Gut Microbiota and Autism Spectrum Disorders: Neurodevelopmental, Behavioral, and Gastrointestinal Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. The Evaluation of Medical Background
2.3. Collecting and Storing Stool Samples
2.4. The Analysis of Microbiome Composition
2.5. Statistics and Biostatistics
2.6. Ethical Approval
3. Results
3.1. Gut Microbiota Sequencing
3.2. The Structure of the Group
3.3. Diet and Food Selectivity
3.4. Functional Gastrointestinal Disorders
3.5. Vineland Adaptive Behavioral Scale Results
3.5.1. Domain: Communication
Subdomain: Receptive Communication
Subdomain: Expressive Communication
Subdomain: Writing Skills
3.5.2. Domain: Daily Living Skills
Subdomain: Personal Skills
Subdomain: Domestic Skills
Subdomain: Community Skills
3.5.3. Domain: Socialization
Subdomain: Interpersonal Skills
Subdomain: Play and Leisure Skills
Subdomain: Coping Skills
3.5.4. Domain: Motor Skills
Subdomain: Large Muscle Skills
Subdomain: Small Muscle Skills
4. Discussion
4.1. The Structure of the Study Group and Microbiota Composition
4.2. The Influence of Diet on Gut Microbiota Composition
4.3. The Role of Gut Microbiota in Functional Gastrointestinal Disorders
4.4. The Influence of Microbiota Composition on Communication Skills
4.5. Microbiota Composition and Daily Living Skills
4.6. The Role of Microbiota in the Development of Socialization Skills
4.7. Microbiota and the Development of Motor Skills
4.8. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ASD | Autism Spectrum Disorder |
| ChBD | carbohydrate-based diet |
| CNS | central nervous system |
| FGID | Functional Gastrointestinal Disorder |
| FS | food selectivity |
| GABA | γ-aminobutyric acid |
| HCD | high-calorie diet |
| IBS | irritable bowel syndrome |
| ICD-10 | International Statistical Classification of Diseases and Related Health Problems—10th Revision |
| LCD | low-calorie diet |
| MDD | major depressive disorder |
| MS | multiple sclerosis |
| MSA | multiple system atrophy |
| PBD | protein-based diet |
| PD | Parkinson’s disease |
| RS | Rett syndrome |
| SCFA | short-chain fatty acid |
| SMA | spinal muscular atrophy |
| VABS | Vineland Adaptive Behavior Scales |
References
- World Health Organization. Autism. Available online: https://www.who.int/news-room/fact-sheets/detail/autism-spectrum-disorders (accessed on 10 July 2025).
- Zeidan, J.; Fombonne, E.; Scorah, J.; Ibrahim, A.; Durkin, M.S.; Saxena, S.; Yusuf, A.; Shih, A.; Elsabbagh, M. Global prevalence of autism: A systematic review update. Autism Res. 2022, 15, 778–790. [Google Scholar] [CrossRef]
- ICD-10 Version: 2019. Pervasive Developmental Disorders. Available online: https://icd.who.int/browse10/2019/en#/F84 (accessed on 2 May 2025).
- Tye, C.; Runicles, A.K.; Whitehouse, A.J.O.; Alvares, G.A. Characterizing the Interplay Between Autism Spectrum Disorder and Comorbid Medical Conditions: An Integrative Review. Front. Psychiatry 2019, 23, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Krajmalnik-Brown, R.; Lozupone, C.; Kang, D.W.; Adams, J.B. Gut Bacteria in Children with Autism Spectrum Disorders: Challenges and Promise of Studying How a Complex Community Influences a Complex Disease. Microb. Ecol. Health Dis. 2015, 26, 26914. [Google Scholar] [CrossRef] [PubMed]
- Arnold, L.E.; Luna, R.A.; Williams, K.; Chan, J.; Parker, R.A.; Wu, Q.; Hollway, J.A.; Jeffs, A.; Lu, F.; Hayes, C.; et al. Probiotics for Gastrointestinal Symptoms and Quality of Life in Autism: A Placebo-Controlled Pilot Trial. J. Child. Adolesc. Psychopharmacol. 2019, 29, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska-Pietruszka, Z.; Figlerowicz, M.; Mazur-Melewska, K. The History of the Intestinal Microbiota and the Gut-Brain Axis. Pathogens 2022, 11, 1540. [Google Scholar] [CrossRef]
- Lewandowska-Pietruszka, Z.; Figlerowicz, M.; Mazur-Melewska, K. Microbiota in Autism Spectrum Disorder: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 16660. [Google Scholar] [CrossRef]
- West, K.A.; Yin, X.; Rutherford, E.M.; Wee, B.; Choi, J.; Chrisman, B.S.; Dunlap, K.L.; Hannibal, R.L.; Hartono, W.; Lin, M.; et al. Multi-angle meta-analysis of the gut microbiome in Autism Spectrum Disorder: A step toward understanding patient subgroups. Sci. Rep. 2022, 12, 17034. [Google Scholar] [CrossRef]
- Wu, T.; Wang, H.; Lu, W.; Zhai, Q.; Zhang, Q.; Yuan, W.; Gu, Z.; Zhao, J.; Zhang, H.; Chen, W. Potential of gut microbiome for detection of autism spectrum disorder. Microb. Pathog. 2020, 149, 104568. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Son, J.S.; Zheng, L.J.; Rowehl, L.M.; Tian, X.; Zhang, Y.; Zhu, W.; Litcher-Kelly, L.; Gadow, K.D.; Gathungu, G.; Robertson, C.E.; et al. Comparison of Fecal Microbiota in Children with Autism Spectrum Disorders and Neurotypical Siblings in the Simons Simplex Collection. PLoS ONE 2015, 10, e0137725. [Google Scholar] [CrossRef]
- Grimaldi, R.; Gibson, G.R.; Vulevic, J.; Giallourou, N.; Castro-Mejía, J.L.; Hansen, L.H.; Leigh Gibson, E.; Nielsen, D.S.; Costabile, A. A prebiotic intervention study in children with autism spectrum disorders (ASDs). Microbiome 2018, 6, 133. [Google Scholar] [CrossRef]
- Tomova, A.; Soltys, K.; Kemenyova, P.; Karhanek, M.; Babinska, K. The Influence of Food Intake Specificity in Children with Autism on Gut Microbiota. Int. J. Mol. Sci. 2020, 21, 2797. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska-Pietruszka, Z.; Figlerowicz, M.; Mazur-Melewska, K. Oral Microbiota Composition and Its Association with Gastrointestinal and Developmental Abnormalities in Children with Autism Spectrum Disorder. Microorganisms 2025, 13, 1822. [Google Scholar] [CrossRef]
- Paudel, D.; Uehara, O.; Giri, S.; Yoshida, K.; Morikawa, T.; Kitagawa, T.; Matsuoka, H.; Miura, H.; Toyofuku, A.; Kuramitsu, Y.; et al. Effect of psychological stress on the oral-gut microbiota and the potential oral-gut-brain axis. Jpn. Dent. Sci. Rev. 2022, 58, 365–375. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Klepacz, N.; Rabęda, A.; Grzesik, K.; Pilarczyk, K.; Adamska, H.; Kaus, M.; Nowak, W.E.; Sawczuk, H.; Cudziło, Z.; Malicka, M. The microbiome-mind connection: Exploring gut health’s impact on depression. The microbiome-mind connection: Exploring gut health’s impact on depression. J. Med. Sci. 2025, 94, e1173. [Google Scholar]
- Lewandowska, Z.; Mazur-Melewska, K.; Figlerowicz, M. Cortisol in individuals with autism spectrum disorders—Review. Neurol. Dziecięca 2020, 29, 36–44. [Google Scholar] [CrossRef]
- Dao, M.C.; Subar, A.F.; Warthon-Medina, M.; Cade, J.E.; Burrows, T.; Golley, R.K.; Forouhi, N.G.; Pearce, M.; Holmes, B.A. Dietary assessment toolkits: An overview. Public Health Nutr. 2018, 22, 404–418. [Google Scholar] [CrossRef]
- Pearce, M.; Powell, R.; Imamura, F.; De Lucia Rolfe, E.; Brage, S.; Forouhi, N. Estimated Food Diaries. Measurement Toolkit. Available online: https://www.measurement-toolkit.org/diet/subjective-methods/estimated-food-diaries (accessed on 21 June 2025).
