Lactiplantibacillus plantarum Lp20 Alleviates High Fat Diet-Induced Obesity in Mice via Its Bile Salt Hydrolase Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Condition
2.2. BSH Activity Assays of L. plantarum
2.3. Draft Genome Sequencing and Comparative Analysis
2.4. Construction of bsh1 Deletion Mutant and Complemented Strains
2.5. Animal Experiment
2.6. Biochemical Assay of Blood Serum
2.7. Liver Histological Analysis
2.8. RT-qPCR
2.9. Microbiome Analysis
2.10. Statistical Analysis
3. Results
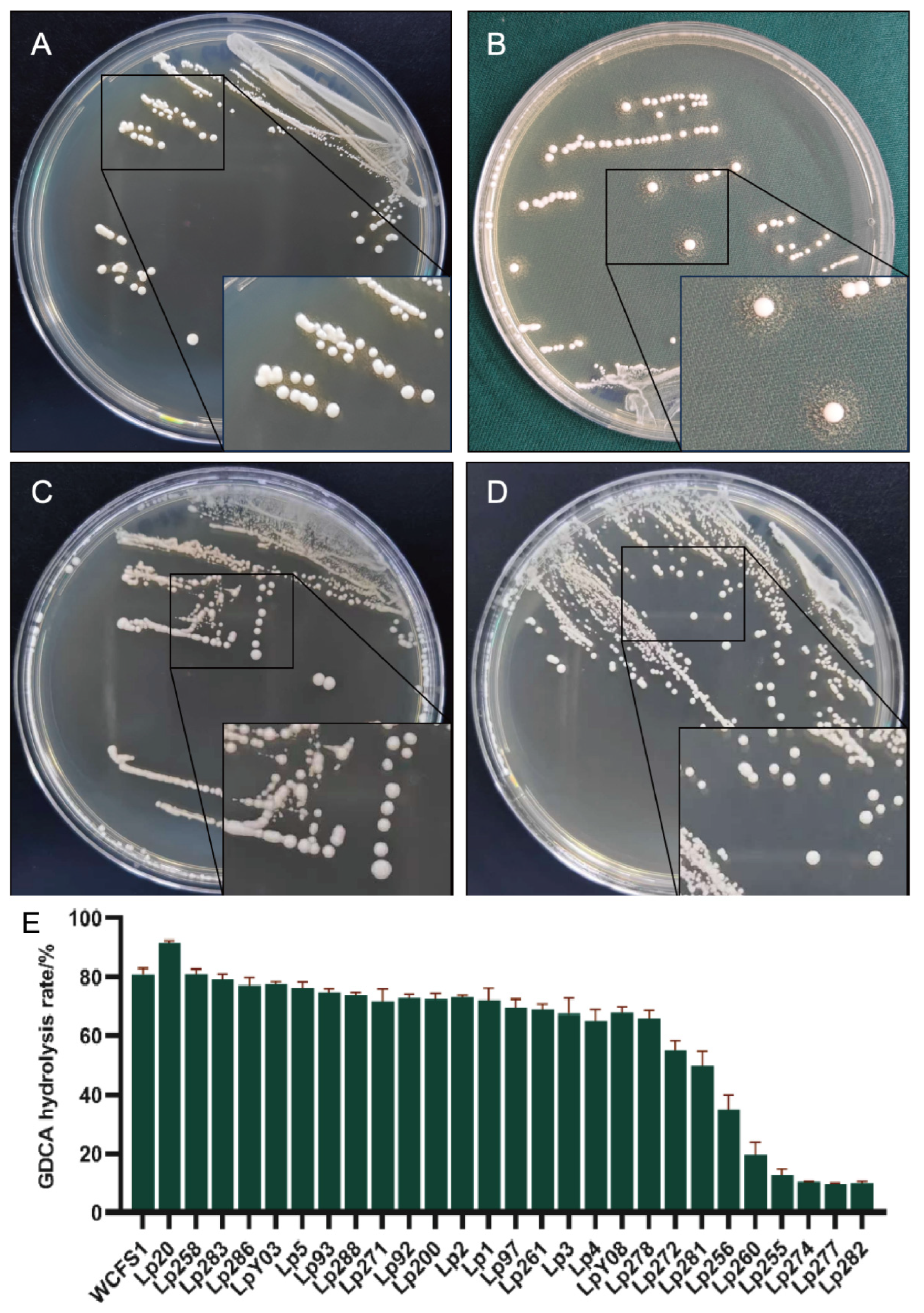
3.1. L. plantarum Lp20 Exhibited the Highest GDCA Hydrolase Activity
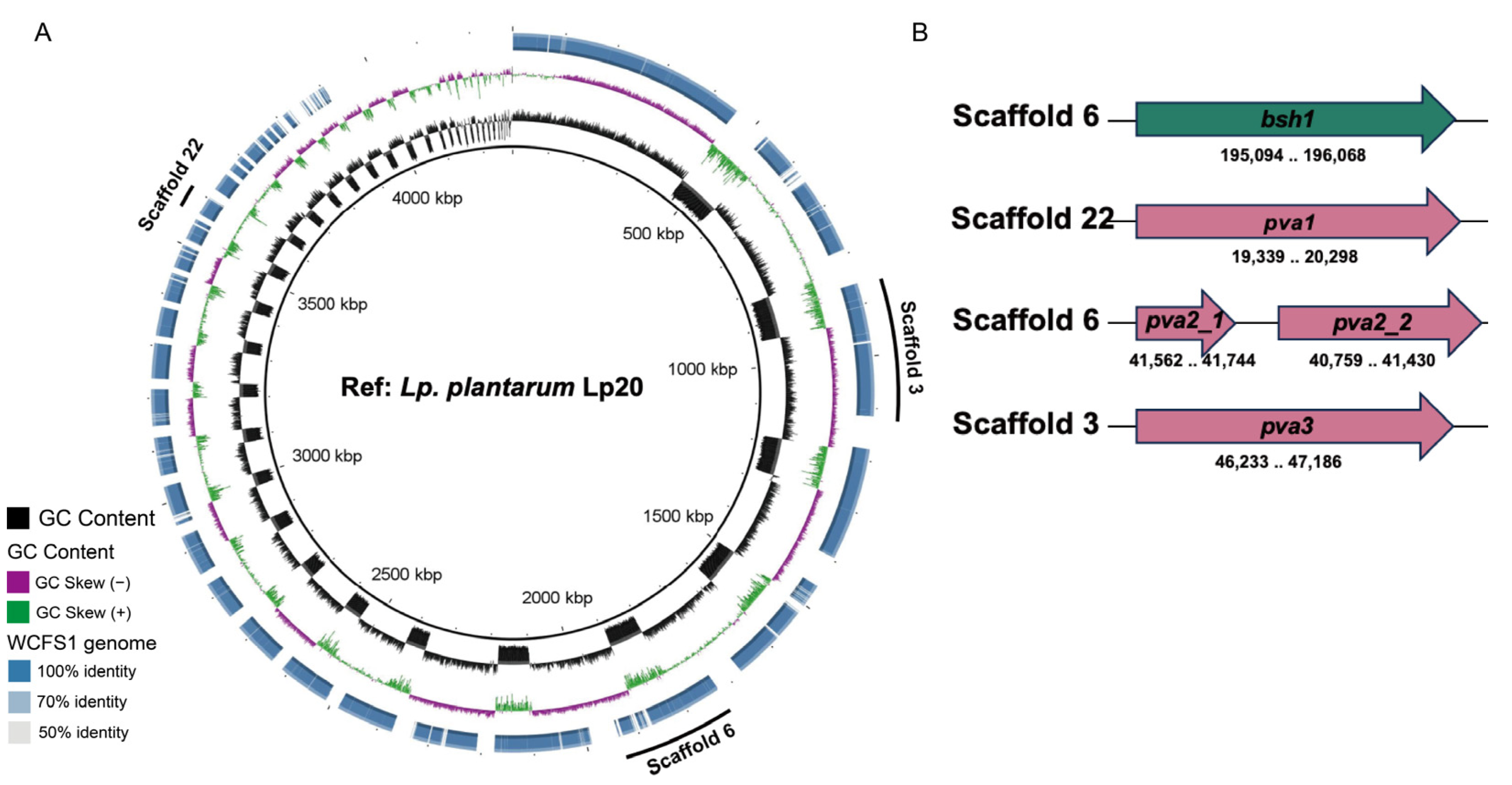
3.2. BSH-like Genes Were Located in the Chromosome of L. plantarum Lp20
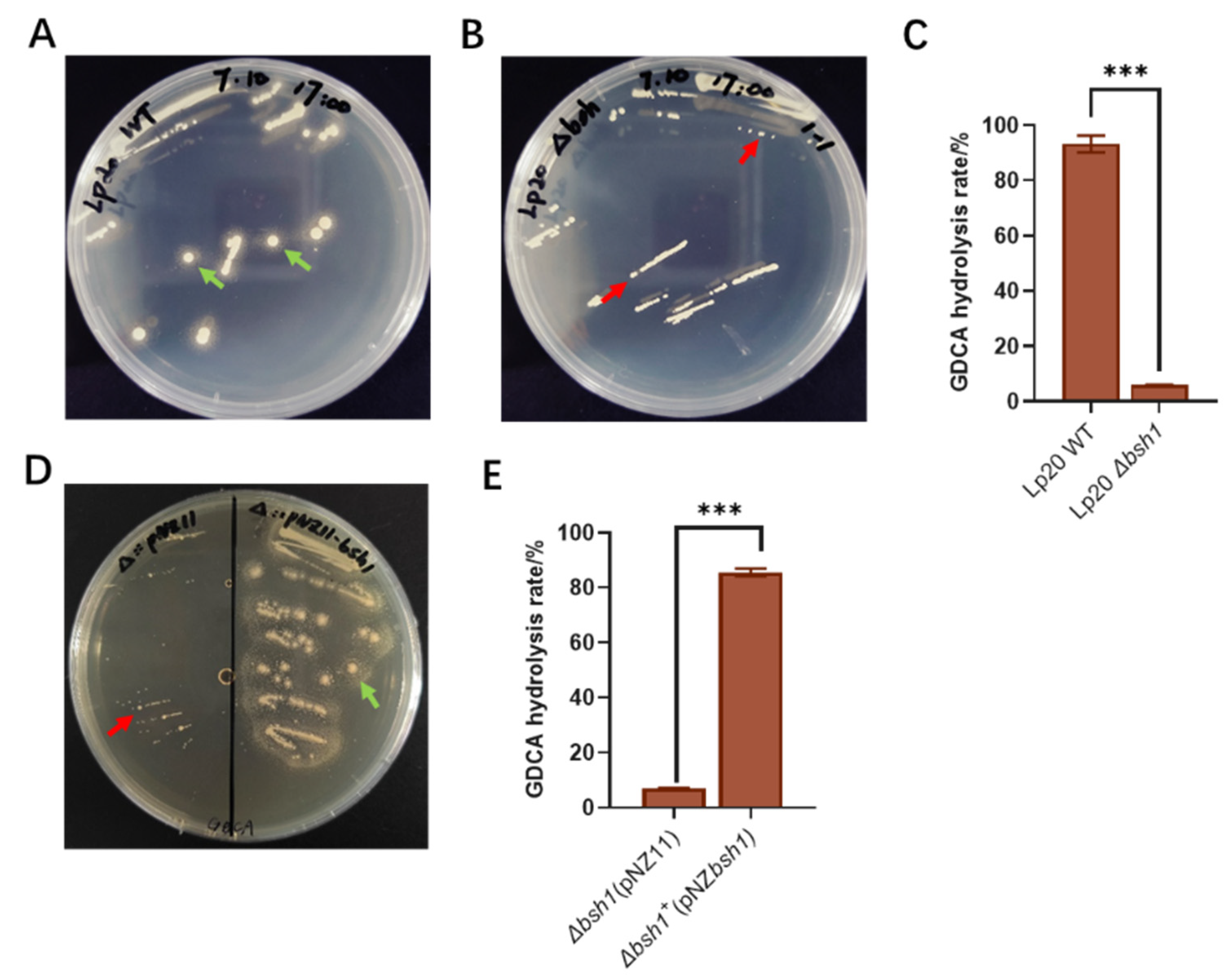
3.3. Bsh1 Played a Key Role in the GDCA Hydrolysis Activity of L. plantarum Lp20
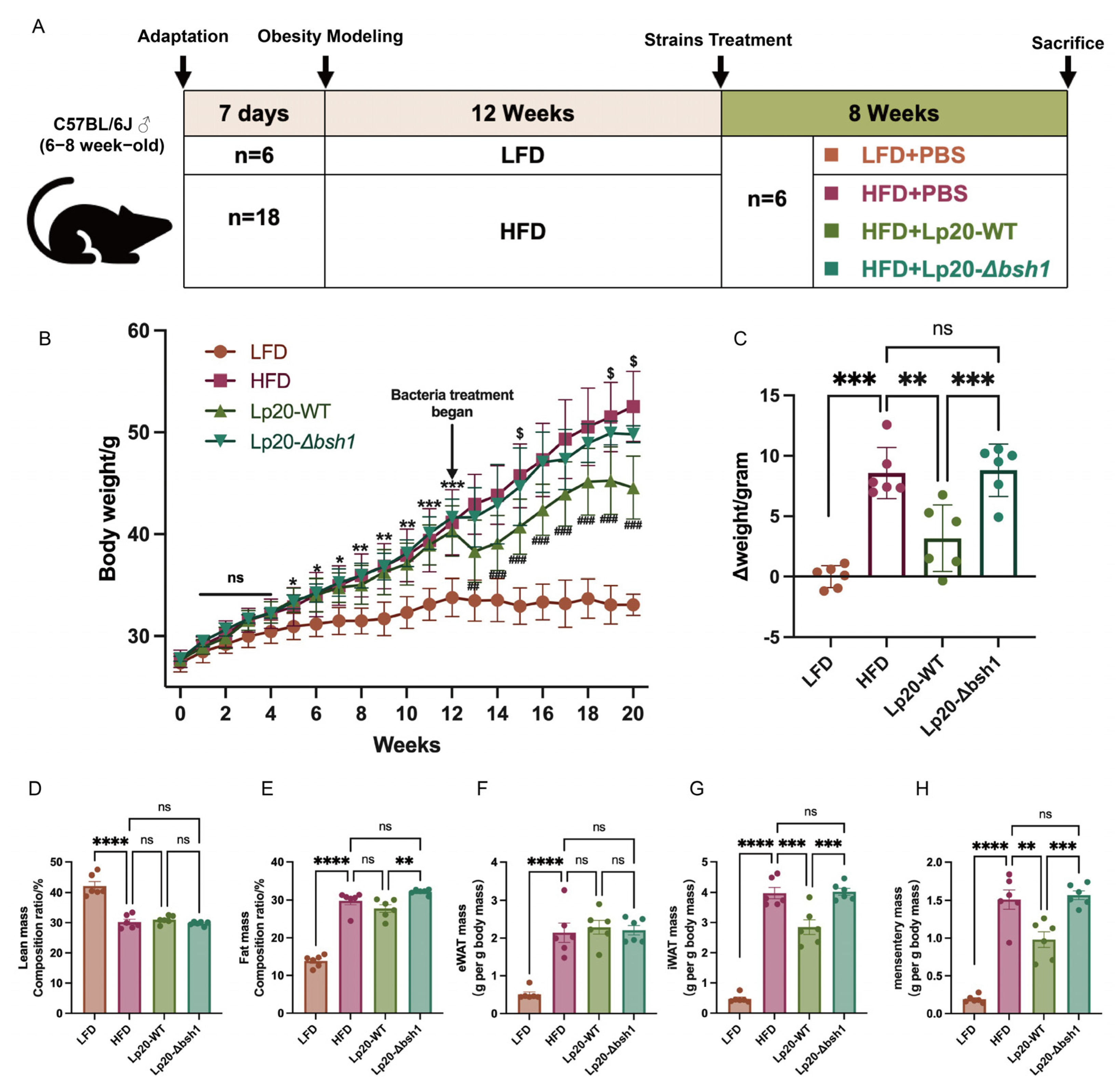
3.4. L. plantarum Lp20 Decreased Weight Gain and Fat Accumulation in Obese Mice
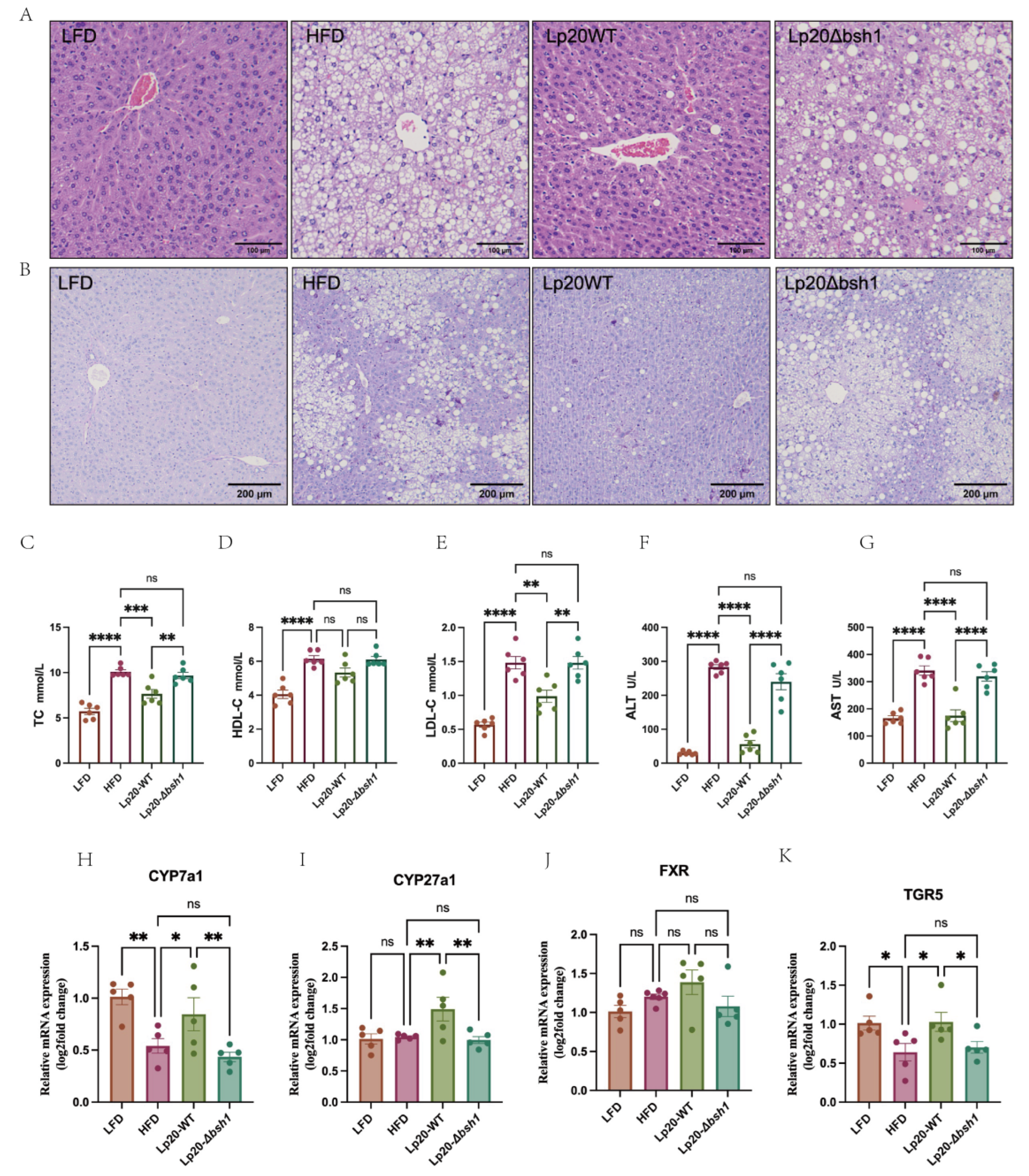
3.5. Bsh1 Alleviated Liver Steatosis and Decreased Serum Cholesterol in Obese Mice
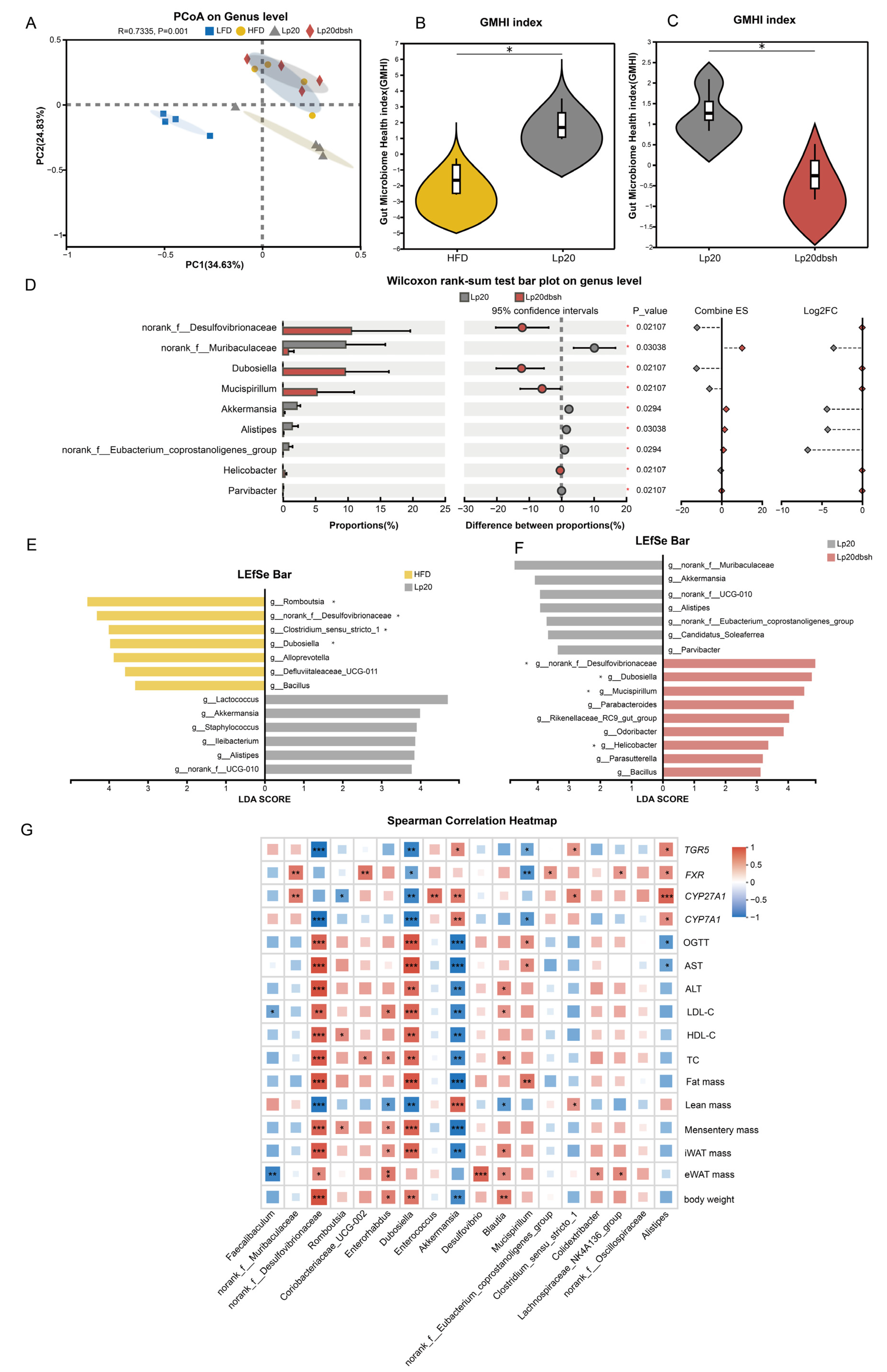
3.6. L. plantarum Lp20 Restored the Intestinal Microbial Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef]
- Shaikh, S.R.; Beck, M.A.; Alwarawrah, Y.; MacIver, N.J. Emerging mechanisms of obesity-associated immune dysfunction. Nat. Rev. Endocrinol. 2024, 20, 136–148. [Google Scholar] [CrossRef]
- Lobstein, T.; Jackson-Leach, R.; Powis, J.; Brinsden, H.; Gray, M. World Obesity Atlas 2023; World Obesity Federation: London, UK, 2023. [Google Scholar]
- Cox, A.J.; West, N.P.; Cripps, A.W. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015, 3, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed]
