High Consumption of Ultra-Processed Foods Is Associated with Genome-Wide DNA Methylation Differences in Women: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. UPF Consumption
2.3. Collect of Anthropometric Information
2.4. DNA Extraction
2.5. Methylome Analyses
2.6. Statistical Analyses
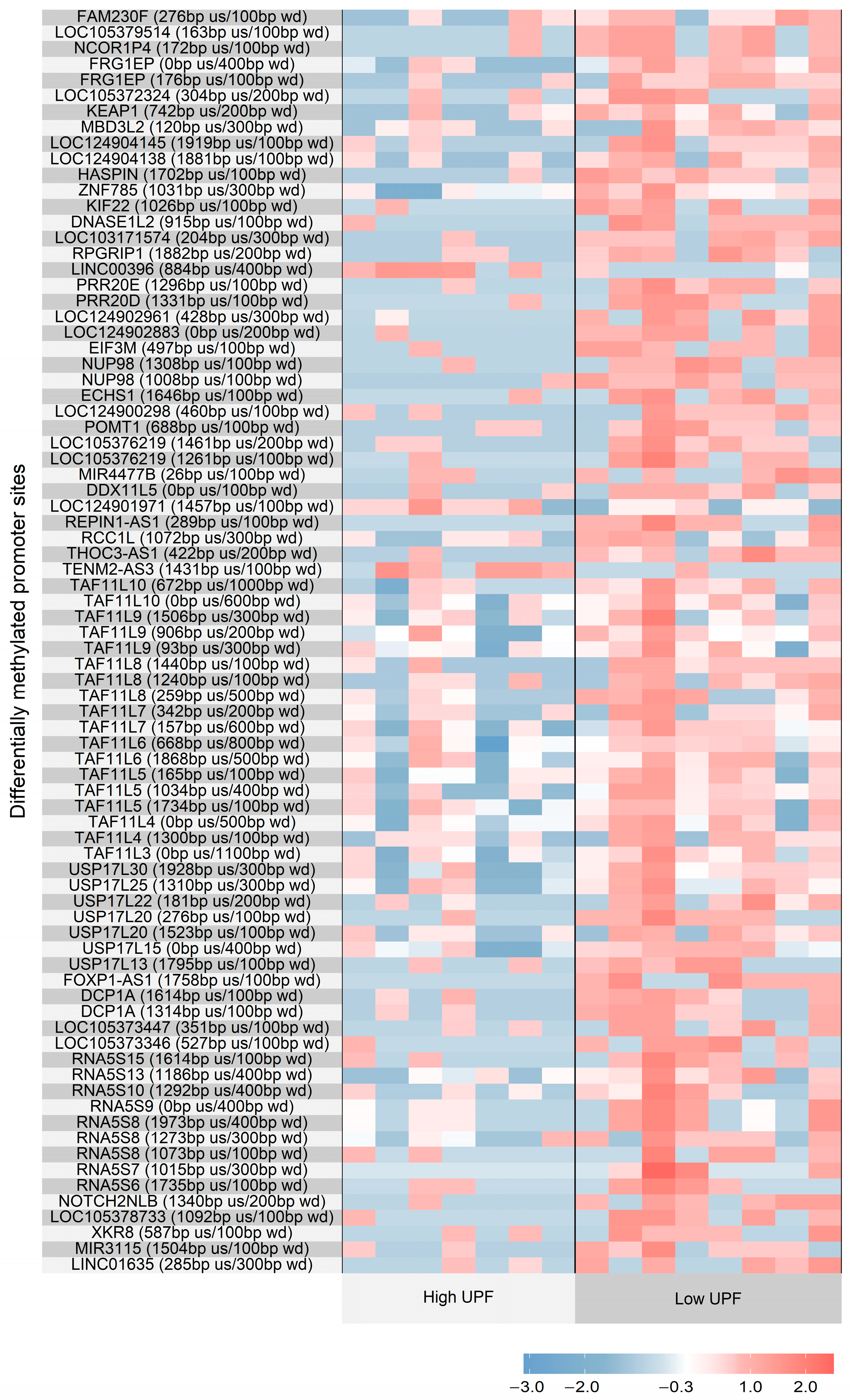
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UPF | Ultra-processed food |
| TEI | Total energy intake |
| NGS | Next-generation sequencing |
| IQR | Interquartile Range |
| DMRs | Differentially methylated regions |
References
- Monteiro, C.A.; Cannon, G.; Moubarac, J.C.; Levy, R.B.; Louzada, M.L.C.; Jaime, P.C. The UN Decade of Nutrition, the NOVA food classification and the trouble with ultra-processing. Public Health Nutr. 2018, 21, 5–17. [Google Scholar] [CrossRef]
- Monteiro, C.A.; Cannon, G.; Levy, R.B.; Moubarac, J.C.; Louzada, M.L.; Rauber, F.; Jaime, P.C.; Khandpur, N.; Cediel, G.; Neri, D.; et al. Ultra-processed foods: What they are and how to identify them. Public Health Nutr. 2019, 22, 936–941. [Google Scholar] [CrossRef] [PubMed]
- GBD 2021 Adult BMI Collaborators. Global, regional, and national prevalence of adult overweight and obesity, 1990–2021, with forecasts to 2050: A forecasting study for the Global Burden of Disease Study 2021. Lancet 2025, 405, 813–838. [Google Scholar] [CrossRef] [PubMed]
- Budreviciute, A.; Damiati, S.; Sabir, D.K.; Onder, K.; Schuller-Goetzburg, P.; Plakys, G.; Kodzius, R.; Katileviciute, A.; Khoja, S. Management and prevention strategies for non-communicable diseases (NCDs) and their risk factors. Front. Public Health 2020, 8, 574111. [Google Scholar] [CrossRef]
- Juul, F.; Martinez-Steele, E.; Parekh, N.; Monteiro, C.A.; Chang, V.W. Ultra-processed food consumption and excess weight among US adults. Br. J. Nutr. 2018, 120, 90–100. [Google Scholar] [CrossRef]
- Baker, P.; Machado, P.; Santos, T.; Sievert, K.; Backholer, K.; Hadjikakou, M.; Lawrence, M.; Russell, C.; Huse, O.; Bell, C.; et al. Ultra-processed foods and the nutrition transition: Global, regional and national trends, food systems transformations and political economy drivers. Obes. Rev. 2020, 21, e13126. [Google Scholar] [CrossRef]