- Sawicka-Gutaj, N.; Gruszczyński, D.; Guzik, P.; Mostowska, A.; Walkowiak, J. Publication ethics of human studies in the light of the Declaration of Helsinki—A mini-review. J. Med. Sci. 2022, 91, e700. [Google Scholar] [CrossRef]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted microbiota and opportunistic pathogen colonisation in caesarean section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Nakhal, M.M.; Yassin, L.K.; Alyaqoubi, R.; Saeed, S.; Alderei, A.; Alhammadi, A.; Alshehhi, M.; Almehairbi, A.; Al Houqani, S.; BaniYas, S.; et al. The Microbiota-Gut-Brain Axis and Neurological Disorders: A Comprehensive Review. Life 2024, 14, 1234. [Google Scholar] [CrossRef]
- Winiarska-Mieczan, A.; Tomaszewska, E.; Donaldson, J.; Jachimowicz, K. The Role of Nutritional Factors in the Modulation of the Composition of the Gut Microbiota in People with Autoimmune Diabetes. Nutrients 2022, 14, 2498. [Google Scholar] [CrossRef]
- Ioannou, P.; Kampanieri, E.; Koukias, S.; Baliou, S.; Tsantes, A.G.; Kofteridis, D. Kytococcus Species Infections in Humans-A Narrative Review. Microorganisms 2025, 13, 1072. [Google Scholar] [CrossRef]
- Marples, R.R.; Richardson, J.F. Micrococcus in the blood. J. Med. Microbiol. 1980, 13, 355–362. [Google Scholar] [CrossRef]
- Fatahi-Bafghi, M. Characterization of the Rothia spp. and their role in human clinical infections. Infect. Genet. Evol. 2021, 93, 104877. [Google Scholar] [CrossRef] [PubMed]
- Radjabzadeh, D.; Bosch, J.A.; Uitterlinden, A.G.; Zwinderman, A.H.; Ikram, M.A.; van Meurs, J.B.J.; Luik, A.I.; Nieuwdorp, M.; Lok, A.; van Duijn, C.M.; et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 2022, 13, 7128. [Google Scholar] [CrossRef] [PubMed]
- Barandouzi, Z.A.; Starkweather, A.R.; Henderson, W.A.; Gyamfi, A.; Cong, X.S. Altered Composition of Gut Microbiota in Depression: A Systematic Review. Front. Psychiatry 2020, 11, 541. [Google Scholar] [CrossRef]
- Li, P.; Wang, S.; Li, J.; Xiao, Z.; Zhu, H.; Sheng, D.; Liu, W.; Xiao, B.; Zhou, L. Appraising the Effects of Gut Microbiota on Insomnia Risk Through Genetic Causal Analysis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2025, 198, e33021. [Google Scholar] [CrossRef]
- Gao, M.; Wang, J.; Liu, P.; Tu, H.; Zhang, R.; Zhang, Y.; Sun, N.; Zhang, K. Gut microbiota composition in depressive disorder: A systematic review, meta-analysis, and meta-regression. Transl. Psychiatry 2023, 13, 379. [Google Scholar] [CrossRef] [PubMed]
- Oñate, F.P.; Chamignon, C.; Burz, S.D.; Lapaque, N.; Monnoye, M.; Philippe, C.; Bredel, M.; Chêne, L.; Farin, W.; Paillarse, J.M.; et al. Adlercreutzia equolifaciens is an Anti-Inflammatory Commensal Bacterium with Decreased Abundance in Gut Microbiota of Patients with Metabolic Liver Disease. Int. J. Mol. Sci. 2023, 24, 12232. [Google Scholar] [CrossRef]
- Martinez-Guryn, K.; Leone, V.; Chang, E.B. Regional Diversity of the Gastrointestinal Microbiome. Cell Host Microbe 2019, 26, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Winpenny, E.M.; van Sluijs, E.M.F.; White, M.; Klepp, K.I.; Wold, B.; Lien, N. Changes in diet through adolescence and early adulthood: Longitudinal trajectories and association with key life transitions. Int. J. Behav. Nutr. Phys. Act. 2018, 15, 86. [Google Scholar] [CrossRef]
- Jiang, S.; Cai, L.; Lv, L.; Li, L. Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell Fact. 2021, 20, 45. [Google Scholar] [CrossRef]
- Iseppi, R.; Messi, P.; Camellini, S.; Sabia, C. Bacteriocin activity of Lactobacillus brevis and Lactobacillus paracasei ssp. paracasei. J. Med. Microbiol. 2019, 68, 1359–1366. [Google Scholar] [CrossRef]
- Zou, X.; Pan, L.; Xu, M.; Wang, X.; Wang, Q.; Han, Y. Probiotic potential of Lactobacillus sakei L-7 in regulating gut microbiota and metabolism. Microbiol. Res. 2023, 274, 127438. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Musmeci, E.; Candeliere, F.; Amaretti, A.; Rossi, M. Identification of mucin degraders of the human gut microbiota. Sci. Rep. 2021, 11, 11094. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lan, Q.; Zhou, M.; Liu, F. From gut to kidney: Microbiota modulates stone risk through inflammation–a mediated Mendelian randomization study. Mamm. Genome 2025, 36, 250–261. [Google Scholar] [CrossRef]
- Shen, Y.; Li, C.; Zhang, X.; Wang, Y.; Zhang, H.; Yu, Z.; Gui, B.; Hu, R.; Li, Q.; Gao, A.; et al. Gut microbiota linked to hydrocephalus through inflammatory factors: A Mendelian randomization study. Front. Immunol. 2024, 15, 1372051. [Google Scholar] [CrossRef]
- Ma, J.; Zhu, Z.; Yishajiang, Y.; Alarjani, K.M.; Hong, L.; Luo, L. Role of gut microbiota and inflammatory factors in acute respiratory distress syndrome: A Mendelian randomization analysis. Front. Microbiol. 2023, 14, 1294692. [Google Scholar] [CrossRef]
- Wen, J.; He, J.Q. The Causal Impact of the Gut Microbiota on Respiratory Tuberculosis Susceptibility. Infect. Dis. Ther. 2023, 12, 2535–2544. [Google Scholar] [CrossRef] [PubMed]
- Crost, E.H.; Coletto, E.; Bell, A.; Juge, N. Ruminococcus gnavus: Friend or foe for human health. FEMS Microbiol Rev. 2023, 47, fuad014. [Google Scholar] [CrossRef]
- Genseke, S.; Berisha, M.; Teerstegen, A.; Meyer, B.; Kaasch, A.J.; Färber, J.; Schalk, E.; Zautner, A.E.; Esser, T.; Kahlfuß, S. Lautropia mirabilis sepsis in immunodeficiency: First report and genomic features. Infection 2025, 53, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Tap, J.; Derrien, M.; Törnblom, H.; Brazeilles, R.; Cools-Portier, S.; Doré, J.; Störsrud, S.; Le Nevé, B.; Öhman, L.; Simrén, M. Identification of an Intestinal Microbiota Signature Associated with Severity of Irritable Bowel Syndrome. Gastroenterology 2017, 152, 111–123.e8. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, C.; Huang, S.; Wang, X.; Cao, M.; Gu, T.; Ou, X.; Pan, S.; Lin, Z.; Wang, X.; et al. Multi-omics analyses demonstrate the modulating role of gut microbiota on the associations of unbalanced dietary intake with gastrointestinal symptoms in children with autism spectrum disorder. Gut Microbes 2023, 15, 2281350. [Google Scholar] [CrossRef] [PubMed]
- Verhaar, B.J.H.; Hendriksen, H.M.A.; de Leeuw, F.A.; Doorduijn, A.S.; van Leeuwenstijn, M.; Teunissen, C.E.; Barkhof, F.; Scheltens, P.; Kraaij, R.; van Duijn, C.M.; et al. Gut Microbiota Composition Is Related to AD Pathology. Front. Immunol. 2022, 12, 794519. [Google Scholar] [CrossRef]
- Yang, Y.; Wen, C.; Zheng, S.; Song, F.; Liu, Y.; Yao, X.; Tang, Y.; Feng, X.; Chen, J.; Yang, F. Lactobacillus fermentum Alleviates the Colorectal Inflammation Induced by Low-Dose Sub-Chronic Microcystin-LR Exposure. Toxins 2023, 15, 579. [Google Scholar] [CrossRef]
- Lee, J.; d’Aigle, J.; Atadja, L.; Quaicoe, V.; Honarpisheh, P.; Ganesh, B.P.; Hassan, A.; Graf, J.; Petrosino, J.; Putluri, N.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ. Res. 2020, 127, 453–465. [Google Scholar] [CrossRef]
- Moriki, D.; León, E.D.; García-Gamero, G.; Jiménez-Hernández, N.; Artacho, A.; Pons, X.; Koumpagioti, D.; Dinopoulos, A.; Papaevangelou, V.; Priftis, K.N.; et al. Specific Gut Microbiome Signatures in Children with Cow’s Milk Allergy. Nutrients 2024, 16, 2752. [Google Scholar] [CrossRef]
