Gut Microbiota and Food Allergy: A Review of Mechanisms and Microbiota-Targeted Interventions
Abstract
1. Introduction
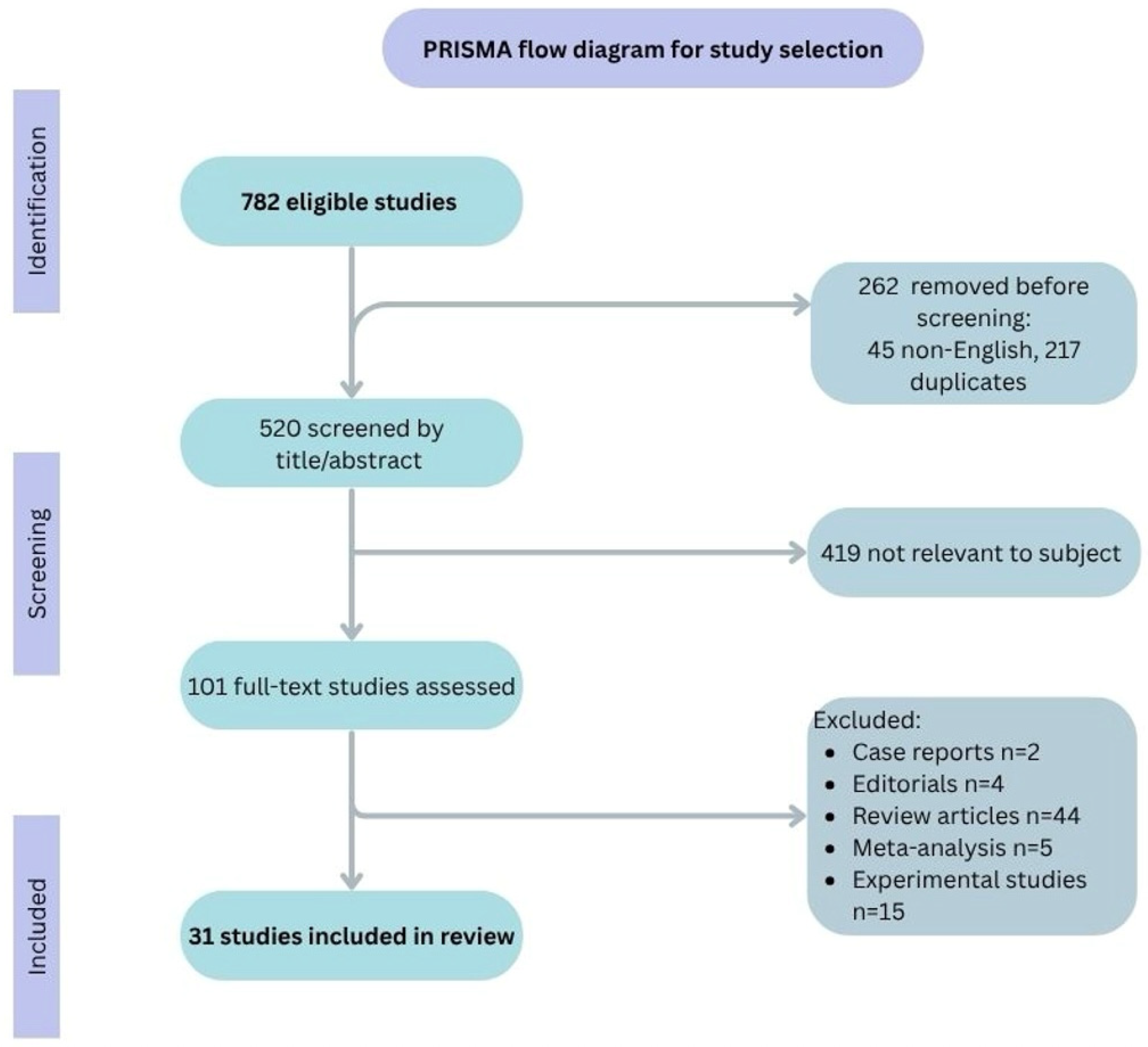
2. Materials and Methods
Quality Assessment
3. Results
4. Discussion
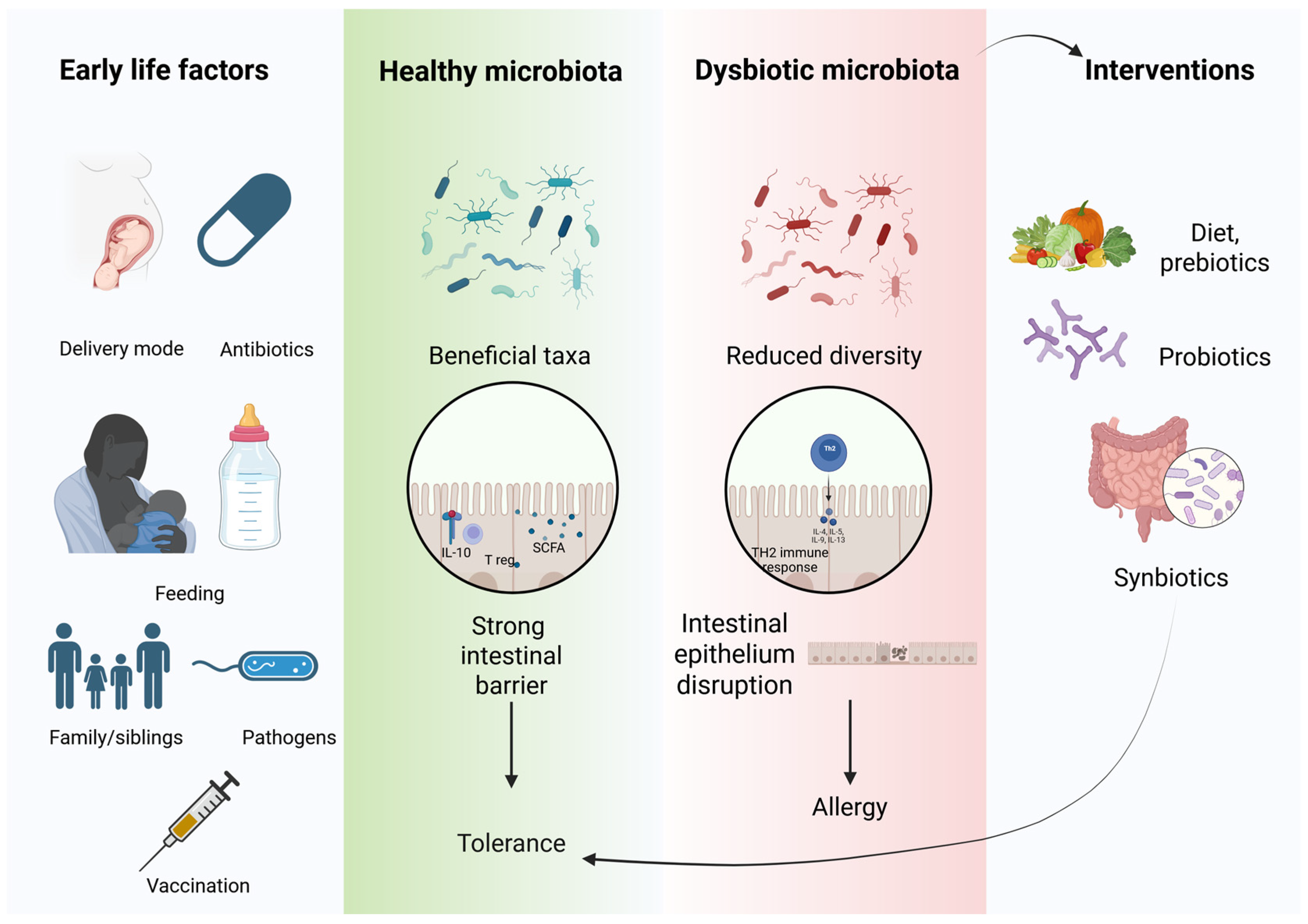
4.1. Hygiene Hypothesis
4.2. Development of Gut Microbiota in Children and Adults
4.3. Pathophysiological Mechanisms Linking Gut Microbiota to Food Allergy Development
4.4. Experimental Evidence from Animal Models
4.5. The Gut Microbiota–Food Allergy Axis: Evidence from Human Studies
4.6. Dietary Modulation of Gut Microbiota in Allergic Individuals: The Role of Prebiotics and Probiotics
4.6.1. Diet and Prebiotics
4.6.2. Probiotics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Calvani, M.; Anania, C.; Caffarelli, C.; Martelli, A.; Miraglia del Giudice, M.; Cravidi, C.; Duse, M.; Manti, S.; Tosca, M.A.; Cardinale, F.; et al. Food Allergy: An Updated Review on Pathogenesis, Diagnosis, Prevention and Management. Acta Bio Medica Atenei Parm. 2020, 91, e2020012. [Google Scholar] [CrossRef]
- Warren, C.M.; Jiang, J.; Gupta, R.S. Epidemiology and Burden of Food Allergy. Curr. Allergy Asthma Rep. 2020, 20, 6. [Google Scholar] [CrossRef]
- Wong, G.W.-K. Food Allergies around the World. Front. Nutr. 2024, 11, 1373110. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Warren, C.M.; Smith, B.M.; Blumenstock, J.A.; Jiang, J.; Davis, M.M.; Nadeau, K.C. The Public Health Impact of Parent-Reported Childhood Food Allergies in the United States. Pediatrics 2018, 142, e20181235, Erratum in: Pediatrics 2019, 143, e20183835. [Google Scholar] [CrossRef]
- Le, T.T.K.; Nguyen, D.H.; Vu, A.T.L.; Ruethers, T.; Taki, A.C.; Lopata, A.L. A Cross-Sectional, Population-Based Study on the Prevalence of Food Allergies among Children in Two Different Socio-Economic Regions of Vietnam. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2019, 30, 348–355. [Google Scholar] [CrossRef]
- Tham, E.H.; Leung, A.S.Y.; Pacharn, P.; Lee, S.; Ebisawa, M.; Lee, B.W.; Wong, G.W.K.; APAPARI Anaphylaxis Study Group. Anaphylaxis—Lessons Learnt When East Meets West. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2019, 30, 681–688. [Google Scholar] [CrossRef]
- Prescott, S.; Allen, K.J. Food Allergy: Riding the Second Wave of the Allergy Epidemic. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2011, 22, 155–160. [Google Scholar] [CrossRef]
- Turner, P.J.; Gowland, M.H.; Sharma, V.; Ierodiakonou, D.; Harper, N.; Garcez, T.; Pumphrey, R.; Boyle, R.J. Increase in Anaphylaxis-Related Hospitalizations but No Increase in Fatalities: An Analysis of United Kingdom National Anaphylaxis Data, 1992–2012. J. Allergy Clin. Immunol. 2015, 135, 956–963.e1. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Lee, E.; Kim, B.J.; Kang, M.J.; Choi, K.Y.; Cho, H.J.; Kim, Y.; Yang, S.I.; Jung, Y.H.; Kim, H.Y.; Seo, J.H.; et al. Dynamics of Gut Microbiota According to the Delivery Mode in Healthy Korean Infants. Allergy Asthma Immunol. Res. 2016, 8, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.E.; Lange, N.E.; Zhou, Y.; O’Connor, G.T.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E.; et al. Diet during Pregnancy and Infancy and the Infant Intestinal Microbiome. J. Pediatr. 2018, 203, 47–54.e4. [Google Scholar] [CrossRef]