- Lane, M.M.; Gamage, E.; Du, S.; Ashtree, D.N.; McGuinness, A.J.; Gauci, S.; Marx, W.; Baker, S.; Lawrence, M.; Srour, B.; et al. Ultra-processed food exposure and adverse health outcomes: Umbrella review of epidemiological meta-analyses. BMJ 2024, 384, e077310. [Google Scholar] [CrossRef]
- Hall, K.D.; Ayuketah, A.; Brychta, R.; Cai, H.; Cassimatis, T.; Chen, K.Y.; Zhou, M.; Chung, S.T.; Costa, E.; Cassimatis, T.; et al. Ultra-processed diets cause excess calorie intake and weight gain: An inpatient randomized controlled trial of ad libitum food intake. Cell Metab. 2019, 30, 67–77.e3. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Gialluisi, A.; Di Castelnuovo, A.; Costanzo, S.; Pepe, A.; Ruggiero, E.; de Curtis, A.; Persichillo, M.; Cerletti, C.; Iacoviello, L.; et al. Ultra-processed food consumption is associated with the acceleration of biological aging in the Moli-sani Study. Am. J. Clin. Nutr. 2024, 120, 1432–1440. [Google Scholar] [CrossRef]
- Cardoso, B.R.; Liu, J.; Machado, P.; Kwon, D.; Belsky, D.W.; Steele, E.M. Association between ultra-processed food intake and biological ageing in US adults: Findings from National Health and Nutrition Examination Survey (NHANES) 2003–2010. Age Ageing 2024, 53, afae268. [Google Scholar] [CrossRef]
- Lane, M.M.; Davis, J.A.; Beattle, S.; Gómez-Donoso, C.; Loughman, A.; O’Neil, A.; Rocks, T.; Jacka, F.; Berk, M.; Page, R.; et al. Ultraprocessed food and chronic noncommunicable diseases: A systematic review and meta-analysis of 43 observational studies. Obes. Rev. 2021, 22, e13146. [Google Scholar] [CrossRef]
- Domínguez-Barragán, J.; Fernández-Sanlés, A.; Hernáez, Á.; Llauradó-Pont, J.; Marrugat, J.; Robinson, O.; Lassale, C.; Tzoulaki, I.; Elosua, R. Blood DNA methylation signature of diet quality and association with cardiometabolic traits. Eur. J. Prev. Cardiol. 2024, 31, 191–202. [Google Scholar] [CrossRef]