- Michael, D.R.; Jack, A.A.; Masetti, G.; Davies, T.S.; Loxley, K.E.; Kerry-Smith, J.; Plummer, J.F.; Marchesi, J.R.; Mullish, B.H.; McDonald, J.A.K.; et al. A randomised controlled study shows supplementation of overweight and obese adults with lactobacilli and bifidobacteria reduces bodyweight and improves well-being. Sci. Rep. 2020, 10, 4183. [Google Scholar] [CrossRef]
- Hsieh, F.C.; Lan, C.C.; Huang, T.Y.; Chen, K.W.; Chai, C.Y.; Chen, W.T.; Fang, A.H.; Chen, Y.H.; Wu, C.S. Heat-killed and live Lactobacillus reuteri GMNL-263 exhibit similar effects on improving metabolic functions in high-fat diet-induced obese rats. Food Funct. 2016, 7, 2374–2388. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Y.; Ma, F.; Sun, M.; Mu, G.; Tuo, Y. The ameliorative effect of Lactobacillus plantarum Y44 oral administration on inflammation and lipid metabolism in obese mice fed with a high fat diet. Food Funct. 2020, 11, 5024–5039. [Google Scholar] [CrossRef]
- Fan, J.; Jin, W.L.; Su, S.C.; Khan, A.; Wu, Y.; Chen, Y.Y.; Feng, P.Y.; Jeon, B.-H.; Salama, E.-S.; Ling, Z.M.; et al. A probiotic targets bile acids metabolism to alleviate ulcerative colitis by reducing conjugated bile acids. Mol. Nutr. Food Res. 2024, 68, 2300731. [Google Scholar] [CrossRef]
- Li, L.; Liu, T.; Gu, Y.; Wang, X.; Xie, R.; Sun, Y.; Wang, B.; Cao, H. Regulation of gut microbiota-bile acids axis by probiotics in inflammatory bowel disease. Front. Immunol. 2022, 13, 974305. [Google Scholar] [CrossRef]
- Huang, C.H.; Ho, C.Y.; Chen, C.T.; Hsu, H.F.; Lin, Y.H. Probiotic BSH Activity and Anti-Obesity Potential of Lactobacillus plantarum Strain TCI378 Isolated from Korean Kimchi. Prev. Nutr. Food Sci. 2019, 24, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhou, X.H.; Gong, P.M.; Niu, H.Y.; Lyu, L.Z.; Wu, Y.F.; Han, X.; Zhang, L.W. Lactiplantibacillus plantarum H-87 prevents high-fat diet-induced obesity by regulating bile acid metabolism in C57BL/6J mice. Food Funct. 2021, 12, 4315–4324. [Google Scholar] [CrossRef]