- Keskitalo, A.; Munukka, E.; Toivonen, R.; Hollmén, M.; Kainulainen, H.; Huovinen, P.; Jalkanen, S.; Pekkala, S. Enterobacter cloacae administration induces hepatic damage and subcutaneous fat accumulation in high-fat diet fed mice. PLoS ONE 2018, 13, e0198262. [Google Scholar] [CrossRef]
- Yan, H.; Fei, N.; Wu, G.; Zhang, C.; Zhao, L.; Zhang, M. Regulated Inflammation and Lipid Metabolism in Colon mRNA Expressions of Obese Germfree Mice Responding to Enterobacter cloacae B29 Combined with the High Fat Diet. Front. Microbiol. 2016, 7, 1786. [Google Scholar] [CrossRef]
- Bai, X.B.; Xu, S.; Zhou, L.J.; Meng, X.Q.; Li, Y.L.; Chen, Y.L.; Jiang, Y.H.; Lin, W.Z.; Chen, B.Y.; Du, L.J.; et al. Oral pathogens exacerbate Parkinson’s disease by promoting Th1 cell infiltration in mice. Microbiome 2023, 11, 254. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Ueyama, J.; Kashihara, K.; Ito, M.; Hamaguchi, T.; Maeda, T.; Tsuboi, Y.; Katsuno, M.; Hirayama, M.; Ohno, K. Gut microbiota in dementia with Lewy bodies. NPJ Park. Dis. 2022, 8, 169. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Song, Y.X.; Wu, X.D.; Luo, Y.G.; Miao, R.; Yu, X.M.; Guo, X.; Wu, D.Z.; Bao, R.; Mi, W.D.; et al. Gut microbiota and cognitive performance: A bidirectional two-sample Mendelian randomization. J. Affect. Disord. 2024, 353, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Akinkunmi, E.O.; Adeyemi, O.I.; Igbeneghu, O.A.; Olaniyan, E.O.; Omonisi, A.E.; Lamikanra, A. The pathogenicity of Staphylococcus epidermidis on the intestinal organs of rats and mice: An experimental investigation. BMC Gastroenterol. 2014, 14, 126. [Google Scholar] [CrossRef]
- Zhu, Z.; Ma, X.; Wu, J.; Xiao, Z.; Wu, W.; Ding, S.; Zheng, L.; Liang, X.; Luo, J.; Ding, D.; et al. Altered Gut Microbiota and Its Clinical Relevance in Mild Cognitive Impairment and Alzheimer’s Disease: Shanghai Aging Study and Shanghai Memory Study. Nutrients 2022, 14, 3959. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Huang, S.Y.; Chen, S.D.; Zhang, Y.R.; Huang, Y.Y.; Yu, J.T. Investigating Casual Associations Among Gut Microbiota, Metabolites, and Neurodegenerative Diseases: A Mendelian Randomization Study. J. Alzheimer’s Dis. 2022, 87, 211–222. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Pindo, M.; Renzi, D.; et al. Altered gut microbiota in Rett syndrome. Microbiome 2016, 4, 41. [Google Scholar] [CrossRef]
- Lee, S.H.; Han, C.; Shin, C. IUPHAR review: Microbiota-gut-brain axis and its role in neuropsychiatric disorders. Pharmacol. Res. 2025, 216, 107749. [Google Scholar] [CrossRef]
- Aboushaala, K.; Chee, A.V.; Adnan, D.; Toro, S.J.; Singh, H.; Savoia, A.; Dhillon, E.S.; Yuh, C.; Dourdourekas, J.; Patel, I.K.; et al. Gut microbiome dysbiosis is associated with lumbar degenerative spondylolisthesis in symptomatic patients. JOR Spine 2024, 7, e70005. [Google Scholar] [CrossRef]
- Teixeira, C.G.; Fusieger, A.; Milião, G.L.; Martins, E.; Drider, D.; Nero, L.A.; de Carvalho, A.F. Weissella: An Emerging Bacterium with Promising Health Benefits. Probiotics Antimicrob. Proteins 2021, 13, 915–925. [Google Scholar] [CrossRef]
- Molina-López, J.; Leiva-García, B.; Planells, E.; Planells, P. Food selectivity, nutritional inadequacies, and mealtime behavioral problems in children with autism spectrum disorder compared to neurotypical children. Int. J. Eat. Disord. 2021, 54, 2155–2166. [Google Scholar] [CrossRef]
- Huang, H.; Cheng, S.; Yang, X.; Liu, L.; Cheng, B.; Meng, P.; Pan, C.; Wen, Y.; Jia, Y.; Liu, H.; et al. Dissecting the Association between Gut Microbiota and Brain Structure Change Rate: A Two-Sample Bidirectional Mendelian Randomization Study. Nutrients 2023, 15, 4227. [Google Scholar] [CrossRef]
- Nilholm, C.; Manoharan, L.; Roth, B.; D’Amato, M.; Ohlsson, B. A starch- and sucrose-reduced dietary intervention in irritable bowel syndrome patients produced a shift in gut microbiota composition along with changes in phylum, genus, and amplicon sequence variant abundances, without affecting the micro-RNA levels. United Eur. Gastroenterol. J. 2022, 10, 363–375. [Google Scholar] [CrossRef]
- Marizzoni, M.; Mirabelli, P.; Mombelli, E.; Coppola, L.; Festari, C.; Lopizzo, N.; Luongo, D.; Mazzelli, M.; Naviglio, D.; Blouin, J.L.; et al. A peripheral signature of Alzheimer’s disease featuring microbiota-gut-brain axis markers. Alzheimer’s Res. Ther. 2023, 15, 101. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, C.; Wang, J.; Cheng, Y.; Wang, L.; Tong, Y.; Yan, D. Identification of host gene-microbiome associations in colorectal cancer patients using mendelian randomization. J. Transl. Med. 2023, 21, 535. [Google Scholar] [CrossRef]
- Liang, L.D.; Li, S.; Huang, M.J.; Peng, H.X.; Lu, Z.J.; Zhang, Z.H.; Su, L.Y.; Sooranna, S.R.; Liu, Y.; Huang, Z.H. Causal relationship between gut microbiota and puerperal sepsis: A 2-sample Mendelian randomization study. Front. Microbiol. 2024, 15, 1407324. [Google Scholar] [CrossRef]
- Lynch, J.B.; Gonzalez, E.L.; Choy, K.; Faull, K.F.; Jewell, T.; Arellano, A.; Liang, J.; Yu, K.B.; Paramo, J.; Hsiao, E.Y. Gut microbiota Turicibacter strains differentially modify bile acids and host lipids. Nat. Commun. 2023, 14, 3669. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Buhman, K.K.; Hartman, P.A.; Beitz, D.C. Hypocholesterolemic effect of Eubacterium coprostanoligenes ATCC 51222 in rabbits. Lett. Appl. Microbiol. 1995, 20, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Telle-Hansen, V.H.; Gaundal, L.; Bastani, N.; Rud, I.; Byfuglien, M.G.; Gjøvaag, T.; Retterstøl, K.; Holven, K.B.; Ulven, S.M.; Myhrstad, M.C.W. Replacing saturated fatty acids with polyunsaturated fatty acids increases the abundance of Lachnospiraceae and is associated with reduced total cholesterol levels-a randomized controlled trial in healthy individuals. Lipids Health Dis. 2022, 21, 92. [Google Scholar] [CrossRef] [PubMed]
- Udayappan, S.; Manneras-Holm, L.; Chaplin-Scott, A.; Belzer, C.; Herrema, H.; Dallinga-Thie, G.M.; Duncan, S.H.; Stroes, E.S.G.; Groen, A.K.; Flint, H.J.; et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes 2016, 2, 16009. [Google Scholar] [CrossRef]
- Bonnechère, B.; Amin, N.; van Duijn, C. What Are the Key Gut Microbiota Involved in Neurological Diseases? A Systematic Review. Int. J. Mol. Sci. 2022, 23, 13665. [Google Scholar] [CrossRef]
- Saulnier, D.M.; Riehle, K.; Mistretta, T.A.; Diaz, M.A.; Mandal, D.; Raza, S.; Weidler, E.M.; Qin, X.; Coarfa, C.; Milosavljevic, A.; et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011, 141, 1782–1791. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Milagro, F.I.; Guruceaga, E.; Cuervo, M.; Goni, L.; García-Granero, M.; Martinez, J.A.; Riezu-Boj, J.I. A weight-loss model based on baseline microbiota and genetic scores for selection of dietary treatments in overweight and obese population. Clin. Nutr. 2022, 41, 1712–1723. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.E.; Min, S.W. Amelioration of colitis in mice by Leuconostoc lactis EJ-1 by M1 to M2 macrophage polarization. Microbiol. Immunol. 2020, 64, 133–142. [Google Scholar] [CrossRef]
- Muthusamy, K.; Han, H.S.; Soundharrajan, I.; Jung, J.S.; Valan Arasu, M.; Choi, K.C. A Novel Strain of Probiotic Leuconostoc citreum Inhibits Infection-Causing Bacterial Pathogens. Microorganisms 2023, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.C.; Gao, X.; Xu, J.; Holder, M.; Petrosino, J.; Kumar, R.; Liu, W.; Höök, M.; Mackenzie, C.; Hillhouse, A.; et al. A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog. 2021, 17, e1009182. [Google Scholar]