- Sergeeva, A.; Davydova, K.; Perenkov, A.; Vedunova, M. Mechanisms of human DNA methylation, alteration of methylation patterns in physiological processes and oncology. Gene 2023, 875, 147487. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Vieira, T.; Freitas, F.V.; Silva Neto, L.C.B.; Borçoi, A.R.; Mendes, S.O.; Olinda, A.S.; da Silva, A.M.A.; Quaioto, B.R.; de Souza, M.L.M.; Barbosa, W.M.; et al. An industrialized diet as a determinant of methylation in the 1F region of the NR3C1 gene promoter. Front. Nutr. 2024, 11, 1168715. [Google Scholar] [CrossRef]
- Fernandes, A.E.; Rosa, P.W.L.; Melo, M.E.; Martins, R.C.; Santin, F.G.; Moura, A.M.; Mancini, M.C.; Sabino, E.C.; Coelho, G.S.M.A.; Cercato, C.; et al. Differences in the gut microbiota of women according to ultra-processed food consumption. Nutr. Metab. Cardiovasc. Dis. 2023, 33, 84–89. [Google Scholar] [CrossRef]
- Freedman, L.S.; Guenther, P.M.; Krebs-Smith, S.M.; Dodd, K.W.; Midthune, D. A population’s distribution of healthy eating index-2005 component scores can be estimated when more than one 24-hour recall is available. J. Nutr. 2010, 140, 1529–1534. [Google Scholar] [CrossRef]
- Núcleo de Estudos e Pesquisas em Alimentação da Universidade Estadual de Campinas—UNICAMP. Tabela Brasileira de Composição de Alimentos, 4th ed.; UNICAMP: Campinas, Brazil, 2011. [Google Scholar]
- Harttig, U.; Haubrock, J.; Knüppel, S.; Boeing, H.; EFCOVAL Consortium. The MSM program: Web-based statistics package for estimating usual dietary intake using the Multiple Source Method. Eur. J. Clin. Nutr. 2011, 65 (Suppl. S1), S87–S91. [Google Scholar] [CrossRef]
- Invitrogen. MethylMiner™ Methylated DNA Enrichment kit: For the Enrichment of Fragmented DNA Based on the Degree of Methylation; Invitrogen: Carlsbad, CA, USA, 2009; Available online: https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2Fmethylminer_man.pdf (accessed on 25 April 2025).
- Thermo Fisher Scientific. Bisulfite Methylation Library Production and Analysis Using the Ion AmpliSeq™ Kit for Chef DL8; Thermo Fisher Scientific: Carlsbad, CA, USA, 2019; Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0017892_Bisulfite_AmpliSeq_Library_Kit_DL8_UB.pdf (accessed on 25 April 2025).