- Joyce, S.A.; MacSharry, J.; Casey, P.G.; Kinsella, M.; Murphy, E.F.; Shanahan, F.; Hill, C.; Gahan, C.G. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. USA 2014, 111, 7421–7426. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, S.; Zhang, Z.; Zhang, T.; Teng, X.; Xiao, G.; Huang, S. Isolation of Lactobacillus acidophilus strain and its anti-obesity effect in a diet induced obese murine model. Lett. Appl. Microbiol. 2024, 77, ovae021. [Google Scholar] [CrossRef]
- Jia, B.; Zou, Y.; Han, X.; Bae, J.W.; Jeon, C.O. Gut microbiome-mediated mechanisms for reducing cholesterol levels: Implications for ameliorating cardiovascular disease. Trends Microbiol. 2023, 31, 76–91. [Google Scholar] [CrossRef]
- Joyce, S.A.; Shanahan, F.; Hill, C.; Gahan, C.G. Bacterial bile salt hydrolase in host metabolism: Potential for influencing gastrointestinal microbe-host crosstalk. Gut Microbes 2014, 5, 669–674. [Google Scholar] [CrossRef]
- Li, T.; Owsley, E.; Matozel, M.; Hsu, P.; Novak, C.M.; Chiang, J.Y. Transgenic expression of cholesterol 7α-hydroxylase in the liver prevents high-fat diet–induced obesity and insulin resistance in mice. Hepatology 2010, 52, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Degirolamo, C.; Rainaldi, S.; Bovenga, F.; Murzilli, S.; Moschetta, A. Microbiota modification with probiotics induces hepatic bile acid synthesis via downregulation of the Fxr-Fgf15 axis in mice. Cell Rep. 2014, 7, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.B.; Lew, L.C.; Yeo, S.K.; Nair Parvathy, S.; Liong, M.T. Probiotics and the BSH-related cholesterol lowering mechanism: A Jekyll and Hyde scenario. Crit. Rev. Biotechnol. 2015, 35, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Huang, W.; Young, R.L.; Jones, K.L.; Horowitz, M.; Rayner, C.K.; Wu, T. Role of Bile Acids in the Regulation of Food Intake, and Their Dysregulation in Metabolic Disease. Nutrients 2021, 13, 1104. [Google Scholar] [CrossRef]