- Van Samkar, A.; Brouwer, M.C.; Pannekoek, Y.; van der Ende, A.; van de Beek, D. Streptococcus gallolyticus meningitis in adults: Report of five cases and review of the literature. Clin. Microbiol. Infect. 2015, 21, 1077–1083. [Google Scholar] [CrossRef]
- Xie, H.; Chen, J.; Chen, Q.; Zhao, Y.; Liu, J.; Sun, J.; Hu, X. The Diagnostic Value of Gut Microbiota Analysis for Post-Stroke Sleep Disorders. Diagnostics 2023, 13, 2970. [Google Scholar] [CrossRef] [PubMed]
- LiYa, L.; XinSheng, Z.; Xiang, H.; Zhao, L.; Lu, L.; XiuMing, L.; Ye, L.; Jing, C.; KeMing, Z.; HongChi, W.; et al. A cross-sectional survey study on the correlation analysis of nutritional status and intestinal flora in patients with esophageal cancer. Front. Nutr. 2024, 11, 1424039. [Google Scholar] [CrossRef]
- Mao, R.; Yu, Q.; Li, J. The causal relationship between gut microbiota and inflammatory dermatoses: A Mendelian randomization study. Front. Immunol. 2023, 14, 1231848. [Google Scholar] [CrossRef]
- Shi, S.; Zhang, Q.; Sang, Y.; Ge, S.; Wang, Q.; Wang, R.; He, J. Probiotic Bifidobacterium longum BB68S Improves Cognitive Functions in Healthy Older Adults: A Randomized, Double-Blind, Placebo-Controlled Trial. Nutrients 2022, 15, 51. [Google Scholar] [CrossRef]
- Ortega, M.A.; Álvarez-Mon, M.A.; García-Montero, C.; Fraile-Martínez, Ó.; Monserrat, J.; Martinez-Rozas, L.; Rodríguez-Jiménez, R.; Álvarez-Mon, M.; Lahera, G. Microbiota-gut-brain axis mechanisms in the complex network of bipolar disorders: Potential clinical implications and translational opportunities. Mol. Psychiatry 2023, 28, 2645–2673. [Google Scholar] [CrossRef]
- Allaart, J.G.; van Asten, A.J.; Gröne, A. Predisposing factors and prevention of Clostridium perfringens-associated enteritis. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 449–464. [Google Scholar] [CrossRef]
- Choi, M.G.; Jung, H.K. Health related quality of life in functional gastrointestinal disorders in Asia. J. Neurogastroenterol. Motil. 2011, 17, 245–251. [Google Scholar] [CrossRef]
- Sundas, A.; Sampath, H.; Lamtha, S.C.; Soohinda, G.; Dutta, S. Psychosocial quality-of-life correlates in functional gastrointestinal disorders. Rev. Gastroenterol. Méx. 2024, 89, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.Q.; Shen, L.L.; Li, W.W.; Fu, X.; Zeng, F.; Gui, L.; Lü, Y.; Cai, M.; Zhu, C.; Tan, Y.L.; et al. Gut Microbiota is Altered in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 2018, 63, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Quan, M.; Zhao, H.; Jia, J. Gut Microbiota Changes and Their Correlation with Cognitive and Neuropsychiatric Symptoms in Alzheimer’s Disease. J. Alzheimers Dis. 2021, 81, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, T.; Liu, X.; Wang, Q.; Zhang, H. Causal effect between gut microbiota and gastroesophageal reflux disease: A bidirectional two-sample Mendelian randomization study. Eur. J. Gastroenterol. Hepatol. 2024, 36, 875–883. [Google Scholar] [CrossRef]
- Miyamoto, J.; Shimizu, H.; Hisa, K.; Matsuzaki, C.; Inuki, S.; Ando, Y.; Nishida, A.; Izumi, A.; Yamano, M.; Ushiroda, C.; et al. Host metabolic benefits of prebiotic exopolysaccharides produced by Leuconostoc mesenteroides. Gut Microbes 2023, 15, 2161271. [Google Scholar] [CrossRef]
- Peng, X.; Yi, X.; Deng, N.; Liu, J.; Tan, Z.; Cai, Y. Zhishi Daozhi decoction alleviates constipation induced by a high-fat and high-protein diet via regulating intestinal mucosal microbiota and oxidative stress. Front. Microbiol. 2023, 14, 1214577. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.; Thijs, C.; Mommers, M.; Endika, M.; Klostermann, C.; Schols, H.; Smidt, H.; Nauta, A.; Arts, I.; Penders, J. Why do babies cry? Exploring the role of the gut microbiota in infantile colic, constipation, and cramps in the KOALA birth cohort study. Gut Microbes 2025, 17, 2485326. [Google Scholar] [CrossRef]
- Bhattacharjee, D.; Flores, C.; Woelfel-Monsivais, C.; Seekatz, A.M. Diversity and Prevalence of Clostridium innocuum in the Human Gut Microbiota. mSphere 2023, 8, e0056922. [Google Scholar] [CrossRef]
- Al-Fakhrany, O.M.; Elekhnawy, E. Next-generation probiotics: The upcoming biotherapeutics. Mol. Biol. Rep. 2024, 51, 505. [Google Scholar] [CrossRef]
- Zou, B.; Liu, S.; Dong, C.; Shen, H.; Lv, Y.; He, J.; Li, X.; Ruan, M.; Huang, Z.; Shu, S. Fecal microbiota transplantation restores gut microbiota diversity in children with active Crohn’s disease: A prospective trial. J. Transl. Med. 2025, 23, 288. [Google Scholar] [CrossRef]
- Gotoh, Y.; Nanba, F.; Shioya, N.; Sugimura, H.; Suzuki, T. A dose-finding study for a supplement containing Lactococcus lactis subsp. cremoris FC in healthy adults with mild constipation. Biosci. Microbiota Food Health 2020, 39, 19–22. [Google Scholar] [CrossRef]
- Duncan, S.H.; Richardson, A.J.; Kaul, P.; Holmes, R.P.; Allison, M.J.; Stewart, C.S. Oxalobacter formigenes and its potential role in human health. Appl. Environ. Microbiol. 2002, 68, 3841–3847. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lai, J.; Zhang, P.; Ding, J.; Jiang, J.; Liu, C.; Huang, H.; Zhen, H.; Xi, C.; Sun, Y.; et al. Multi-omics analyses of serum metabolome, gut microbiome and brain function reveal dysregulated microbiota-gut-brain axis in bipolar depression. Mol. Psychiatry 2022, 27, 4123–4135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Jiang, S.J.; Cao, J.J.; Xu, Y.; Wang, X.Y.; Li, R.; Miao, Z.W. Investigating the causal relationship between gut microbiota and gastroenteropancreatic neuroendocrine neoplasms: A bidirectional Mendelian randomization study. Front. Microbiol. 2024, 15, 1420167. [Google Scholar] [CrossRef]
- Kim, W.J.; Hyun, J.H.; Lee, N.K.; Paik, H.D. Protective Effects of a Novel Lactobacillus brevis Strain with Probiotic Characteristics against Staphylococcus aureus Lipoteichoic Acid-Induced Intestinal Inflammatory Response. J. Microbiol. Biotechnol. 2022, 32, 205–211. [Google Scholar] [CrossRef]
- Acosta-Rodríguez-Bueno, C.P.; Abreu y Abreu, A.T.; Guarner, F.; Guno, M.J.V.; Pehlivanoğlu, E.; Perez, M., III. Bacillus clausii for Gastrointestinal Disorders: A Narrative Literature Review. Adv. Ther. 2020, 39, 4854–4874. [Google Scholar] [CrossRef] [PubMed]
- Novau-Ferré, N.; Papandreou, C.; Rojo-Marticella, M.; Canals-Sans, J.; Bulló, M. Gut microbiome differences in children with Attention Deficit Hyperactivity Disorder and Autism Spectrum Disorder and effects of probiotic supplementation: A randomized controlled trial. Res. Dev. Disabil. 2025, 161, 105003. [Google Scholar] [CrossRef]
- He, J.; Gong, X.; Hu, B.; Lin, L.; Lin, X.; Gong, W.; Zhang, B.; Cao, M.; Xu, Y.; Xia, R.; et al. Altered Gut Microbiota and Short-chain Fatty Acids in Chinese Children with Constipated Autism Spectrum Disorder. Sci. Rep. 2023, 13, 19103. [Google Scholar] [CrossRef]
- Kok, C.R.; Hutkins, R. Yogurt, and other fermented foods as sources of health-promoting bacteria. Nutr Rev. 2018, 76 (Suppl. 1), 4–15. [Google Scholar] [CrossRef] [PubMed]