- Marrs, T.; Jo, J.-H.; Perkin, M.R.; Rivett, D.W.; Witney, A.A.; Bruce, K.D.; Logan, K.; Craven, J.; Radulovic, S.; Versteeg, S.A.; et al. Gut Microbiota Development during Infancy: Impact of Introducing Allergenic Foods. J. Allergy Clin. Immunol. 2021, 147, 613–621.e9. [Google Scholar] [CrossRef]
- Davis, E.C.; Castagna, V.P.; Sela, D.A.; Hillard, M.A.; Lindberg, S.; Mantis, N.J.; Seppo, A.E.; Järvinen, K.M. Gut Microbiome and Breast-Feeding: Implications for Early Immune Regulation. J. Allergy Clin. Immunol. 2022, 150, 523–534. [Google Scholar] [CrossRef]
- Herrera-Quintana, L.; Vázquez-Lorente, H.; Hinojosa-Nogueira, D.; Plaza-Diaz, J. Relationship between Infant Feeding and the Microbiome: Implications for Allergies and Food Intolerances. Children 2024, 11, 1030. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayama, J. Development of the Gut Microbiota in Infancy and Its Impact on Health in Later Life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, J.; Zhang, X.; Gu, Q.; Wu, Y.; Tao, X.; Tian, T.; Pan, G.; Chu, M. The Potential Impact of Antibiotic Exposure on the Microbiome and Human Health. Microorganisms 2025, 13, 602. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P. The Immunological Interplay between Vaccination and the Intestinal Microbiota. Npj Vaccines 2023, 8, 24. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Sperandio, V. Interactions between the Microbiota and Pathogenic Bacteria in the Gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant Gut Microbiota and Food Sensitization: Associations in the First Year of Life. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2015, 45, 632–643. [Google Scholar] [CrossRef]
- Ojo, O. Recent Advances in Nutrition and Diabetes. Nutrients 2021, 13, 1573. [Google Scholar] [CrossRef]
- Mennini, M.; Arasi, S.; Artesani, M.C.; Fiocchi, A.G. Probiotics in Food Allergy. Curr. Opin. Allergy Clin. Immunol. 2021, 21, 309–316. [Google Scholar] [CrossRef]
- Fiocchi, A.; Cabana, M.D.; Mennini, M. Current Use of Probiotics and Prebiotics in Allergy. J. Allergy Clin. Immunol. Pract. 2022, 10, 2219–2242. [Google Scholar] [CrossRef]
- Kang, Y.; Cai, Y. Future Prospect of Faecal Microbiota Transplantation as a Potential Therapy in Asthma. Allergol. Immunopathol. 2018, 46, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Study Quality Assessment Tools|NHLBI, NIH. Available online: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools (accessed on 12 June 2025).