- Foley, M.H.; O’Flaherty, S.; Barrangou, R.; Theriot, C.M. Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019, 15, e1007581. [Google Scholar] [CrossRef]
- Karlov, D.S.; Long, S.L.; Zeng, X.; Xu, F.; Lal, K.; Cao, L.; Hayoun, K.; Lin, J.; Joyce, S.A.; Tikhonova, I.G. Characterization of the mechanism of bile salt hydrolase substrate specificity by experimental and computational analyses. Structure 2023, 31, 629–638.e625. [Google Scholar] [CrossRef] [PubMed]
- Pushpass, R.G.; Alzoufairi, S.; Jackson, K.G.; Lovegrove, J.A. Circulating bile acids as a link between the gut microbiota and cardiovascular health: Impact of prebiotics, probiotics and polyphenol-rich foods. Nutr. Res. Rev. 2022, 35, 161–180. [Google Scholar] [CrossRef]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile acids and the gut microbiota: Metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2023, 21, 236–247. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Gege, C.; Hambruch, E.; Hambruch, N.; Kinzel, O.; Kremoser, C. Nonsteroidal FXR Ligands: Current Status and Clinical Applications. Handb. Exp. Pharmacol. 2019, 256, 167–205. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sun, K.; Wu, Z.; Yao, J.; Guo, B. All 4 bile salt hydrolase proteins are responsible for the hydrolysis activity in Lactobacillus plantarum ST-III. J. Food Sci. 2011, 76, M622–M628. [Google Scholar] [CrossRef]
- Kumar, R.; Grover, S.; Batish, V.K. Bile Salt Hydrolase (Bsh) Activity Screening of Lactobacilli: In Vitro Selection of Indigenous Lactobacillus Strains with Potential Bile Salt Hydrolysing and Cholesterol-Lowering Ability. Probiotics Antimicrob. Proteins 2012, 4, 162–172. [Google Scholar] [CrossRef]