- Olorocisimo, J.P.; Diaz, L.A.; Co, D.E.; Carag, H.M.; Ibana, J.A.; Velarde, M.C. Lactobacillus delbrueckii reduces anxiety-like behavior in zebrafish through a gut microbiome—Brain crosstalk. Neuropharmacology 2023, 225, 109401. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, Y.; Shen, Y.; Li, Q.; Lan, J.; Wu, Y.; Zhang, R.; Cao, G.; Yang, C. Effects of Bacillus subtilis and Bacillus licheniformis on growth performance, immunity, short chain fatty acid production, antioxidant capacity, and cecal microflora in broilers. Poult. Sci. 2021, 100, 101358. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, F.; Fan, B.; Wang, R.; Ren, J.; Jia, S.; Wang, L.; Chen, Z.; Liu, X.A. The Molecular Gut-Brain Axis in Early Brain Development. Int. J. Mol. Sci. 2022, 23, 15389. [Google Scholar] [CrossRef]
- Marangelo, C.; Vernocchi, P.; Del Chierico, F.; Scanu, M.; Marsiglia, R.; Petrolo, E.; Fucà, E.; Guerrera, S.; Valeri, G.; Vicari, S.; et al. Stratification of Gut Microbiota Profiling Based on Autism Neuropsychological Assessments. Microorganisms 2024, 12, 2041. [Google Scholar] [CrossRef]
- Takewaki, D.; Kiguchi, Y.; Masuoka, H.; Manu, M.S.; Raveney, B.J.E.; Narushima, S.; Kurokawa, R.; Ogata, Y.; Kimura, Y.; Sato, N.; et al. Tyzzerella nexilis strains enriched in mobile genetic elements are involved in progressive multiple sclerosis. Cell Rep. 2024, 43, 114785. [Google Scholar] [CrossRef]
- Luo, S.; Zhao, Y.; Zhu, S.; Liu, L.; Cheng, K.; Ye, B.; Han, Y.; Fan, J.; Xia, M. Flavonifractor plautii Protects Against Elevated Arterial Stiffness. Circ. Res. 2023, 132, 167–181. [Google Scholar] [CrossRef]
- iMSMS Consortium. Gut microbiome of multiple sclerosis patients and paired household healthy controls reveal associations with disease risk and course. Cell 2022, 185, 3467–3486.e16. [Google Scholar] [CrossRef]
- Hunter, S.; Flaten, E.; Petersen, C.; Gervain, J.; Werker, J.F.; Trainor, L.J.; Finlay, B.B. Babies, bugs, and brains: How the early microbiome associates with infant brain and behavior development. PLoS ONE 2023, 18, e0288689. [Google Scholar] [CrossRef]
- Feng, Y.; Cui, Y.; Jin, J.; Huang, S.; Wei, J.; Yao, M.; Zhou, D.; Mao, S. The Alterations of Gut Microbiome and Lipid Metabolism in Patients with Spinal Muscular Atrophy. Neurol. Ther. 2023, 12, 961–976. [Google Scholar] [CrossRef]
- Socała, K.; Doboszewska, U.; Szopa, A.; Serefko, A.; Włodarczyk, M.; Zielińska, A.; Poleszak, E.; Fichna, J.; Wlaź, P. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol. Res. 2021, 172, 105840. [Google Scholar] [CrossRef]
- Bojović, K.; Ignjatović, Ð.I.; Soković Bajić, S.; Vojnović Milutinović, D.; Tomić, M.; Golić, N.; Tolinački, M. Gut Microbiota Dysbiosis Associated with Altered Production of Short Chain Fatty Acids in Children with Neurodevelopmental Disorders. Front. Cell Infect. Microbiol. 2020, 10, 223. [Google Scholar] [CrossRef]
- Li, N.; Chen, H.; Cheng, Y.; Xu, F.; Ruan, G.; Ying, S.; Tang, W.; Chen, L.; Chen, M.; Lv, L.; et al. Fecal Microbiota Transplantation Relieves Gastrointestinal and Autism Symptoms by Improving the Gut Microbiota in an Open-Label Study. Front. Cell Infect. Microbiol. 2021, 11, 759435. [Google Scholar]
- Bronzini, M.; Maglione, A.; Rosso, R.; Matta, M.; Masuzzo, F.; Rolla, S.; Clerico, M. Feeding the gut microbiome: Impact on multiple sclerosis. Front. Immunol. 2023, 14, 1176016. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Chen, L.; Zhang, Y.; Zhang, J.; Zhang, H. Bifidobacterium animalis subsp. lactis Probio-M8 alleviates abnormal behavior and regulates gut microbiota in a mouse model suffering from autism. Msystems 2024, 9, e0101323. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Meng, F.-H.; Zhang, M.-Y.; Li, F.-X.; Lei, Y.-X.; Ma, Z.-G.; Li, J.-Q.; Lou, Y.-N.; Chu, Y.-F.; Ma, K.; et al. Gut Lactococcus garvieae promotes protective immunity to foodborne Clostridium perfringens infection. Microbiol. Spectr. 2024, 12, e0402523. [Google Scholar] [CrossRef]
- Messaoudi, S.; Manai, M.; Kergourlay, G.; Prévost, H.; Connil, N.; Chobert, J.M.; Dousset, X. Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiol. 2013, 36, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Andreozzi, V.; Cuoco, S.; Balestrieri, M.; Fierro, F.; Ferrara, N.; Erro, R.; Di Filippo, M.; Barbella, G.; Memoli, M.C.; Silvestri, A.; et al. Synbiotic supplementation may globally improve non-motor symptoms in patients with stable Parkinson’s disease: Results from an open label single-arm study. Sci. Rep. 2024, 14, 23095. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Gong, X.; Ma, Y.; Deng, X.; Li, A.; Li, X.; Kong, X.; Liu, Y.; Liu, X.; Guo, K.; Yang, Y.; et al. Intestinal dysbiosis exacerbates susceptibility to the anti-NMDA receptor encephalitis-like phenotype by changing blood brain barrier permeability and immune homeostasis. Brain Behav. Immun. 2024, 116, 34–51. [Google Scholar] [CrossRef]
- Thompson, R.S.; Gaffney, M.; Hopkins, S.; Kelley, T.; Gonzalez, A.; Bowers, S.J.; Vitaterna, M.H.; Turek, F.W.; Foxx, C.L.; Lowry, C.A.; et al. Ruminiclostridium 5, Parabacteroides distasonis, and bile acid profile are modulated by prebiotic diet and associate with facilitated sleep/clock realignment after chronic disruption of rhythms. Brain Behav. Immun. 2021, 97, 150–166. [Google Scholar] [CrossRef]
- Mohammadi, F.; Green, M.; Tolsdorf, E.; Greffard, K.; Leclercq, M.; Bilodeau, J.F.; Droit, A.; Foster, J.; Bertrand, N.; Rudkowska, I. Industrial and Ruminant Trans-Fatty Acids-Enriched Diets Differentially Modulate the Microbiome and Fecal Metabolites in C57BL/6 Mice. Nutrients 2023, 15, 1433. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Jin, H.; Yulug, B.; Altay, O.; Li, X.; Hanoglu, L.; Cankaya, S.; Coskun, E.; Idil, E.; Nogaylar, R.; et al. Multi-omics analysis reveals the key factors involved in the severity of the Alzheimer’s disease. Alzheimer’s Res. Ther. 2024, 16, 213. [Google Scholar] [CrossRef] [PubMed]
- Langmajerová, M.; Ježková, J.; Kreisinger, J.; Semerád, J.; Titov, I.; Procházková, P.; Cajthaml, T.; Jiřička, V.; Vevera, J.; Roubalová, R. Gut Microbiome in Impulsively Violent Female Convicts. Neuropsychobiology 2025, 84, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mulder, D.; Jakobi, B.; Shi, Y.; Mulders, P.; Kist, J.D.; Collard, R.M.; Vrijsen, J.N.; van Eijndhoven, P.; Tendolkar, I.; Bloemendaal, M.; et al. Gut microbiota composition links to variation in functional domains across psychiatric disorders. Brain Behav. Immun. 2024, 120, 275–287. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, H.; Xu, J.; Guo, X.; Zhao, H.; Chen, Y.; Zhou, Y.; Nie, Y.F. prausnitzii and its supernatant increase SCFAs-producing bacteria to restore gut dysbiosis in TNBS-induced colitis. AMB Express 2021, 11, 33. [Google Scholar] [CrossRef]
- Islam, T.; Xu, B.; Bian, Z. Anti-inflammatory and gut microbiota regulatory effects of ultrasonic degraded polysaccharides from Auricularia auricula-judae in DSS-induced colitis mice. Ultrason. Sonochem. 2025, 117, 107339. [Google Scholar] [CrossRef]
- Kim, H.S.; Oh, S.J.; Kim, B.K.; Kim, J.E.; Kim, B.-H.; Park, Y.-K.; Yang, B.-G.; Lee, J.-E.; Bae, J.-W.; Lee, C.K. Dysbiotic signatures and diagnostic potential of gut microbial markers for inflammatory bowel disease in Korean population. Sci. Rep. 2024, 14, 23701. [Google Scholar] [CrossRef] [PubMed]
- Stubbendieck, R.M.; Hurst, J.H.; Kelly, M.S. Dolosigranulum pigrum: A promising nasal probiotic candidate. PLoS Pathog. 2024, 20, e1011955. [Google Scholar] [CrossRef] [PubMed]