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Tischler, G.; Leonard, S. Biobambam: Tools for read pair collation based algorithms on BAM files. Source Code Biol. Med. 2014, 9, 13. [Google Scholar] [CrossRef]
- Maksimovic, J.; Phipson, B.; Oshlack, A. A cross-package Bioconductor workflow for analysing methylation array data. F1000Research 2016, 5, 1281. [Google Scholar] [CrossRef]
- Zhu, L.J.; Gazin, C.; Lawson, N.D.; Pagès, H.; Lin, S.M.; Lapointe, D.S.; Green, M.R. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 2010, 11, 237. [Google Scholar] [CrossRef]
- Lienhard, M.; Grimm, C.; Morkel, M.; Herwig, R.; Chavez, L. MEDIPS: Genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 2014, 30, 284–286. [Google Scholar] [CrossRef] [PubMed]
- The R Foundation. The comprehensive R archive network. Version R-4.5.0.tar.gz. Available online: https://cran.r-project.org/ (accessed on 5 August 2025).
- Ding, X.; Zhang, Y.; You, S. A novel prognostic model based on telomere-related lncRNAs in gastric cancer. Transl. Cancer Res. 2024, 13, 4608–4624. [Google Scholar] [CrossRef]
- Abshagen, K.; Berger, C.; Dietrich, A.; Schütz, T.; Wittekind, C.; Stumvoll, M.; Blüher, M.; Klöting, N. A human REPIN1 gene variant: Genetic risk factor for the development of nonalcoholic fatty liver disease. Clin. Transl. Gastroenterol. 2020, 11, e00114. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Berger, C.; Weidle, K.; Schleinitz, D.; Tönjes, A.; Stumvoll, M.; Blüher, M.; Kovacs, P.; Klöting, N. Metabolic effects of genetic variation in the human REPIN1 gene. Int. J. Obes. 2019, 43, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Hesselbarth, N.; Kunath, A.; Kern, M.; Gericke, M.; Mejhert, N.; Rydén, M.; Stumvoll, M.; Blüher, M.; Klöting, N. Repin1 deficiency in adipose tissue improves whole-body insulin sensitivity, and lipid metabolism. Int. J. Obes. 2017, 41, 1815–1823. [Google Scholar] [CrossRef]
- Kunath, A.; Hesselbarth, N.; Gericke, M.; Kern, M.; Dommel, S.; Kovács, P.; Stumvoll, M.; Blüher, M.; Klöting, N. Repin1 deficiency improves insulin sensitivity and glucose metabolism in db/db mice by reducing adipose tissue mass and inflammation. Biochem. Biophys. Res. Commun. 2016, 478, 398–402. [Google Scholar] [CrossRef]
- Sender, S.; Sultan, A.W.; Palmer, D.; Koczan, D.; Sekora, A.; Beck, J.; Schuetz, E.; Taher, L.; Brenig, B.; Murua Escobar, H.; et al. Evaluation of the synergistic potential of simultaneous pan- or isoform-specific BET and SYK inhibition in B-cell lymphoma: An in vitro approach. Cancers 2022, 14, 4691. [Google Scholar] [CrossRef]
- Wen, X.M.; Xu, Z.J.; Ni, H.X.; Liu, S.W.; Jin, Y.; Zhao, W.; Luo, S.; Fang, Y.; Lin, J.; Qian, J.; et al. FOXP1 is associated with oncogenesis and clinical outcomes in hematologic malignancies. Front. Immunol. 2025, 16, 1569641. [Google Scholar] [CrossRef]