- Liong, M.T.; Shah, N.P. Bile salt deconjugation ability, bile salt hydrolase activity and cholesterol co-precipitation ability of lactobacilli strains. Int. Dairy J. 2005, 15, 391–398. [Google Scholar] [CrossRef]
- Zhu, H.; Zhao, F.; Zhang, W.; Xia, W.; Chen, Y.; Liu, Y.; Fan, Z.; Zhang, Y.; Yang, Y. Cholesterol-lowering effect of bile salt hydrolase from a Lactobacillus johnsonii strain mediated by FXR pathway regulation. Food Funct. 2022, 13, 725–736. [Google Scholar] [CrossRef]
- Zhao, M.; Kuang, W.; Yang, J.; Liu, Y.; Yang, M.; Chen, Y.; Zhu, H.; Yang, Y. Cholesterol lowering in diet-induced hypercholesterolemic mice using Lactobacillus bile salt hydrolases with different substrate specificities. Food Funct. 2024, 15, 1340–1354. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H.; Reddy, V.S.; Moreno-Hagelsieb, G.; Hendargo, K.J.; Zhang, Y.; Iddamsetty, V.; Lam, K.J.K.; Tian, N.; Russum, S.; Wang, J.; et al. The Transporter Classification Database (TCDB): 2021 update. Nucleic Acids Res. 2021, 49, D461–D467. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Huo, Y.; Lu, X.; Wang, X.; Wang, X.; Chen, L.; Guo, H.; Zhang, M.; Li, Y. Bifidobacterium animalis subsp. lactis A6 Alleviates Obesity Associated with Promoting Mitochondrial Biogenesis and Function of Adipose Tissue in Mice. Molecules 2020, 25, 1490. [Google Scholar] [CrossRef]
- Tian, A.; Yang, Y. Diagnosis of nonalcoholic fatty liver disease: The importance of pathology. J. Clin. Hepatol. 2023, 39, 491–497. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2017, 2, 10–1128. [Google Scholar] [CrossRef]