- Han, H.S.; Soundharrajan, I.; Valan Arasu, M.; Kim, D.; Choi, K.C. Leuconostoc Citreum Inhibits Adipogenesis and Lipogenesis by Inhibiting p38 MAPK/Erk 44/42 and Stimulating AMPKα Signaling Pathways. Int. J. Mol. Sci. 2023, 24, 7367. [Google Scholar] [CrossRef]
- Korona-Glowniak, I.; Skawinska-Bednarczyk, A.; Wrobel, R.; Pietrak, J.; Tkacz-Ciebiera, I.; Maslanko-Switala, M.; Krawczyk, D.; Bakiera, A.; Borek, A.; Malm, A.; et al. Streptococcus sobrinus as a Predominant Oral Bacteria Related to the Occurrence of Dental Caries in Polish Children at 12 Years Old. Int. J. Env. Res. Public Health 2022, 19, 15005. [Google Scholar] [CrossRef]
- Mochon, A.B.; Sussland, D.; Saubolle, M.A. Aerobic Actinomycetes of Clinical Significance. Microbiol Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Corvec, S. Clinical and Biological Features of Cutibacterium (Formerly Propionibacterium) avidum, an Underrecognized Microorganism. Clin. Microbiol. Rev. 2018, 31, e00064-17. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Lo, S.C.; Hung, G.C.; Li, B.; Lei, H.; Li, T.; Nagamine, K.; Tsai, S.; Zucker, M.J.; Olesnicky, L. Mixed group of Rhizobiales microbes in lung and blood of a patient with fatal pulmonary illness. Int. J. Clin. Exp. Pathol. 2015, 8, 13834–13852. [Google Scholar] [PubMed]
- Chen, Y.; Tang, S. Gut microbiota and immune mediation: A Mendelian randomization study on granulomatosis with polyangiitis. Front. Immunol. 2023, 14, 1296016. [Google Scholar] [CrossRef]
- Babar, S.; Liu, E.; Kaur, S.; Hussain, J.; Danaher, P.J.; Anstead, G.M. Pseudopropionibacterium propionicum as a Cause of Empyema; A Diagnosis with Next-Generation Sequencing. Pathogens 2024, 13, 165. [Google Scholar] [CrossRef]
- Speirs, G.; Warren, R.E.; Rampling, A. Clostridium tertium septicemia in patients with neutropenia. J. Infect. Dis. 1988, 158, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Pappas, G.; Liberopoulos, E.; Tsianos, E.; Elisaf, M. Enterococcus casseliflavus bacteremia. Case report and literature review. J. Infect. 2004, 48, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Archambaud, C.; Nunez, N.; da Silva, R.A.G.; Kline, K.A.; Serror, P. Enterococcus faecalis: An overlooked cell invader. Microbiol. Mol. Biol. Rev. 2024, 88, e0006924. [Google Scholar] [CrossRef] [PubMed]
- Dunalska, A.; Saramak, K.; Szejko, N. The Role of Gut Microbiome in the Pathogenesis of Multiple Sclerosis and Related Disorders. Cells 2023, 12, 1760. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, L.; Li, Q.; Wang, P.; Wang, H.; Shi, H.; Lu, W.; Zhang, Y. Prenatal PFAS exposure, gut microbiota dysbiosis, and neurobehavioral development in childhood. J. Hazard. Mater. 2024, 469, 133920. [Google Scholar] [CrossRef]
- Suganya, K.; Koo, B.S. Gut-Brain Axis: Role of Gut Microbiota on Neurological Disorders and How Probiotics/Prebiotics Beneficially Modulate Microbial and Immune Pathways to Improve Brain Functions. Int. J. Mol. Sci. 2020, 21, 7551. [Google Scholar] [CrossRef]
- Jones, J.; Reinke, S.N.; Mousavi-Derazmahalleh, M.; Palmer, D.J.; Christophersen, C.T. Changes to the Gut Microbiome in Young Children Showing Early Behavioral Signs of Autism. Front. Microbiol. 2022, 13, 905901. [Google Scholar] [CrossRef]
- Chesdachai, S.; Yetmar, Z.A.; Tabaja, H.; Comba, I.Y.; Go, J.R.; Challener, D.W.; Misra, A.; Abu Saleh, O.M. Contemporary experience of Abiotrophia, Granulicatella and Gemella bacteremia. J. Infect. 2022, 84, 511–517. [Google Scholar] [CrossRef]
- Ryan, P.M.; Shin, C.P. Native joint infections caused by Parvimonas micra. Anaerobe 2021, 71, 102412. [Google Scholar] [CrossRef]
- Li, J.; Xu, J.; Guo, X.; Xu, H.; Huang, C.; Nie, Y.; Zhou, Y. Odoribacter splanchnicus-A Next-Generation Probiotic Candidate. Microorganisms 2025, 13, 815. [Google Scholar] [CrossRef]
- Hadjiyannis, Y.; Ali, S.; Wang, Q.; Crawford, E.C.; Scholz, S.; Waltz, P.K.; Alissa, F.; Ayers, M.H.; Reyes-Múgica, M.; Salgado, C.M. The Spectrum of Sarcina Colonization in the Gastrointestinal Tract of Pediatric and Adolescent Patients. Int. J. Surg. Pathol. 2025, 33, 65–75. [Google Scholar] [CrossRef]
- Liu, S.; Men, X.; Guo, Y.; Cai, W.; Wu, R.; Gao, R.; Zhong, W.; Guo, H.; Ruan, H.; Chou, S.; et al. Gut microbes exacerbate systemic inflammation and behavior disorders in neurologic disease CADASIL. Microbiome 2023, 11, 202. [Google Scholar] [CrossRef]
- Boucher, M.B.; Bedotto, M.; Couderc, C.; Gomez, C.; Reynaud-Gaubert, M.; Drancourt, M. Haemophilus pittmaniae respiratory infection in a patient with siderosis: A case report. J. Med. Case Rep. 2012, 6, 120. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.H.; Yeom, M.J.; Ahn, S.; Oh, J.Y.; Ji, S.; Kim, T.H.; Park, H.J. Acupuncture inhibits neuroinflammation and gut microbial dysbiosis in a mouse model of Parkinson’s disease. Brain Behav. Immun. 2020, 89, 641–655. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, D.; Wu, D.; Gao, X.; Shao, F.; Zhao, M.; Wang, J.; Ma, J.; Wang, W.; Qin, X.; et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe 2023, 31, 418–432.e8. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Mahurkar-Joshi, S.; Liu, C.; Jaffe, N.; Labus, J.S.; Dong, T.S.; Gupta, A.; Patel, S.; Mayer, E.A.; Chang, L. The Association Between a Mediterranean Diet and Symptoms of Irritable Bowel Syndrome. Clin. Gastroenterol. Hepatol. 2024, 22, 164–172.e6. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, Z.; Ling, Y.; Teng, W.; Cui, J.; Yan, Z.; Hou, X.; Cen, W.; Long, N.; Li, W.; et al. Causal effect of air pollution on the risk of brain health and potential mediation by gut microbiota. Ecotoxicol. Environ. Saf. 2024, 285, 117080. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, S.; Wei, F.; Hu, J. The effects of Qingchang Ligan formula on hepatic encephalopathy in mouse model: Results from gut microbiome-metabolomics analysis. Front. Cell Infect. Microbiol. 2024, 14, 1381209. [Google Scholar] [CrossRef]
- Raygoza Garay, J.A.; Turpin, W.; Lee, S.H.; Smith, M.I.; Goethel, A.; Griffiths, A.M.; Moayyedi, P.; Espin-Garcia, O.; Abreu, M.; Aumais, G.L.; et al. Gut Microbiome Composition Is Associated with Future Onset of Crohn’s Disease in Healthy First-Degree Relatives. Gastroenterology 2023, 165, 670–681. [Google Scholar] [CrossRef] [PubMed]
- Kenna, J.E.; Chua, E.G.; Bakeberg, M.; Tay, A.; McGregor, S.; Gorecki, A.; Horne, M.; Marshall, B.; Mastaglia, F.L.; Anderton, R.S. Changes in the Gut Microbiome and Predicted Functional Metabolic Effects in an Australian Parkinson’s Disease Cohort. Front. Neurosci. 2021, 15, 756951. [Google Scholar] [CrossRef]
- Bai, J.; Bruner, D.W.; Fedirko, V.; Beitler, J.J.; Zhou, C.; Gu, J.; Zhao, H.; Lin, I.H.; Chico, C.E.; Higgins, K.A.; et al. Gut Microbiome Associated with the Psychoneurological Symptom Cluster in Patients with Head and Neck Cancers. Cancers 2020, 12, 2531. [Google Scholar] [CrossRef]
- Fregatto, L.F.; Costa, I.B.; De Bortoli Teixeira, D.; Duarte, J.C.M.; Mascarin, A.M.N.; da Silveira, S.B., Jr.; Serva, B.E.B.M.; da Silva, R.G.; Junior, F.A.; Cola, P.C. Oral hygiene and oral microbiota in children and young people with neurological impairment and oropharyngeal dysphagia. Sci. Rep. 2021, 11, 18090. [Google Scholar] [CrossRef]