- Stewart, J.; Cho, G.H.Y. Autism-like features and FOXP1 syndrome: A scoping review. Brain Dev. 2025, 47, 104346. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mojica, J.E.; Lillycrop, K.A.; Cooper, C.; Calder, P.C.; Burdge, G.C. Docosahexaenoic acid and oleic acid induce altered DNA methylation of individual CpG loci in Jurkat T cells. Prostaglandins Leukot. Essent. Fat. Acids 2020, 158, 102128. [Google Scholar] [CrossRef] [PubMed]
- Dicken, S.J.; Jassil, F.C.; Brown, A.; Kalis, M.; Stanley, C.; Ranson, C.; Batterham, R.L.; Ruwona, T.; Qamar, S.; Buck, C.; et al. Ultraprocessed or minimally processed diets following healthy dietary guidelines on weight and cardiometabolic health: A randomized, crossover trial. Nat. Med. 2025, 31, 3297–3308. [Google Scholar] [CrossRef] [PubMed]
- Llauradó-Pont, J.; Stratakis, N.; Fiorito, G.; Handakas, E.; Neumann, A.; Barros, H.; Lise Brantsæter, A.; Chang, K.; Chatzi, L.; Grazuleviciene, L.; et al. A meta-analysis of epigenome-wide association studies of ultra-processed food consumption with DNA methylation in European children. Clin. Epigenetics 2025, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Crawford, P.B.; Obarzanek, E.; Morrison, J.; Sabry, Z.I. Comparative advantage of 3-day food records over 24-hour recall and 5-day food frequency validated by observation of 9- and 10-year-old girls. J. Am. Diet. Assoc. 1994, 94, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.J.; Maxwell, A.P.; Benson, K.A.; Killner, J.; McKay, G.J.; McKnight, A.J. Validation of differentially methylated microRNAs identified from an epigenome-wide association study; Sanger and next generation sequencing approaches. BMC Res. Notes 2018, 11, 767. [Google Scholar] [CrossRef]
- Flanagan, J.M. Epigenome-wide association studies (EWAS): Past, present, and future. In Cancer Epigenetics; Verma, M., Ed.; Methods in Molecular, Biology; Humana Press: New York, NY, USA, 2015; Volume 1238, pp. 51–70. [Google Scholar] [CrossRef]
| Variable | Low-UPF Group (n = 7) | High-UPF Group (n = 8) | p-Value |
|---|---|---|---|
| Age (years) | 29 (27–34) | 36 (24–37) | 0.954 |
| Weight (kg) | 75.0 ± 19 | 75.5 ± 21.4 | 0.962 |
| BMI (kg/m2) | 28.9 (23.7–36.5) | 24.7 (23–35.2) | 0.779 |
| Waist circumference (cm) | 90.7 ± 17.3 | 91.4 ± 21.6 | 0.947 |
| Lean mass (kg) | 40.5 ± 5.2 | 41.8 ± 4.7 | 0.639 |
| Fat mass (%) | 40.5 ± 9.5 | 38.8 ± 10.7 | 0.757 |
| Glucose (mg/dL) | 77 ± 5.4 | 79.3 ± 9.0 | 0.555 |
| Insulin (μIU/mL) | 13.1 ± 7.5 | 14.1 ± 8.7 | 0.817 |
| Glycated hemoglobin (%) | 5.3 ± 0.3 | 5.3 ± 0.5 | 0.952 |
| Total cholesterol (mg/dL) | 195 ± 35 | 144.7 ± 21.5 | 0.005 |
| LDL cholesterol (mg/dL) | 119.6 ± 28.6 | 68.9 ± 14.9 | 0.001 |
| HDL cholesterol (mg/dL) | 56.8 ± 11.6 | 59.9 ± 15.3 | 0.670 |
| Non-HDL cholesterol (mg/dL) | 138.3 ± 33 | 84.9 ± 17.3 | 0.002 |
| Triglycerides (mg/dL) | 85.6 ± 34.5 | 82.3 ± 40.6 | 0.882 |