- Lambert, J.M.; Bongers, R.S.; de Vos, W.M.; Kleerebezem, M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 2008, 74, 4719–4726. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, R.; Odjidja, E.N.; Scott, D.; Shivappa, N.; Hébert, J.R.; Hodge, A.; de Courten, B. The dietary inflammatory index, obesity, type 2 diabetes, and cardiovascular risk factors and diseases. Obes. Rev. 2022, 23, e13349. [Google Scholar] [CrossRef]
- Auerbach, N.; Liu, V.N.; Huang, D.R.; Clift, A.K.; Al-Ammouri, M.; El-Osta, A. What are community perspectives and experiences around GLP-1 receptor agonist medications for weight loss? A cross-sectional survey study in the UK. BMJ Public Health 2025, 3, e002519. [Google Scholar] [CrossRef]
- Lin, J.; Nie, Q.; Cheng, J.; Zhong, Y.N.; Zhang, T.; Zhang, X.; Ge, X.; Ding, Y.; Niu, C.; Gao, Y.; et al. A microbial amino-acid-conjugated bile acid, tryptophan-cholic acid, improves glucose homeostasis via the orphan receptor MRGPRE. Cell 2025, 188, P4530–P4548.E25. [Google Scholar] [CrossRef]
- Guzior, D.V.; Okros, M.; Shivel, M.; Armwald, B.; Bridges, C.; Fu, Y.; Martin, C.; Schilmiller, A.L.; Miller, W.M.; Ziegler, K.M.; et al. Bile salt hydrolase acyltransferase activity expands bile acid diversity. Nature 2024, 626, 852–858. [Google Scholar] [CrossRef]
- Prete, R.; Long, S.L.; Gallardo, A.L.; Gahan, C.G.; Corsetti, A.; Joyce, S.A. Beneficial bile acid metabolism from Lactobacillus plantarum of food origin. Sci. Rep. 2020, 10, 1165. [Google Scholar] [CrossRef]
- Pasolli, E.; De Filippis, F.; Mauriello, I.E.; Cumbo, F.; Walsh, A.M.; Leech, J.; Cotter, P.D.; Segata, N.; Ercolini, D. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020, 11, 2610. [Google Scholar] [CrossRef]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, H.; Feng, X.; Tang, H.; Xiong, Z.; Xia, Y.; Ai, L.; Song, X. Specific bile salt hydrolase genes in Lactobacillus plantarum AR113 and relationship with bile salt resistance. LWT 2021, 145, 111208. [Google Scholar] [CrossRef]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef]