- Wei, L.; Singh, R.; Ro, S.; Ghoshal, U.C. Gut microbiota dysbiosis in functional gastrointestinal disorders: Underpinning the symptoms and pathophysiology. JGH Open 2021, 5, 976–987. [Google Scholar] [CrossRef]
- Chen, F.; Guo, Z.; Chen, Y.; Li, S.; Chen, P. Non-alcoholic fatty liver disease enhances the beneficial effect of renal denervation on gut microbiota aberrations in rats with heart failure. BMC Microbiol. 2025, 25, 311. [Google Scholar] [CrossRef]
- Liu, Z.C.; Wu, D.; Qu, A.N.; Wang, L.L. Diversity and functional prediction of gut microbiota in children with autism spectrum disorder. Chin. J. Contemp. Pediatr. 2022, 24, 1356–1364. [Google Scholar]
- Krzyściak, W.; Jurczak, A.; Kościelniak, D.; Bystrowska, B.; Skalniak, A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Baranova, A.; Cao, H.; Zhang, F. Gut microbiome and major depressive disorder: Insights from two-sample Mendelian randomization. BMC Psychiatry 2024, 24, 493. [Google Scholar] [CrossRef] [PubMed]
- Kushak, R.I.; Winter, H.S.; Buie, T.M.; Cox, S.B.; Phillips, C.D.; Ward, N.L. Analysis of the Duodenal Microbiome in Autistic Individuals: Association with Carbohydrate Digestion. J. Pediatr. Gastroenterol. Nutr. 2017, 64, e110–e116. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Wu, H.; Wu, S.D.; Lu, N.; Wang, Y.T.; Liu, H.N.; Dong, L.; Liu, T.T.; Shen, X.Z. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J. Gastroenterol. Hepatol. 2018, 33, 1844–1852. [Google Scholar] [CrossRef]
- Chu, Y.W.; Wong, C.H.; Chu, M.Y.; Cheung, C.P.; Cheung, T.K.; Tse, C.; Luk, W.K.; Lo, J.Y. Varibaculum cambriense infections in Hong Kong, China, 2006. Emerg. Infect. Dis. 2009, 15, 1137–1139. [Google Scholar] [CrossRef]
- Eltwisy, H.O.; Twisy, H.O.; Hafez, M.H.; Sayed, I.M.; El-Mokhtar, M.A. Clinical Infections, Antibiotic Resistance, and Pathogenesis of Staphylococcus haemolyticus. Microorganisms 2022, 10, 1130. [Google Scholar] [CrossRef]
- Al-Akel, F.C.; Chiperi, L.E.; Eszter, V.K.; Bacârea, A. Streptococcus salivarius Role as a Probiotic in Children’s Health and Disease Prophylaxis—A Systematic Review. Life 2024, 14, 1613. [Google Scholar] [CrossRef]
- Ojha, S.; Patil, N.; Jain, M.; Kole, C.; Kaushik, P. Probiotics for Neurodegenerative Diseases: A Systemic Review. Microorganisms 2023, 11, 1083. [Google Scholar] [CrossRef]
- Deutschmann, M.W.; Livingstone, D.; Cho, J.J.; Vanderkooi, O.G.; Brookes, J.T. The significance of Streptococcus anginosus group in intracranial complications of pediatric rhinosinusitis. JAMA Otolaryngol. Head Neck Surg. 2013, 139, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L.; et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 12015. [Google Scholar] [CrossRef] [PubMed]
- Bor, B.; Bedree, J.K.; Shi, W.; McLean, J.S.; He, X. Saccharibacteria (TM7) in the Human Oral Microbiome. J. Dent. Res. 2019, 98, 500–509. [Google Scholar] [CrossRef] [PubMed]


























| Total Number of Participants: n = 45 | ||
|---|---|---|
| Sex | Female | n = 7 |
| Male | n = 38 | |
| Age (distribution: 7.9 ± 3.35) | 2–6 years | n = 12 |
| 6–14 years | n = 28 | |
| 15–18 years | n = 5 | |
| Diagnosis according to ICD-10 | Childhood autism | n = 30 (67%) |
| Other diagnosis | n = 15 (33%) | |
| Mode of delivery | Vaginal delivery | n = 31 (69%) |
| Cesarean section | n = 14 (31%) | |
| Gastrointestinal symptoms | The presence of any FGID | n = 18 (40%) |
| Functional constipation | n = 8 (18%) | |
| Functional diarrhea | n = 9 (20%) | |
| Functional bloating | n = 7 (16%) | |
| Diet | Type of diet (macroelements) | Protein-based: n = 31 (6%) |
| Carbohydrate-based: n = 14 (31%) | ||
| Type of diet (calorie intake) | Low-calorie diet: n = 23 (51%) | |
| High-calorie diet: n = 12 (49%) | ||
| Food selectivity | n = 16 (36%) | |
| Mean daily protein consumption (g/day) | 90.05 ± 31.06 | |
| Mean protein intake normalized to body weight (g/kg BW) | 3.81 ± 1.71 | |
| Mean daily carbohydrate consumption (g/day) | 251.02 ± 62.66 | |
| Percentage of energy from carbohydrates (%) | 51.41 ± 9.31 | |
| Mean proportion of simple carbohydrates in total carbohydrate intake (%) | 41.31 ± 7.82 | |
| Mean daily fat consumption (g/day) | 79.51 ± 31.32 | |
| Mean fat intake normalized to body weight (g/kg BW) | 3.3 ± 1.7 | |
| Fraction of unsaturated fatty acids within total fat intake (%) | 59.89 ± 5.30 |
| Feature | Bacteria | p-Value ≤ |
|---|---|---|
| Class | ||
| high-calorie diet | Saccharimonadia | 0.05 |
| no bloating | Bacilli | 0.05 |
| communication—expressive skills (adequate > low) | Clostridia | 0.05 |
| communication—expressive skills (high > low) | Negativicutes | 0.05 |
| communication—writing skills (low > high) | Coriobacteria | 0.05 |
| communication—total (high > adequate) | Bacilli | 0.05 |
| communication—total (high > low) | Alphaproteobacteria | 0.05 |
| Gammaproteobacteria | 0.05 | |
| daily living skills—domestic skills (adequate > low) | Negativicutes | 0.05 |
| daily living skills—domestic skills (low > high) | Gammaproteobacteria | 0.05 |
| daily living skills—community skills (adequate > low) | Negativicutes | 0.01 |
| daily living skills—total (high > low) | Gammaproteobacteria | 0.05 |
| daily living skills—total (high > adequate) | Gammaproteobacteria | 0.05 |
| socialization—coping skills (adequate > low) | Bacilli | 0.05 |
| Saccharimonadia | 0.05 | |
| socialization—coping skills (high > low) | Saccharimonadia | 0.05 |
| using large muscles (high > low) | Actinobacteria | 0.05 |
| using small muscles (adequate > low) | Negativicutes | 0.05 |
| Order | ||
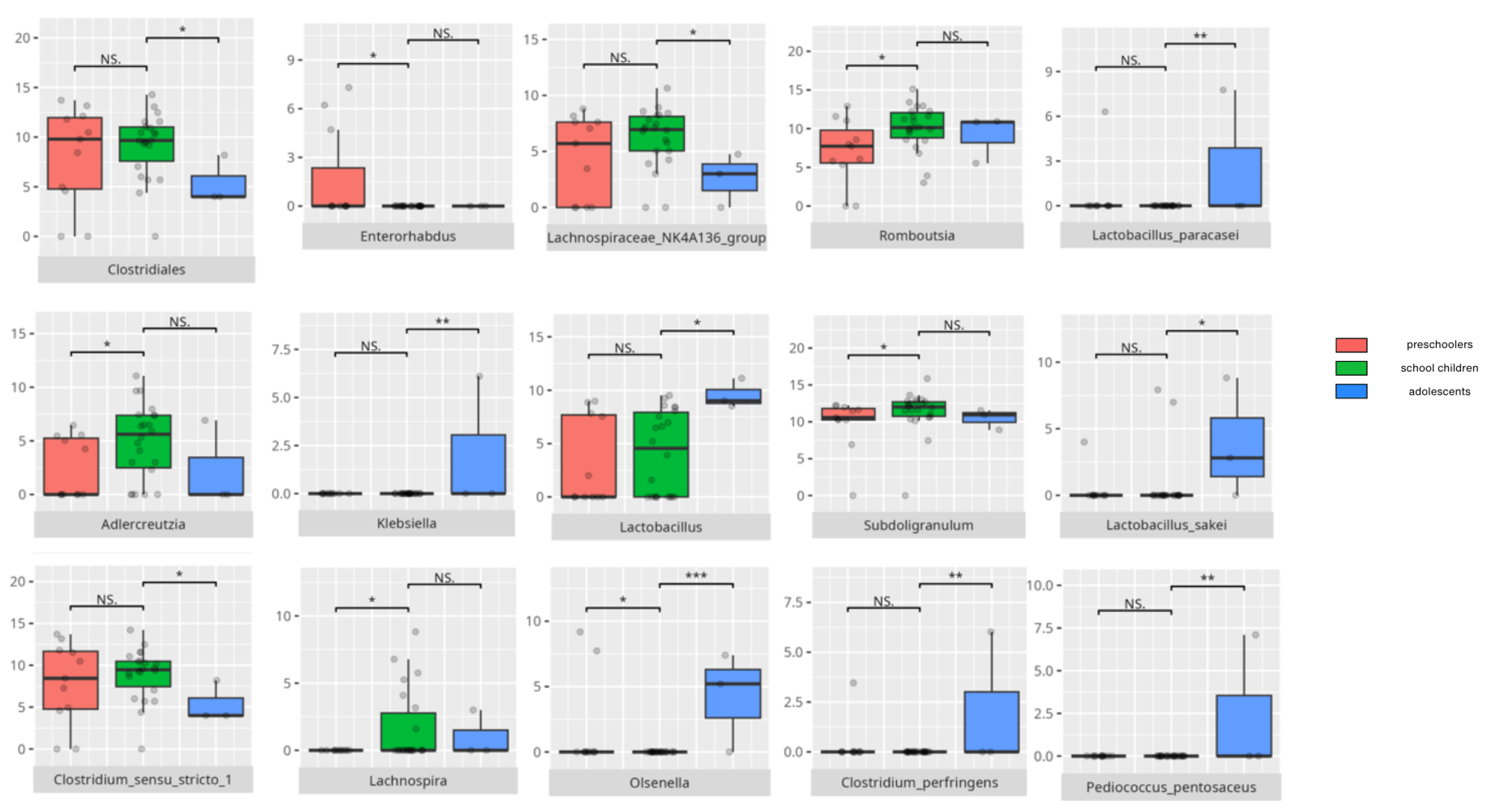
| age (school children > adolescents) | Clostridiales | 0.05 |
| sex: female | Rhodospirillales | 0.05 |
| sex: male | Peptococcales | 0.05 |
| Caesarean’s section | Clostridiales | 0.05 |
| high-calorie diet | Saccharimonadales | 0.05 |
| Staphylococcales | 0.01 | |
| no food selectivity | Peptostreptococcales/Tissierellales | 0.05 |
| communication—expressive skills (adequate > low) | Clostridiales | 0.001 |
| Oscillospirales | 0.05 | |
| Peptostreptococcales/Tissierellales | 0.05 | |
| communication—expressive skills (high > low) | Clostridia | 0.05 |
| Peptostreptococcales/Tissierellales | 0.01 | |
| Veillonellales/Selenomonadales | 0.05 | |
| communication—receptive skills (high > low) | Clostridia | 0.01 |
| Peptostreptococcales/Tissierellales | 0.05 | |
| communication—receptive skills (high > adequate) | Veillonellales/Selenomonadales | 0.05 |
| communication—writing skills (low > high) | Coriobacteriales | 0.05 |
| communication—writing skills (low > adequate) | Lachnospirales | 0.05 |
| communication—writing skills (adequate > low) | Pseudomonadales | 0.001 |
| Rhizobiales | 0.001 | |
| communication—total (low > adequate) | Actinomycetales | 0.05 |
| Lactobacillales | 0.05 | |
| communication—total (low > high) | Clostridia | 0.05 |
| Propionibacteriales | 0.05 | |
| Pseudomonadales | 0.05 | |
| Rhizobiales | 0.05 | |
| communication—total (adequate > low) | Acidaminococcales | 0.05 |
| Clostridia | 0.05 | |
| communication—total (adequate > high) | Acidaminococcales | 0.05 |
| communication—total (high > low) | Peptostreptococcales/Tissierellales | 0.05 |
| communication—total (high > adequate) | Lactobacillales | 0.05 |
