, 15:30] d | <0.001 |
| Breakfast-Lunch (h) | 8.2 [7.5, 9.0] a | 10.0 [9.6, 10.8] b | 6.0 [5.0, 6.5] c | 6.0 [5.5, 6.8] c | <0.001 |
| Lunch-Dinner (h) | 6.0 [5.3, 6.5] a | 5.0 [4.8, 6.0] b | 5.5 [4.6, 6.0] b | 6.0 [5.0, 6.8] a | <0.001 |
| Chronotype, n (%) | |||||
| Morning | 27 (34.2) a | 4 (25.0) ab | 1 (1.7) b | 58 (25.1) a | 0.001 |
| Intermediate | 49 (62.0) a | 12 (75.0) ab | 51 (86.4) b | 153 (66.2) a | |
| Evening | 3 (3.8) a | 0 (0.0) a | 7 (11.9) a | 20 (8.7) a | |
| Spectral | C1 Early n = 86 (22.2%) | C2 Late-Intermediate n = 138 (35.6%) | C3 Early-Intermediate n = 125 (32.2%) | C4 Late with Early Breakfast n = 39 (10.1%) | p-Value |
| Eating Window (h) | 14.1 [13.5, 15.0] a | 12.0 [11.0, 12.5] b | 12.0 [11.0, 13.0] b | 13.0 [12.1, 15.4] c | <0.001 |
| Overnight Fasting (h) | 9.9 [9.0, 10.5] a | 12.0 [11.5, 13.0] b | 12.0 [11.0, 13.0] b | 11.0 [8.6, 11.9] c | <0.001 |
| Eating Midpoint | 13:45 [13:30, 14:00] a | 15:51 [15:30, 16:15] b | 15:00 [14:45, 15:25] c | 15:15 [14:45, 17:00] bc | <0.001 |
| Breakfast–Lunch (h) | 8.2 [7.5, 9.0] a | 6.0 [5.5, 7.0] b | 6.0 [5.0, 6.0] b | 9.0 [7.0, 10.0] a | <0.001 |
| Lunch–Dinner (h) | 6.0 [5.5, 6.5] a | 5.5 [5.0, 6.5] ab | 6.0 [5.5, 6.8] a | 5.0 [4.6, 6.0] b | <0.001 |
| Chronotype, n (%) | |||||
| Morning | 32 (37.2) a | 19 (13.8) b | 35 (28.0) a | 4 (10.3) b | <0.001 |
| Intermediate | 52 (60.5) a | 99 (71.7) ab | 82 (65.6) ab | 32 (82.1) b | |
| Evening | 2 (2.3) a | 19 (13.8) b | 7 (5.6) a | 2 (5.1) ab | |
| K-means | |||||
| Variable | C1 Early n = 94 (24.2%) | C2 Early–Intermediate n = 68 (17.5%) | C3 Late n = 59 (15.2%) | C4 Late–Intermediate n = 167 (43.0%) | p-Value |
| BMI (kg/m2) | 22.9 [20.8, 26.4] | 23.0 [21.1, 25.2] | 22.5 [21.0, 27.0] | 22.7 [20.1, 25.6] | 0.949 |
| Waist circumference (cm) | 71.5 [66.4, 78.7] | 72.9 [66.5, 79.9] | 70.3 [66.7, 80.3] | 72.4 [66.2, 78.4] | 0.955 |
| Food Intake Quality, n (%) | |||||
| Healthy food intake | 39 (41.5) a | 31 (45.6) a | 9 (15.3) b | 36 (21.6) b | <0.001 |
| Habits in need of improvement | 28 (29.8) a | 18 (26.5) a | 20 (33.9) a | 50 (29.9) a | |
| Unhealthy food intake | 26 (27.7) a | 18 (26.5) a | 30 (50.8) b | 80 (47.9) b | |
| Hierarchical | |||||
| Variable | C1 Early n = 90 (23.2%) | C2 Late-Intermediate n = 60 (15.5%) | C3 Late n = 142 (36.6%) | C4 Early-Intermediate n = 96 (24.7%) | p-Value |
| BMI (kg/m2) | 22.9 [20.6, 25.6] | 22.8 [21.2, 25.5] | 22.8 [20.5, 26.5] | 22.1 [20.4, 25.6] | 0.521 |
| Waist circumference (cm) | 72.1 [66.3, 79.2] | 72.9 [68.2, 80.0] | 72.2 [66.6, 79.2] | 71.1 [65.4, 77.2] | 0.537 |
| Food Intake Quality, n (%) | |||||
| Healthy food intake | 39 (43.3) a | 16 (26.7) ab | 26 (18.3) b | 34 (35.4) a | <0.001 |
| Habits in need of improvement | 25 (27.8) a | 17 (28.3) a | 42 (29.6) a | 32 (33.3) a | |
| Unhealthy food intake | 26 (28.9) a | 27 (45.0) ab | 73 (51.4) b | 28 (29.2) a | |
| Gaussian Mixture Models | |||||
| Variable | C1 Early n = 79 (20.4%) | C2 Late with Early Breakfast n = 17 (4.4%) | C3 Late–Intermediate n = 59 (15.2%) | C4 Early–Intermediate n = 233 (60.1%) | p-Value |
| BMI (kg/m2) | 23.0 [20.6, 25.6] | 22.8 [21.8, 24.6] | 23.0 [20.7, 27.0] | 22.5 [20.6, 25.6] | 0.605 |