- García-Ruiz, I.; Solís-Muñoz, P.; Fernández-Moreira, D.; Grau, M.; Colina, F.; Muñoz-Yagüe, T.; Solís-Herruzo, J.A. High-fat diet decreases activity of the oxidative phosphorylation complexes and causes nonalcoholic steatohepatitis in mice. Dis. Model. Mech. 2014, 7, 1287–1296. [Google Scholar] [CrossRef]
- Pratt, D.S.; Kaplan, M.M. Evaluation of abnormal liver-enzyme results in asymptomatic patients. N. Engl. J. Med. 2000, 342, 1266–1271. [Google Scholar] [CrossRef]
- Kong, B.; Wang, L.; Chiang, J.Y.; Zhang, Y.; Klaassen, C.D.; Guo, G.L. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 2012, 56, 1034–1043. [Google Scholar] [CrossRef]
- Stanimirov, B.; Stankov, K.; Mikov, M. Bile acid signaling through farnesoid X and TGR5 receptors in hepatobiliary and intestinal diseases. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 18–33. [Google Scholar] [CrossRef] [PubMed]
| Product | D12450J (LFD) | D12492 (HFD) * | ||
|---|---|---|---|---|
| gm% | kcal% | gm% | kcal% | |
| Protein | 19.2 | 20.0 | 26 | 20.0 |
| Carbohydrate | 67.3 | 70.0 | 26 | 20.0 |
| Fat | 4.3 | 10.0 | 35 | 60.0 |
| Ingredient | gm | kcal | gm | kcal |
| Casein, 30 Mesh | 200 | 800 | 200 | 800 |
| L-Cystine | 3 | 12 | 3 | 12 |
| Corn Starch | 506.2 | 2024.8 | 0 | 0 |
| Maltodextrin 10 | 125 | 500 | 125 | 500 |
| Sucrose | 68.8 | 275.2 | 68.8 | 275.2 |
| Cellulose, BW 200 | 50 | 0 | 50 | 0 |
| Soybean Oil | 25 | 225 | 25 | 225 |
| Lard | 20 | 180 | 245 | 2205 |
| Mineral Mix S10026 | 10 | 0 | 10 | 0 |
| Dicalcium Phosphate | 13 | 0 | 13 | 0 |
| Calcium Carbonate | 5.5 | 0 | 5.5 | 0 |
| Potassium Citrate·1H2O | 16.5 | 0 | 16.5 | 0 |
| Vitamin Mix V10001 | 10 | 40 | 10 | 40 |