- Berni Canani, R.; Sangwan, N.; Stefka, A.T.; Nocerino, R.; Paparo, L.; Aitoro, R.; Calignano, A.; Khan, A.A.; Gilbert, J.A.; Nagler, C.R. Lactobacillus Rhamnosus GG-Supplemented Formula Expands Butyrate-Producing Bacterial Strains in Food Allergic Infants. ISME J. 2016, 10, 742–750. [Google Scholar] [CrossRef]
- Dzidic, M.; Abrahamsson, T.R.; Artacho, A.; Björkstén, B.; Collado, M.C.; Mira, A.; Jenmalm, M.C. Aberrant IgA Responses to the Gut Microbiota during Infancy Precede Asthma and Allergy Development. J. Allergy Clin. Immunol. 2017, 139, 1017–1025.e14. [Google Scholar] [CrossRef]
- Berni Canani, R.; Di Costanzo, M.; Bedogni, G.; Amoroso, A.; Cosenza, L.; Di Scala, C.; Granata, V.; Nocerino, R. Extensively Hydrolyzed Casein Formula Containing Lactobacillus Rhamnosus GG Reduces the Occurrence of Other Allergic Manifestations in Children with Cow’s Milk Allergy: 3-Year Randomized Controlled Trial. J. Allergy Clin. Immunol. 2017, 139, 1906–1913.e4. [Google Scholar] [CrossRef]
- Candy, D.C.A.; Van Ampting, M.T.J.; Oude Nijhuis, M.M.; Wopereis, H.; Butt, A.M.; Peroni, D.G.; Vandenplas, Y.; Fox, A.T.; Shah, N.; West, C.E.; et al. A Synbiotic-Containing Amino-Acid-Based Formula Improves Gut Microbiota in Non-IgE-Mediated Allergic Infants. Pediatr. Res. 2018, 83, 677–686. [Google Scholar] [CrossRef]
- Nocerino, R.; Di Costanzo, M.; Bedogni, G.; Cosenza, L.; Maddalena, Y.; Di Scala, C.; Della Gatta, G.; Carucci, L.; Voto, L.; Coppola, S.; et al. Dietary Treatment with Extensively Hydrolyzed Casein Formula Containing the Probiotic Lactobacillus Rhamnosus GG Prevents the Occurrence of Functional Gastrointestinal Disorders in Children with Cow’s Milk Allergy. J. Pediatr. 2019, 213, 137–142.e2. [Google Scholar] [CrossRef]
- Aparicio, M.; Alba, C.; Rodríguez, J.M.; Fernández, L. Microbiological and Immunological Markers in Milk and Infant Feces for Common Gastrointestinal Disorders: A Pilot Study. Nutrients 2020, 12, 634. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Liu, Q.; Wang, W. Bifidobacterium Bifidum TMC3115 Ameliorates Milk Protein Allergy in by Affecting Gut Microbiota: A Randomized Double-Blind Control Trial. J. Food Biochem. 2020, 44, e13489. [Google Scholar] [CrossRef] [PubMed]
- Bao, R.; Hesser, L.A.; He, Z.; Zhou, X.; Nadeau, K.C.; Nagler, C.R. Fecal Microbiome and Metabolome Differ in Healthy and Food-Allergic Twins. J. Clin. Investig. 2021, 131, e141935. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.L.; Mohebbi, M.; Craig, J.M.; Dawson, P.; Clarke, G.; Tang, M.L.; Jacka, F.N. Targeting the Perinatal Diet to Modulate the Gut Microbiota Increases Dietary Variety and Prebiotic and Probiotic Food Intakes: Results from a Randomised Controlled Trial. Public Health Nutr. 2021, 24, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Nocerino, R.; Bedogni, G.; Carucci, L.; Cosenza, L.; Cozzolino, T.; Paparo, L.; Palazzo, S.; Riva, L.; Verduci, E.; Berni Canani, R. The Impact of Formula Choice for the Management of Pediatric Cow’s Milk Allergy on the Occurrence of Other Allergic Manifestations: The Atopic March Cohort Study. J. Pediatr. 2021, 232, 183–191.e3. [Google Scholar] [CrossRef]
- Homann, C.-M.; Rossel, C.A.J.; Dizzell, S.; Bervoets, L.; Simioni, J.; Li, J.; Gunn, E.; Surette, M.G.; de Souza, R.J.; Mommers, M.; et al. Infants’ First Solid Foods: Impact on Gut Microbiota Development in Two Intercontinental Cohorts. Nutrients 2021, 13, 2639. [Google Scholar] [CrossRef]
- De Filippis, F.; Paparo, L.; Nocerino, R.; Della Gatta, G.; Carucci, L.; Russo, R.; Pasolli, E.; Ercolini, D.; Berni Canani, R. Specific Gut Microbiome Signatures and the Associated Pro-Inflamatory Functions Are Linked to Pediatric Allergy and Acquisition of Immune Tolerance. Nat. Commun. 2021, 12, 5958. [Google Scholar] [CrossRef]
- Boulangé, C.L.; Pedersen, H.K.; Martin, F.-P.; Siegwald, L.; Pallejà Caro, A.; Eklund, A.C.; Jia, W.; Zhang, H.; Berger, B.; Sprenger, N.; et al. An Extensively Hydrolyzed Formula Supplemented with Two Human Milk Oligosaccharides Modifies the Fecal Microbiome and Metabolome in Infants with Cow’s Milk Protein Allergy. Int. J. Mol. Sci. 2023, 24, 11422. [Google Scholar] [CrossRef]
- Yamamoto-Hanada, K.; Sato, M.; Toyokuni, K.; Irahara, M.; Hiraide-Kotaki, E.; Harima-Mizusawa, N.; Morita, H.; Matsumoto, K.; Ohya, Y. Combination of Heat-Killed Lactiplantibacillus Plantarum YIT 0132 (LP0132) and Oral Immunotherapy in Cow’s Milk Allergy: A Randomised Controlled Trial. Benef. Microbes 2023, 14, 17–30. [Google Scholar] [CrossRef]