| Waist circumference (cm) | 71.8 [66.2, 79.5] | 74.8 [69.8, 80.0] | 72.0 [67.1, 79.3] | 71.6 [66.1, 78.8] | 0.396 |
| Food Intake Quality, n (%) | |||||
| Healthy food intake | 37 (46.8) a | 3 (17.6) ab | 12 (20.3) b | 63 (27.0) b | 0.017 |
| Habits in need of improvement | 19 (24.1) a | 5 (29.4) a | 21 (35.6) a | 71 (30.5) a | |
| Unhealthy food intake | 23 (29.1) a | 8 (47.1) a | 26 (44.1) a | 97 (41.6) a | |
| Spectral | |||||
| Variable | C1 Early n = 86 (22.2%) | C2 Late–Intermediate n = 138 (35.6%) | C3 Early–Intermediate n = 125 (32.2%) | C4 Late with Early Breakfast n = 39 (10.1%) | p-Value |
| BMI (kg/m2) | 22.8 [20.4, 25.5] | 22.6 [20.0, 26.3] | 22.6 [20.8, 25.6] | 23.8 [21.4, 26.6] | 0.717 |
| Waist circumference (cm) | 71.5 [66.3, 78.9] | 72.2 [66.0, 80.0] | 71.6 [66.3, 78.0] | 72.4 [68.4, 81.2] | 0.697 |
| Food Intake Quality, n (%) | |||||
| Healthy food intake | 38 (44.2) a | 26 (18.8) b | 43 (34.4) a | 8 (20.5) b | <0.001 |
| Habits in need of improvement | 23 (26.7) a | 37 (26.8) a | 41 (32.8) a | 15 (38.5) a | |
| Unhealthy food intake | 25 (29.1) a | 74 (53.6) b | 40 (32.0) a | 15 (38.5) ab | |
| Method | Silhouette a | Dunn a | Calinski-Harabasz a | Hubert’s Gamma a | Davies-Bouldin b | Entropy b |
| K-means | 0.250 | 0.045 | 149.8 | 0.477 | 1.396 | 1.298 |
| Hierarchical | 0.195 | 0.039 | 124.5 | 0.355 | 1.411 | 1.341 |
| GMM | 0.247 | 0.038 | 112.1 | 0.493 | 1.262 | 1.054 |
| Spectral | 0.215 | 0.019 | 108.1 | 0.446 | 1.870 | 1.297 |
| Concordance Matrix (Adjusted Rand Index) | ||||||
| K-means | Hierarchical | GMM | Spectral | |||
| K-means | — | |||||
| Hierarchical | 0.281 * | — | ||||
| GMM | 0.408 ** | 0.271 * | — | |||
| Spectral | 0.402 ** | 0.485 *** | 0.411 ** | — | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Mora-Almanza, J.G.; Betancourt-Núñez, A.; Nava-Amante, P.A.; Bernal-Orozco, M.F.; Díaz-López, A.; Martínez, J.A.; Vizmanos, B. Traditional and Non-Traditional Clustering Techniques for Identifying Chrononutrition Patterns in University Students. Nutrients 2026, 18, 190. https://doi.org/10.3390/nu18020190
Mora-Almanza JG, Betancourt-Núñez A, Nava-Amante PA, Bernal-Orozco MF, Díaz-López A, Martínez JA, Vizmanos B. Traditional and Non-Traditional Clustering Techniques for Identifying Chrononutrition Patterns in University Students. Nutrients. 2026; 18(2):190. https://doi.org/10.3390/nu18020190
Chicago/Turabian StyleMora-Almanza, José Gerardo, Alejandra Betancourt-Núñez, Pablo Alejandro Nava-Amante, María Fernanda Bernal-Orozco, Andrés Díaz-López, José Alfredo Martínez, and Barbara Vizmanos. 2026. "Traditional and Non-Traditional Clustering Techniques for Identifying Chrononutrition Patterns in University Students" Nutrients 18, no. 2: 190. https://doi.org/10.3390/nu18020190
APA StyleMora-Almanza, J. G., Betancourt-Núñez, A., Nava-Amante, P. A., Bernal-Orozco, M. F., Díaz-López, A., Martínez, J. A., & Vizmanos, B. (2026). Traditional and Non-Traditional Clustering Techniques for Identifying Chrononutrition Patterns in University Students. Nutrients, 18(2), 190. https://doi.org/10.3390/nu18020190