| Choline Bitartrate | 2 | 0 | 2 | 0 |
| FD&C Yellow Dye #5 | 0.04 | 0 | 0 | 0 |
| FD&C Red Dye #40 | 0 | 0 | 0 | 0 |
| FD&C Blue Day #1 | 0.01 | 0 | 0.05 | 0 |
| Total | 1055.05 | 4057 | 773.85 | 4057 |
| kcal/gm | 3.85 | 5.24 | ||
| Gene | Primer Sequence (5′-3′) |
|---|---|
| CYP7a1 | Forward: GGGATTGCTGTGGTAGTGAGC |
| Reverse: GGTATGGAATCAACCCGTTGTC | |
| CYP27a1 | Forward: CCAGGCACAGGAGAGTACG |
| Reverse: GGGCAAGTGCAGCACATAG | |
| FXR | Forward: GCTTGATGTGCTACAAAAGCTG |
| Reverse: CGTGGTGATGGTTGAATGTCC | |
| TGR5 | Forward: CCTGGCAAGCCTCATCGTC |
| Reverse: AGCAGCCCGGCTAGTAGTAG | |
| β-actin | Forward: GTGACGTTGACATCCGTAAAGA |
| Reverse: GCCGGACTCATCGTACTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, X.; Lu, F.; Jing, Y.; Wang, H.; Qian, H.; Zhang, M.; Zhai, Z.; Hao, Y. Lactiplantibacillus plantarum Lp20 Alleviates High Fat Diet-Induced Obesity in Mice via Its Bile Salt Hydrolase Activity. Nutrients 2025, 17, 3555. https://doi.org/10.3390/nu17223555
Bai X, Lu F, Jing Y, Wang H, Qian H, Zhang M, Zhai Z, Hao Y. Lactiplantibacillus plantarum Lp20 Alleviates High Fat Diet-Induced Obesity in Mice via Its Bile Salt Hydrolase Activity. Nutrients. 2025; 17(22):3555. https://doi.org/10.3390/nu17223555
Chicago/Turabian StyleBai, Xiaoyue, Fangzhou Lu, Yizhi Jing, Hui Wang, Haidong Qian, Ming Zhang, Zhengyuan Zhai, and Yanling Hao. 2025. "Lactiplantibacillus plantarum Lp20 Alleviates High Fat Diet-Induced Obesity in Mice via Its Bile Salt Hydrolase Activity" Nutrients 17, no. 22: 3555. https://doi.org/10.3390/nu17223555
APA StyleBai, X., Lu, F., Jing, Y., Wang, H., Qian, H., Zhang, M., Zhai, Z., & Hao, Y. (2025). Lactiplantibacillus plantarum Lp20 Alleviates High Fat Diet-Induced Obesity in Mice via Its Bile Salt Hydrolase Activity. Nutrients, 17(22), 3555. https://doi.org/10.3390/nu17223555