- Yan, X.; Yan, J.; Xiang, Q.; Dai, H.; Wang, Y.; Fang, L.; Huang, K.; Zhang, W. Early-Life Gut Microbiota in Food Allergic Children and Its Impact on the Development of Allergic Disease. Ital. J. Pediatr. 2023, 49, 148. [Google Scholar] [CrossRef]
- Gao, Y.; Stokholm, J.; O’Hely, M.; Ponsonby, A.-L.; Tang, M.L.K.; Ranganathan, S.; Saffery, R.; Harrison, L.C.; Collier, F.; Gray, L.; et al. Gut Microbiota Maturity Mediates the Protective Effect of Siblings on Food Allergy. J. Allergy Clin. Immunol. 2023, 152, 667–675. [Google Scholar] [CrossRef]
- Súkeníková, L.; Černý, V.; Thon, T.; Roubalová, R.; Jirásková Zákostelská, Z.; Novotná, O.; Petrásková, P.; Boráková, K.; Kocourková, I.; Lodinová-Žádníková, R.; et al. Effect of Early Postnatal Supplementation of Newborns with Probiotic Strain E. coli O83:K24:H31 on Allergy Incidence, Dendritic Cells, and Microbiota. Front. Immunol. 2023, 13, 1038328. [Google Scholar] [CrossRef] [PubMed]
- Shibata, R.; Itoh, N.; Nakanishi, Y.; Kato, T.; Suda, W.; Nagao, M.; J-OIT Group; Iwata, T.; Yoshida, H.; Hattori, M.; et al. Gut Microbiota and Fecal Metabolites in Sustained Unresponsiveness by Oral Immunotherapy in School-Age Children with Cow’s Milk Allergy. Allergol. Int. Off. J. Jpn. Soc. Allergol. 2024, 73, 126–136. [Google Scholar] [CrossRef]
- Hara, M.; Suzuki, H.; Hayashi, D.; Morii, W.; Nakamura, T.; Kiyoki, K.; Hara, H.; Ishii, R.; Noguchi, E.; Takada, H. Gut Microbiota of One-and-a-Half-Year-Old Food-Allergic and Healthy Children. Allergol. Int. Off. J. Jpn. Soc. Allergol. 2024, 73, 550–555. [Google Scholar] [CrossRef]
- Castro, A.M.; Navarro, S.; Carvajal, I.; García, A.; Suárez, M.; Toyos, P.; Rodríguez, S.; Jimenez, S.; González, D.; Molinos, C.; et al. Evolutive Study of Dietary Aspects and Intestinal Microbiota of Pediatric Cohort with Cow’s Milk Protein Allergy. Child. Basel Switz. 2024, 11, 1113. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Hurley, S.; Ford, S.A.; Franklin, R.; Byrne, S.; Lunjani, N.; Forde, B.; Neogi, U.; Venter, C.; Walter, J.; et al. Association between Gut Microbiota Development and Allergy in Infants Born during Pandemic-Related Social Distancing Restrictions. Allergy 2024, 79, 1938–1951. [Google Scholar] [CrossRef] [PubMed]
- Nekrasova, A.I.; Kalashnikova, I.G.; Bobrova, M.M.; Korobeinikova, A.V.; Bakoev, S.Y.; Ashniev, G.A.; Petryaikina, E.S.; Nekrasov, A.S.; Zagainova, A.V.; Lukashina, M.V.; et al. Characteristics of the Gut Microbiota in Regard to Atopic Dermatitis and Food Allergies of Children. Biomedicines 2024, 12, 553. [Google Scholar] [CrossRef]
- Chen, C.-C.; Huang, J.-L.; Chen, K.-J.; Kong, M.-S.; Hua, M.-C.; Yeh, Y.-M.; Chang, H.-J. Comparison of 16S rRNA Gene Sequencing Microbiota among Children with Serological IgE-Mediated Food Hypersensitivity. Pediatr. Res. 2024, 95, 241–250. [Google Scholar] [CrossRef]
- Jones, J.M.; Reinke, S.N.; Mousavi-Derazmahalleh, M.; Garssen, J.; Jenmalm, M.C.; Srinivasjois, R.; Silva, D.; Keelan, J.; Prescott, S.L.; Palmer, D.J.; et al. Maternal Prebiotic Supplementation during Pregnancy and Lactation Modifies the Microbiome and Short Chain Fatty Acid Profile of Both Mother and Infant. Clin. Nutr. Edinb. Scotl. 2024, 43, 969–980. [Google Scholar] [CrossRef]
- Shibata, R.; Nakanishi, Y.; Suda, W.; Nakano, T.; Sato, N.; Inaba, Y.; Kawasaki, Y.; Hattori, M.; Shimojo, N.; Ohno, H. Neonatal Gut Microbiota and Risk of Developing Food Sensitization and Allergy. J. Allergy Clin. Immunol. 2025, 155, 932–946. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Huang, J.; Xie, Y.; Wang, D.; Tan, X.; Wang, Y. Investigation of Gut Microbiota in Pediatric Patients with Peanut Allergy in Outpatient Settings. Front. Pediatr. 2025, 13, 1509275. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, H.; Wang, Y.; Xu, Q.; Qiu, X.; Cheng, L.; Xiao, Q.; Liu, Y.; Zhang, J.; Zhang, H.; et al. Gut Microbiota Signatures in Food Allergy Children without and with Malnutrition: A Cross-Sectional Study. BMC Pediatr. 2025, 25, 220. [Google Scholar] [CrossRef]
- Imoto, N.; Kano, C.; Morita, H.; Hirota, T.; Amanuma, F.; Maruyama, H.; Nojiri, S.; Watanabe, S. Impact of Antimicrobial Exposure at Delivery and Siblings on Early Bifidobacterium Succession and Allergy Development up to 24 Months of Age. BMC Microbiol. 2025, 25, 332. [Google Scholar] [CrossRef]
- Nocerino, R.; Bedogni, G.; Carucci, L.; Aquilone, G.; Oglio, F.; Coppola, S.; Masino, A.; Berni Canani, R. Long Term Impact of Formula Choice in Children with Cow Milk Protein Allergy: 6-Year Follow-up of the Atopic March Cohort Study. Clin. Nutr. Edinb. Scotl. 2025, 48, 134–143. [Google Scholar] [CrossRef]
- PRISMA 2020 Flow Diagram. Available online: https://www.prisma-statement.org/prisma-2020-flow-diagram (accessed on 12 June 2025).