| Aspartate aminotransferase (U/L) | 17.3 ± 4.5 | 15.9 ± 5.6 | 0.61 |
| Alanine aminotransferase (U/L) | 15.6 ± 2.6 | 16,6 ± 7.0 | 0.745 |
| Gamma-glutamyl transferase (U/L) | 16.3 ± 6.1 | 16.3 ± 8,3 | 0.993 |
| Adiponectin (μg/mL) | 4.8 ± 2.8 | 6.5 ± 4.9 | 0.452 |
| Leptin (ng/mL) | 10.5 (8.0–23.9) | 11.5 (6.4–31.6) | 0.955 |
| Variable | Low-UPF Group (n = 7) | High-UPF Group (n = 8) | p-Value |
|---|---|---|---|
| Total energy intake (TEI) | 1469 (1236–1645) | 1391 (1234–1729) | 0.955 |
| Protein (% of TEI) | 21.9 ± 5.0 | 15.2 ± 4.0 | 0.013 |
| Carbohydrate (% of TEI) | 40.5 ± 7.5 | 47.8 ± 7.3 | 0.08 |
| Total fat (% of TEI) | 36.9 ± 6.8 | 33.5 ± 5 | 0.294 |
| Cholesterol (mg) | 308.4 ± 133.4 | 207.8 ± 58 | 0.082 |
| Saturated fat (% of TEI) | 11.6 ± 1.9 | 9.4 ± 2.0 | 0.053 |
| Polyunsaturated fat (% of TEI) | 8.7 ± 2.2 | 6.5 ± 1.2 | 0.034 |
| Monounsaturated fat (% of TEI) | 10.5 ± 2.3 | 9.2 ± 3.1 | 0.352 |
| Fiber (g) | 13.3 (11–17.6) | 7.1 (6.4–14.7) | 0.463 |
| Unprocessed/minimally processed foods (% of TEI) | 61.4 ± 10.4 | 39.7 ± 5.4 | <0.001 |
| Culinary ingredients (% of TEI) | 7.3 ± 4.4 | 6.5 ± 1.5 | 0.646 |
| Processed foods (% of TEI) | 10.3 ± 4.2 | 10.3 ± 6.8 | 0.994 |
| Ultra-processed foods (% of TEI) | 14.3 ± 5.8 | 45.1 ± 2.8 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, A.E.; Fernandes, A.E.; Carrasco, A.G.M.; Pellenz, F.M.; da Rosa, P.W.L.; de Moura, A.M.d.S.H.; Santin, F.G.d.O.; Cercato, C.; de Melo, M.E.; Mancini, M.C. High Consumption of Ultra-Processed Foods Is Associated with Genome-Wide DNA Methylation Differences in Women: A Pilot Study. Nutrients 2025, 17, 3465. https://doi.org/10.3390/nu17213465
Rodrigues AE, Fernandes AE, Carrasco AGM, Pellenz FM, da Rosa PWL, de Moura AMdSH, Santin FGdO, Cercato C, de Melo ME, Mancini MC. High Consumption of Ultra-Processed Foods Is Associated with Genome-Wide DNA Methylation Differences in Women: A Pilot Study. Nutrients. 2025; 17(21):3465. https://doi.org/10.3390/nu17213465
Chicago/Turabian StyleRodrigues, Alessandra Escorcio, Ariana Ester Fernandes, Alexis Germán Murillo Carrasco, Felipe Mateus Pellenz, Paula Waki Lopes da Rosa, Aline Maria da Silva Hourneaux de Moura, Fernanda Galvão de Oliveira Santin, Cintia Cercato, Maria Edna de Melo, and Marcio C. Mancini. 2025. "High Consumption of Ultra-Processed Foods Is Associated with Genome-Wide DNA Methylation Differences in Women: A Pilot Study" Nutrients 17, no. 21: 3465. https://doi.org/10.3390/nu17213465
APA StyleRodrigues, A. E., Fernandes, A. E., Carrasco, A. G. M., Pellenz, F. M., da Rosa, P. W. L., de Moura, A. M. d. S. H., Santin, F. G. d. O., Cercato, C., de Melo, M. E., & Mancini, M. C. (2025). High Consumption of Ultra-Processed Foods Is Associated with Genome-Wide DNA Methylation Differences in Women: A Pilot Study. Nutrients, 17(21), 3465. https://doi.org/10.3390/nu17213465