- Strachan, D.P. Hay Fever, Hygiene, and Household Size. BMJ 1989, 299, 1259–1260. [Google Scholar] [CrossRef]
- Strachan, D.P.; Aït-Khaled, N.; Foliaki, S.; Mallol, J.; Odhiambo, J.; Pearce, N.; Williams, H.C. ISAAC Phase Three Study Group Siblings, Asthma, Rhinoconjunctivitis and Eczema: A Worldwide Perspective from the International Study of Asthma and Allergies in Childhood. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2015, 45, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Noverr, M.C.; Huffnagle, G.B. The “microflora Hypothesis” of Allergic Diseases. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2005, 35, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Kilpeläinen, M.; Terho, E.O.; Helenius, H.; Koskenvuo, M. Farm Environment in Childhood Prevents the Development of Allergies. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2000, 30, 201–208. [Google Scholar] [CrossRef]
- Ownby, D.R.; Johnson, C.C.; Peterson, E.L. Exposure to Dogs and Cats in the First Year of Life and Risk of Allergic Sensitization at 6 to 7 Years of Age. JAMA 2002, 288, 963–972. [Google Scholar] [CrossRef]
- Koplin, J.J.; Dharmage, S.C.; Ponsonby, A.-L.; Tang, M.L.K.; Lowe, A.J.; Gurrin, L.C.; Osborne, N.J.; Martin, P.E.; Robinson, M.N.; Wake, M.; et al. Environmental and Demographic Risk Factors for Egg Allergy in a Population-Based Study of Infants. Allergy 2012, 67, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- Garn, H.; Potaczek, D.P.; Pfefferle, P.I. The Hygiene Hypothesis and New Perspectives-Current Challenges Meeting an Old Postulate. Front. Immunol. 2021, 12, 637087. [Google Scholar] [CrossRef]
- von Mutius, E. The “Hygiene Hypothesis” and the Lessons Learnt From Farm Studies. Front. Immunol. 2021, 12, 635522. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Funkhouser, L.J.; Bordenstein, S.R. Mom Knows Best: The Universality of Maternal Microbial Transmission. PLoS Biol. 2013, 11, e1001631. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Early Development of the Gut Microbiota and Immune Health. Pathogens 2014, 3, 769–790. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Björkstén, B.; Engstrand, L.; Andersson, A.F. Decreased Gut Microbiota Diversity, Delayed Bacteroidetes Colonisation and Reduced Th1 Responses in Infants Delivered by Caesarean Section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Long, G.; Hu, Y.; Tao, E.; Chen, B.; Shu, X.; Zheng, W.; Jiang, M. The Influence of Cesarean Section on the Composition and Development of Gut Microbiota During the First 3 Months of Life. Front. Microbiol. 2021, 12, 691312. [Google Scholar] [CrossRef]
- Biasucci, G.; Benenati, B.; Morelli, L.; Bessi, E.; Boehm, G. Cesarean Delivery May Affect the Early Biodiversity of Intestinal Bacteria. J. Nutr. 2008, 138, 1796S–1800S. [Google Scholar] [CrossRef]
- Yang, X.; Zhou, C.; Guo, C.; Wang, J.; Chen, I.; Wen, S.W.; Krewski, D.; Yue, L.; Xie, R.-H. The Prevalence of Food Allergy in Cesarean-Born Children Aged 0-3 Years: A Systematic Review and Meta-Analysis of Cohort Studies. Front. Pediatr. 2022, 10, 1044954. [Google Scholar] [CrossRef]
- Korpela, K.; de Vos, W.M. Antibiotic Use in Childhood Alters the Gut Microbiota and Predisposes to Overweight. Microb. Cell Graz Austria 2016, 3, 296–298. [Google Scholar] [CrossRef]
- Ahmadizar, F.; Vijverberg, S.J.H.; Arets, H.G.M.; de Boer, A.; Lang, J.E.; Garssen, J.; Kraneveld, A.; Maitland-van der Zee, A.H. Early-Life Antibiotic Exposure Increases the Risk of Developing Allergic Symptoms Later in Life: A Meta-Analysis. Allergy 2018, 73, 971–986. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Gyriki, D.; Nikolaidis, C.G.; Bezirtzoglou, E.; Voidarou, C.; Stavropoulou, E.; Tsigalou, C. The Gut Microbiota and Aging: Interactions, Implications, and Interventions. Front. Aging 2025, 6, 1452917. [Google Scholar] [CrossRef]
- Martens, E.C.; Neumann, M.; Desai, M.S. Interactions of Commensal and Pathogenic Microorganisms with the Intestinal Mucosal Barrier. Nat. Rev. Microbiol. 2018, 16, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal Barrier in Human Health and Disease. Int. J. Environ. Res. Public. Health 2021, 18, 12836. [Google Scholar] [CrossRef] [PubMed]
- Schrander, J.J.; Unsalan-Hooyen, R.W.; Forget, P.P.; Jansen, J. [51Cr]EDTA Intestinal Permeability in Children with Cow’s Milk Intolerance. J. Pediatr. Gastroenterol. Nutr. 1990, 10, 189–192. [Google Scholar] [CrossRef]
- Varricchi, G.; Poto, R.; Ianiro, G.; Punziano, A.; Marone, G.; Gasbarrini, A.; Spadaro, G. Gut Microbiome and Common Variable Immunodeficiency: Few Certainties and Many Outstanding Questions. Front. Immunol. 2021, 12, 712915. [Google Scholar] [CrossRef]
- Secondulfo, M.; Iafusco, D.; Carratù, R.; deMagistris, L.; Sapone, A.; Generoso, M.; Mezzogiomo, A.; Sasso, F.C.; Cartenì, M.; De Rosa, R.; et al. Ultrastructural Mucosal Alterations and Increased Intestinal Permeability in Non-Celiac, Type I Diabetic Patients. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2004, 36, 35–45. [Google Scholar] [CrossRef]
- Chen, T.; Liu, X.; Ma, L.; He, W.; Li, W.; Cao, Y.; Liu, Z. Food Allergens Affect the Intestinal Tight Junction Permeability in Inducing Intestinal Food Allergy in Rats. Asian Pac. J. Allergy Immunol. 2014, 32, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Majamaa, H.; Isolauri, E. Evaluation of the Gut Mucosal Barrier: Evidence for Increased Antigen Transfer in Children with Atopic Eczema. J. Allergy Clin. Immunol. 1996, 97, 985–990. [Google Scholar] [CrossRef]
- Caffarelli, C.; Cavagni, G.; Menzies, I.S.; Bertolini, P.; Atherton, D.J. Elimination Diet and Intestinal Permeability in Atopic Eczema: A Preliminary Study. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 1993, 23, 28–31. [Google Scholar] [CrossRef]
- Kidess, E.; Kleerebezem, M.; Brugman, S. Colonizing Microbes, IL-10 and IL-22: Keeping the Peace at the Mucosal Surface. Front. Microbiol. 2021, 12, 729053. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ Regulatory T-Cell Development by a Commensal Bacterium of the Intestinal Microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg Induction by a Rationally Selected Mixture of Clostridia Strains from the Human Microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.; Sicherer, S.; et al. Early-Life Gut Microbiome Composition and Milk Allergy Resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Luo, Z.; Miao, Z.; Shen, X.; Li, M.; Zhang, X.; Chen, J.; Ze, X.; Chen, Q.; He, F. Lactobacilli and Bifidobacteria Derived from Infant Intestines May Activate Macrophages and Lead to Different IL-10 Secretion. Biosci. Biotechnol. Biochem. 2020, 84, 2558–2568. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, W.; Chen, L.; Yang, W.; Huang, X.; Ma, C.; Chen, F.; Xiao, Y.; Zhao, Y.; Ma, C.; et al. Microbiota-Derived Short-Chain Fatty Acids Promote Th1 Cell IL-10 Production to Maintain Intestinal Homeostasis. Nat. Commun. 2018, 9, 3555. [Google Scholar] [CrossRef]
- Acevedo, N.; Alashkar Alhamwe, B.; Caraballo, L.; Ding, M.; Ferrante, A.; Garn, H.; Garssen, J.; Hii, C.S.; Irvine, J.; Llinás-Caballero, K.; et al. Perinatal and Early-Life Nutrition, Epigenetics, and Allergy. Nutrients 2021, 13, 724. [Google Scholar] [CrossRef]
- Roberts, G.; Bahnson, H.T.; Du Toit, G.; O’Rourke, C.; Sever, M.L.; Brittain, E.; Plaut, M.; Lack, G. Defining the Window of Opportunity and Target Populations to Prevent Peanut Allergy. J. Allergy Clin. Immunol. 2023, 151, 1329–1336. [Google Scholar] [CrossRef]
- Augustine, T.; Kumar, M.; Al Khodor, S.; Van Panhuys, N. Microbial Dysbiosis Tunes the Immune Response Towards Allergic Disease Outcomes. Clin. Rev. Allergy Immunol. 2022, 65, 43–71. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E.V. The Composition of the Gut Microbiota Shapes the Colon Mucus Barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef]
- Kwon, O.; Han, T.-S.; Son, M.-Y. Intestinal Morphogenesis in Development, Regeneration, and Disease: The Potential Utility of Intestinal Organoids for Studying Compartmentalization of the Crypt-Villus Structure. Front. Cell Dev. Biol. 2020, 8, 593969. [Google Scholar] [CrossRef]
- Sudo, N.; Sawamura, S.; Tanaka, K.; Aiba, Y.; Kubo, C.; Koga, Y. The Requirement of Intestinal Bacterial Flora for the Development of an IgE Production System Fully Susceptible to Oral Tolerance Induction. J. Immunol. Baltim. Md 1950 1997, 159, 1739–1745. [Google Scholar] [CrossRef]
- Abdel-Gadir, A.; Stephen-Victor, E.; Gerber, G.K.; Noval Rivas, M.; Wang, S.; Harb, H.; Wang, L.; Li, N.; Crestani, E.; Spielman, S.; et al. Microbiota Therapy Acts via a Regulatory T Cell MyD88/RORγt Pathway to Suppress Food Allergy. Nat. Med. 2019, 25, 1164–1174. [Google Scholar] [CrossRef]
- Rodriguez, B.; Prioult, G.; Hacini-Rachinel, F.; Moine, D.; Bruttin, A.; Ngom-Bru, C.; Labellie, C.; Nicolis, I.; Berger, B.; Mercenier, A.; et al. Infant Gut Microbiota Is Protective against Cow’s Milk Allergy in Mice despite Immature Ileal T-Cell Response. FEMS Microbiol. Ecol. 2012, 79, 192–202. [Google Scholar] [CrossRef]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Choi Hong, S.M.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy Infants Harbor Intestinal Bacteria That Protect against Food Allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Liu, Q.; Jing, W.; Wang, W. Bifidobacterium Lactis Ameliorates the Risk of Food Allergy in Chinese Children by Affecting Relative Percentage of Treg and Th17 Cells. Can. J. Infect. Dis. Med. Microbiol. J. Can. Mal. Infect. Microbiol. Medicale 2018, 2018, 4561038. [Google Scholar] [CrossRef]
- Pacheco-Sandoval, A.; Schramm, Y.; Heckel, G.; Brassea-Pérez, E.; Martínez-Porchas, M.; Lago-Lestón, A. The Pacific Harbor Seal Gut Microbiota in Mexico: Its Relationship with Diet and Functional Inferences. PLoS ONE 2019, 14, e0221770. [Google Scholar] [CrossRef] [PubMed]
- Janczy, A.; Aleksandrowicz-Wrona, E.; Kochan, Z.; Małgorzewicz, S. Impact of Diet and Synbiotics on Selected Gut Bacteria and Intestinal Permeability in Individuals with Excess Body Weight—A Prospective, Randomized Study. Acta Biochim. Pol. 2020, 67, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Leeuwendaal, N.K.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. Fermented Foods, Health and the Gut Microbiome. Nutrients 2022, 14, 1527. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The Role of Short-Chain Fatty Acids in Health and Disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Healey, G.; Murphy, R.; Butts, C.; Brough, L.; Whelan, K.; Coad, J. Habitual Dietary Fibre Intake Influences Gut Microbiota Response to an Inulin-Type Fructan Prebiotic: A Randomised, Double-Blind, Placebo-Controlled, Cross-over, Human Intervention Study. Br. J. Nutr. 2018, 119, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, P.; Chen, M.; Luo, Y.; Prabhakar, M.; Zheng, H.; He, Y.; Qi, Q.; Long, H.; Zhang, Y.; et al. Fructooligosaccharide (FOS) and Galactooligosaccharide (GOS) Increase Bifidobacterium but Reduce Butyrate Producing Bacteria with Adverse Glycemic Metabolism in Healthy Young Population. Sci. Rep. 2017, 7, 11789. [Google Scholar] [CrossRef] [PubMed]
- Vulevic, J.; Juric, A.; Walton, G.E.; Claus, S.P.; Tzortzis, G.; Toward, R.E.; Gibson, G.R. Influence of Galacto-Oligosaccharide Mixture (B-GOS) on Gut Microbiota, Immune Parameters and Metabonomics in Elderly Persons. Br. J. Nutr. 2015, 114, 586–595. [Google Scholar] [CrossRef]
- Kunasegaran, T.; Balasubramaniam, V.R.M.T.; Arasoo, V.J.T.; Palanisamy, U.D.; Ramadas, A. Diet Gut Microbiota Axis in Pregnancy: A Systematic Review of Recent Evidence. Curr. Nutr. Rep. 2023, 12, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Sarita, B.; Samadhan, D.; Hassan, M.Z.; Kovaleva, E.G. A Comprehensive Review of Probiotics and Human Health-Current Prospective and Applications. Front. Microbiol. 2024, 15, 1487641. [Google Scholar] [CrossRef]
- Yang, J.; Kuang, H.; Li, N.; Hamdy, A.M.; Song, J. The Modulation and Mechanism of Probiotic-Derived Polysaccharide Capsules on the Immune Response in Allergic Diseases. Crit. Rev. Food Sci. Nutr. 2023, 63, 8768–8780. [Google Scholar] [CrossRef]
| Author (Year) | Article Type | Quality of Article | Justification-Limitations |
|---|---|---|---|
| Canani et al. (2016) [25] | Controlled intervention study | Fair | Controlled intervention (lack of clear randomization and blinding details), coupled with a very small sample size for the intervention groups. |
| Dzidic et al. (2017) [26] | Prospective observational cohort study | Fair | No explicit reporting of participation rate for this specific sub-study, lack of details for blinding procedures for outcome assessment, no information on loss to follow-up for the selected cohort, small sample size. |
| Canani et al. (2017) [27] | Randomized Controlled Trial | Good | Strong methodology. Limitations: parents of the participants were aware of the assigned treatment, which could potentially introduce reporting bias. The study population was limited to children with immunoglobulin E (IgE) -mediated cow’s milk allergy from specific socioeconomic and urban backgrounds, which may restrict the generalizability of the results. |
| Candy et al. (2018) [28] | Randomized control trial | Good | Good methodology. Limitations: Absence of a standardized diagnostic test or mandatory food challenge for non-IgE cow’s milk allergy, potential baseline imbalances in delivery mode, and the exploratory nature of its clinical outcomes, meaning the study was not powered to show significant clinical benefits. |
| Savage et al. (2018) [11] | Observational cross-sectional study | Fair | The observational design precludes establishing causality. The self-reported nature of dietary data, particularly the retrospective assessment of infant diet at the time of stool collection, introduces a risk of misclassification and recall bias. Specific recruitment criteria (family history of allergy/asthma) may limit the generalizability of the findings to broader populations. |
| Nocerino et al. (2019) [29] | Prospective cohort study | Good | Strong methodological quality, well designed and executed. Observational study—cannot establish causality (a fundamental characteristic of its design, not a flaw in execution). The large sample size and clear presentation of results further support its classification as a high-quality study. |
| Aparicio et al. (2020) [30] | Observational pilot cohort study | Fair | Small sample size of 30 mother–infant pairs, which limits the statistical power. The objectives are clearly stated and the methodology for sample collection, molecular, and immunological analyses is well-described, ensuring internal validity for the conducted tests. |
| Jing et al. (2020) [31] | Randomized double-blind control trial | Good | Good methodology. Conducted at a single center, potentially limiting generalizability. |
| Bao et al. (2021) [32] | Cross-sectional study | Good/ Fair | Lacks explicit details on sample size justification and blinding of outcome assessors. Cannot establish temporality of exposure and outcome due to cross-sectional design. |
| Dawson et al. (2021) [33] | Randomized control trial | Fair | Small sample size. The reliance on self-reported dietary data, lack of full blinding, and per-protocol analysis. |
| Marrs et al. (2021) [12] | Randomized control trial | Good | Good methodology. No sample size justification for microbiome analysis. Randomization concealment methods not described. Not all enrolled participants followed for full duration of microbiome study. Not explicitly stated if microbiome/clinical outcome assessors were blinded. Did not discuss/quantify potential inter-group contamination. Direct clinical importance of microbiota changes in preventing food allergy not established. |
| Nocerino et al. (2021) [34] | Prospective cohort study | Good | Good methodology. Participants not randomized to formula groups, continued prescribed formula. Residual confounding cannot be entirely ruled out. |
| Homann et al. (2021) [35] | Longitudinal cohort study | Good | Good methodology. The comparison of two geographically distinct cohorts adds to the robustness of the findings, and the authors acknowledge potential limitations (the relatively small sample size and differences in dietary introduction approaches between cohort) |
| De Filippis et al. (2021) [36] | Randomized double-blind control trial | Good | Good methodology. Absence of reported participation rate, sample size justification, and explicit statement on outcome assessor blinding. |
| Boulange et al. (2023) [37] | Single-arm, prospective clinical study | Fair | Lack of control group (changes could be due to natural infant gut maturation/environmental factors). No sample size justification, small subject number. Approximately 21% participant dropouts. Limited clinical importance (did not directly assess clinical outcomes). Potential for bias (no explicit blinding for lab analysis, manufacturer funding). |
| Hanada et al. (2023) [38] | Randomized controlled trial | Fair | The study explicitly states it is a pilot study and was not powered for its primary clinical outcome, which was indeed found to be non-significant. The lack of explicit detail on controlling for co-interventions is also a minor concern. |
| Yan et al. (2023) [39] | Longitudinal observational study | Fair | Small sample size, fecal samples were not collected at the 2-year follow-up, which prevented establishing direct connections between microbiota shifts and symptom resolution or persistence post-follow-up. |
| Gao et al. (2023) [40] | Prospective cohort study | Good | Rigorous methodology, combined with a substantial sample size across two distinct populations, enhances the clinical relevance of the conclusions. |
| Sukenikova et al. (2023) [41] | Prospective cohort study | Good | Good methodology, substantial 10-year follow-up. The use of both clinical allergy diagnoses (allergist confirmation) and parental reports for allergy status, combined with in vitro immunological assays and gut microbiota analysis (16S rRNA gene sequencing), demonstrates a multi-faceted approach to evaluating the intervention. |
| Shibata et al. (2024) [42] | Ancillary cohort study | Fair | Small sample size, no multivariable models, no adjustment for confounding factors. Lack of detail regarding participation rate. |
| Hara et al. (2024) [43] | Case–Control | Fair | Clear objectives and a well-defined case–control design appropriate for investigating associations. However, as a case–control study, it cannot establish causality. The sample size of 130 participants is moderate for gut microbiome studies. The study’s focus on a single age group and recruitment from one institution might limit the generalizability of its findings. |
| Castro et al. (2024) [44] | Prospective longitudinal cohort study | Fair | Small sample size, which restricts the generalizability and statistical power of the findings. The absence of a control group of healthy children makes it challenging to draw robust conclusions about the observed gut microbiota changes. |
| Korpela et al. (2024) [45] | Prospective observational study | Good | The sample size is substantial, the methodology is robust. The comparison with pre-pandemic cohorts strengthens the study’s ability to assess the impact of social distancing |
| Nekrasova et al. (2024) [46] | Observational case- control | Good | The study is well-designed as a case–control study. A comprehensive set of statistical analyses were utilized to process the complex metagenomic data. The researchers applied data transformation & quality control measures to mitigate sampling and rarefaction bias. |
| Chen et al. (2024) [47] | Observational cross-sectional | Good | Good methodology. The study focused on children aged 18 to 36 months, which might limit the generalizability of the findings to older children or adults. |
| Jones et al. (2024) [48] | Randomized placebo-controlled trial | Good | The study employed a strong randomized, double-blinded, placebo-controlled design, which minimizes bias. The interventions and outcome measures were clearly defined, and robust methodologies were used for microbiome sequencing and SCFA quantification. Statistical analyses were comprehensive and included appropriate corrections for multiple comparisons, and the laboratory analyses were performed while blinded to treatment allocation. |
| Shibata et al. (2025) [49] | Combined analysis of two longitudinal birth-cohort studies | Good | Robust design by combining data from two prospective longitudinal birth-cohort studies, allowing for comprehensive analysis of gut microbiota over time. Limitations: while the study combined two cohorts, certain relationships between gut microbiota and outcomes showed heterogeneity and were not consistently shared between the two studies, except for Bifidobacterium. |
| Li et al. (2025) [50] | Case- Control | Fair | Cross-sectional nature, preventing the establishment of causal relationships. The relatively small sample size and recruitment from a single center might limit the generalizability of the results. Furthermore, the study did not delve into the functional aspects of the gut microbiota and did not fully control for confounding factors like diet, which could significantly influence gut microbial composition. |
| Zhang et al. (2025) [51] | Cross-Sectional study | Good | Clear research question, an appropriate cross-sectional design for its objectives, well-defined participant groups with matching, and detailed, ethically approved methods, robust statistical analyses. Limitations: cross-sectional design, which prevents establishing causal relationships. It also did not account for confounding factors like daily diet, activity levels, or lifestyle. |
| Imoto et al. (2025) [52] | Prospective cohort study | Good | Robust prospective cohort design, clear objectives, and detailed methodology for sample collection and statistical analyses demonstrate scientific rigor. The study’s primary limitation is the focus on specific genes (16S rRNA), which only reveals composition rather than functional capabilities of the microbiota. |
| Nocerino et al. (2025) [53] | Prospective cohort study | Good | Strong prospective cohort design with a lengthy 6-year follow-up, robust methodology. Limitations: no formal sample size calculation was performed specifically for this 72-month follow-up, as it extended a previous 36-month study |
| Author (Year) | Population Studied | Number of Subjects | Key Findings |
|---|---|---|---|
| Dzidic et al. (2017) [26] | Children from the LISA birth cohort in Sweden followed prospectively for the first 7 years of life | 28 children |
|
| Savage et al. (2018) [11] | Infants participating in a cohort selected based on parental history of asthma or allergy. | 323 infants included in the primary analyses:
|
|
| Aparicio et al. (2020) [30] | Mother–infant pairs where the infants were diagnosed with colic, (CMPA), or proctocolitis, healthy control infants. | 30 mother–infant pairs divided into four groups:
|
|
| Bao et al. (2021) [32] | Twin pairs (discordant/concordant for food allergy) | 18 twin pairs |
|
| Marrs et al. (2021) [12] | Exclusively breastfed infants aged between 12 and 17 weeks at enrollment. | Provided baseline (3-month) stool samples for microbiome analysis: 288 infants Subset for longitudinal microbiome analysis (samples at 3, 6, and 12 months): 70 individuals. |
|
| Homann et al. (2021) [35] | Healthy, full-term, vaginally born infants | 24 infants |
|
| De Filippis et al. (2021) [36] | Children with diagnosed IgE-mediated food allergies (FA) or respiratory allergies (RA) and healthy controls (CT) |
|
|
| Yan et al. (2023) [39] | Children with FA and controls | 10 children aged 0 to 3 years with FA and 10 controls |
|
| Gao et al. (2023) [40] | Infants from the Barwon Infant Study (BIS) cohort and The Copenhagen Prospective Studies on Asthma in Childhood (COPSAC2010) cohort from Denmark |
|
|
| Shibata et al. (2024) [42] | Children 1 week- 7 years old & their mothers from two distinct birth cohorts. The CHIBA study focused on a high-risk cohort with a family history of allergic diseases, while the Katsushika study was originally a randomized trial evaluating skincare +/− synbiotics |
|
|
| Hara et al. (2024) [43] | One-and-a-half-year-old food-allergic and healthy children. | 130 participants:
|
|
| Castro et al. (2024) [44] | Pediatric patients with IgE-mediated or non-IgE-mediated CMA, aged 0–12 months a | 26 pediatric patients diagnosed with CMPA who followed a cow’s milk protein-free (CMPF) diet |
|
| Korpela et al. (2024) [45] | Irish infants born between March and May 2020, during the initial phases of the COVID-19 pandemic & associated social distancing restrictions | 360 infants |
|
| Nekrasova et al. (2024) [46] | Children with atopic dermatitis (AD) and food allergies (FA) | 128 children aged 3 to 12 years, divided into three groups:
|
|
| Chen et al. (2024) [47] | Children with different IgE-mediated food hypersensitivity (FH) | Fecal samples from: 57 children with IgE-mediated hypersensitivity (FH) & 24 healthy children aged 18 to 36 months. |
|
| Zhang et al. (2025) [51] | 80 children with FAs, 40 healthy controls |
|
|
| Imoto et al. (2025) [52] | Cohort of Japanese infants from birth through 24 months of age | 121 infants |
|
| Li et al. (2025) [50] | Pediatric patients with peanut allergy and healthy controls | 97 children were included in the study, comprising: 35 children with peanut allergy (PA group) & 62 healthy children (HC group) |
|
| Author (Year) | Population Studied | Number of Subjects | Key Findings |
|---|---|---|---|
| Canani et al. (2016) [25] | Infants with CMA |
|
|
| Canani et al. (2017) [27] | Children with IgE-mediated CMA, aged 1–12 months |
|
|
| Candy et al. (2018) [28] | Infants with suspected non-IgE-mediated CMA |
|
|
| Nocerino et al. (2019) [29] | Children aged 4–6 years with IgE-mediated CMA in their first year of life who had acquired oral tolerance to cow’s milk proteins for at least 12 months, healthy controls |
|
|
| Jing et al. (2020) [31] | Infants with CMA who could not be exclusively breastfed |
|
|
| Dawson et al. (2020) [33] | Healthy pregnant women from gestation week 26 | 45 women randomized:
|
|
| Nocerino et al. (2021) [34] | Non-breastfed infants (aged 1–12 months) with confirmed IgE-mediated CMA | Enrolled into the study: 365 subjects: 73 subjects in each of the 5 formula cohorts (EHCF + LGG, rice hydrolyzed formula, soy formula, EHWF, or AAF) |
|
| Boulange et al. (2023) [37] | Non-breastfed infants aged between 2 weeks and 6 months with symptoms suggestive of CMA | 194 non-breastfed infants:
|
|
| Hanada et al. (2023) [38] | Children aged 1–18 years with IgE-mediated CMA diagnosed by oral-milk challenge test |
|
|
| Sukenikova et al. (2023) [41] | Pregnant women and their neonates, divided into groups based on maternal allergic status and probiotic supplementation. |
|
|
| Shibata et al. (2024) [49] | School-age children with IgE-mediated CMA undergoing oral OIT |
|
|
| Jones et al. (2024) [48] | Pregnant women (recruited before 21 weeks’ gestation) and their infants up to 12 months of age | 74 mother–infant pairs |
|
| Nocerino et al. (2025) [53] | Non-breastfed infants aged 1–12 months with immunoglobulin E (IgE)-mediated CMA |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mareș, R.C.; Săsăran, M.O.; Mărginean, C.O. Gut Microbiota and Food Allergy: A Review of Mechanisms and Microbiota-Targeted Interventions. Nutrients 2025, 17, 3009. https://doi.org/10.3390/nu17183009
Mareș RC, Săsăran MO, Mărginean CO. Gut Microbiota and Food Allergy: A Review of Mechanisms and Microbiota-Targeted Interventions. Nutrients. 2025; 17(18):3009. https://doi.org/10.3390/nu17183009
Chicago/Turabian StyleMareș, Roxana Cristina, Maria Oana Săsăran, and Cristina Oana Mărginean. 2025. "Gut Microbiota and Food Allergy: A Review of Mechanisms and Microbiota-Targeted Interventions" Nutrients 17, no. 18: 3009. https://doi.org/10.3390/nu17183009
APA StyleMareș, R. C., Săsăran, M. O., & Mărginean, C. O. (2025). Gut Microbiota and Food Allergy: A Review of Mechanisms and Microbiota-Targeted Interventions. Nutrients, 17(18), 3009. https://doi.org/10.3390/nu17183009