| daily living skills—personal skills (adequate > low) | Clostridiales | 0.01 |
| daily living skills—personal skills (high > low) | Enterobacterales | 0.05 |
| Peptostreptococcales/Tissierellales | 0.01 | |
| Veillonellales/Selenomonadales | 0.05 | |
| daily living skills—domestic skills (low > adequate) | Clostridia | 0.05 |
| Clostridia | Lachnospirales | 0.05 |
| Pseudomonadales | 0.05 | |
| Rhizobiales | 0.05 | |
| daily living skills—domestic skills (adequate > low) | Clostridiales | 0.01 |
| Veillonellales/Selenomonadales | 0.05 | |
| daily living skills—domestic skills (high > low) | Enterobacterales | 0.05 |
| Peptostreptococcales/Tissierellales | 0.05 | |
| Veillonellales/Selenomonadales | 0.05 | |
| daily living skills—community skills (adequate > low) | Clostridia | 0.01 |
| Veillonellales/Selenomonadales | 0.05 | |
| daily living skills—community skills (adequate > high) | Pasteurellales | 0.05 |
| daily living skills—community skills (high > low) | Enterobacterales | 0.05 |
| daily living skills—total (low > high) | Lachnospirales | 0.05 |
| Pseudomonadales | 0.05 | |
| Rhizobiales | 0.05 | |
| daily living skills—total (adequate > low) | Clostridia | 0.05 |
| Clostridiales | 0.05 | |
| daily living skills—total (high > low) | Enterobacterales | 0.05 |
| Peptostreptococcales/Tissierellales | 0.05 | |
| Veillonellales/Selenomonadales | 0.05 | |
| socialization—interpersonal skills (low > adequate) | Clostridia | 0.05 |
| socialization—interpersonal skills (adequate > low) | Veillonellales/Selenomonadales | 0.05 |
| socialization—interpersonal skills (adequate > high) | Monoglobales | 0.05 |
| socialization—interpersonal skills (high > low) | Veillonellales/Selenomonadales | 0.05 |
| socialization—play and leisure skills (adequate > low) | Clostridia | 0.05 |
| Veillonellales/Selenomonadales | 0.05 | |
| socialization—play and leisure skills (high > low) | Pseudomonadales | 0.01 |
| Rhizobiales | 0.01 | |
| socialization—coping skills (adequate > low) | Actinomycetales | 0.05 |
| Saccharimonadales | 0.05 | |
| socialization—coping skills (high > low) | Burkholderiales | 0.05 |
| Clostridia | 0.05 | |
| Propionibacteriales | 0.001 | |
| Saccharimonadales | 0.05 | |
| socialization—coping skills (high > adequate) | Burkholderiales | 0.05 |
| socialization—total (low > adequate) | Clostridia | 0.05 |
| socialization—total (low > high) | Desulfovibrionales | 0.01 |
| socialization—total (adequate > low) | Enterobacterales | 0.05 |
| Veillonellales/Selenomonadales | 0.01 | |
| socialization—total (adequate > high) | Desulfovibrionales | 0.01 |
| using large muscles (high > low) | Bifidobacteriales | 0.05 |
| using small muscles (adequate > low) | Clostridiales | 0.01 |
| Peptostreptococcales/Tissierellales | 0.05 | |
| using small muscles (adequate > high) | Monoglobales | 0.05 |
| using small muscles (high > low) | Clostridia | 0.05 |
| Clostridiales | 0.01 | |
| Peptostreptococcales/Tissierellales | 0.05 | |
| motor skills—total (adequate > low) | Monoglobales | 0.05 |
| motor skills—total (high > low) | Clostridiales | 0.05 |
| motor skills—total (high > adequate) | Clostridiales | 0.01 |
| Genus | ||
| age (preschoolers > school children) | Enterorhabdus | 0.05 |
| age (school children > preschoolers) | Adlercreutzia | 0.05 |
| Olsenella | 0.05 | |
| Lachnospira | 0.05 | |
| Subdoligranulum | 0.05 | |
| Romboutsia | 0.05 | |
| age (school children > adolescents) | Clostridium sensu stricte | 0.05 |
| Lachnospiraceae NK4A136 | 0.05 | |
| age (adolescents > school children) | Klebsiella | 0.01 |
| Lactobacillus | 0.05 | |
| Olsenella | 0.001 | |
| diagnosis: childhood autism | Ruminococcus torques | 0.01 |
| GCA.900066575 | 0.05 | |
| diagnosis: other | Enterorhabdus | 0.05 |
| sex: female | Eubacterium ruminantium | 0.05 |
| Coprobacillus | 0.05 | |
| Odoribacter | 0.05 | |
| UCG010 | 0.05 | |
| sex: male | Ruminococcus gravus | 0.05 |
| Lachnospiraceae FCS020 | 0.05 | |
| Lautropia | 0.05 | |
| Peptococcus | 0.05 | |
| Marvinbryantia | 0.05 | |
| vaginal delivery | Faecalitalea | 0.05 |
| Caesarian’s section | Eubacterium eligens | 0.05 |
| F0332 | 0.05 | |
| Sellimonas | 0.05 | |
| Clostridium sensu stricte | 0.05 | |
| high protein diet | Eubacterium brachy | 0.05 |
| low-calorie diet | Weissella | 0.05 |
| high-calorie diet | Anaerostipes | 0.05 |
| Blautia | 0.05 | |
| Slackia | 0.05 | |
| Staphylococcus | 0.05 | |
| TM7x | 0.05 | |
| no food selectivity | Clostridium sensu stricte | 0.05 |
| Eubacterium coprostanoligenes | 0.05 | |
| Barnesiella | 0.05 | |
| Coprococcus | 0.05 | |
| DTU089 | 0.01 | |
| Lachnospiraceae NK4A136 | 0.05 | |
| Turicibacter | 0.05 | |
| FGID | Enterobacter | 0.05 |
| F0332 | 0.01 | |
| Holdemania | 0.05 | |
| Oscillibacter | 0.05 | |
| Clostridium innocuum | 0.05 | |
| Staphylococcus | 0.05 | |
| no FGID | Lachnospiraceae UCG004 | 0.05 |
| bloating | Eubacterium coprostanoligenes | 0.05 |
| no bloating | Dielma | 0.05 |
| Oxalobacter | 0.05 | |
| Eubacterium eligens | 0.05 | |
| diarrhoea | Enterobacter | 0.05 |
| Family XII UCG001 | 0.05 | |
| Oscillibacter | 0.05 | |
| Papillibacter | 0.05 | |
| F0332 | 0.05 | |
| no diarrhoea | Oxalobacter | 0.05 |
| constipation | Clostridium innocuum | 0.05 |
| Oscillibacter | 0.05 | |
| Staphylococcus | 0.01 | |
| Lachnospira | 0.05 | |
| no constipation | Agathobacter | 0.05 |
| Lactococcus | 0.05 | |
| communication—expressive skills (adequate > low) | Clostridium sensu stricto | 0.001 |
| Dolosigranulum | 0.05 | |
| Faecalibacterium | 0.05 | |
| Lachnospiraceae UCG003 | 0.05 | |
| Romboutsia | 0.05 | |
| communication—expressive skills (high > low) | CAG352 | 0.05 |
| Eubacterium brachy | 0.05 | |
| Eubacterium coprostanoligenes | 0.05 | |
| Dialister | 0.01 | |
| Enterococcus | 0.05 | |
| Flavonifractor | 0.05 | |
| Incertae sedis | 0.05 | |
| Intestinibacter | 0.05 | |
| Romboutsia | 0.05 | |
| Ruminiclostridium | 0.05 | |
| Veillonella | 0.05 | |
| communication—receptive skills (low > adequate) | Erysipelotrichaceae UCG003 | 0.05 |
| communication—receptive skills (adequate > low) | GCA.900066575 | 0.01 |
| Tyzzerella | 0.05 | |
| communication—receptive skills (adequate > high) | Faecalitalea | 0.05 |
| communication—receptive skills (high > low) | CAG352 | 0.05 |
| Eisenbergiella | 0.05 | |
| Enorma | 0.05 | |
| Enterococcus | 0.01 | |
| Flavonifractor | 0.01 | |
| GCA.900066575 | 0.01 | |
| Hungatella | 0.05 | |
| Intestinibacter | 0.05 | |
| Ruminiclostridium | 0.01 | |
| Tyzzerella | 0.05 | |
| communication—receptive skills (high > adequate) | Dialister | 0.05 |
| Insertae sedis | 0.05 | |
| communication—writing skills (low > adequate) | Blautia | 0.05 |
| communication—writing skills (low > high) | Ruminococcus gauvreauli | 0.05 |
| Collinsella | 0.05 | |
| Dielma | 0.05 | |
| communication—writing skills (adequate > low) | Catenisphaera | 0.05 |
| Lachnospiraceae UCG.003 | 0.05 | |
| Pseudochrobactrum | 0.001 | |
| Pseudomonas | 0.001 | |
| Tyzzerella | 0.01 | |
| communication—writing skills (high > low) | Hungatella | 0.05 |
| communication—total (low > adequate) | Actinomyces | 0.05 |
| Gemella | 0.05 | |
| Granullicatella | 0.05 | |
| Streptococcus | 0.05 | |
| communication—total (low > high) | Defluviitaleaceae UCG.011 | 0.05 |
| GCA.900066575 | 0.05 | |
| Pseudochrobactrum | 0.05 | |
| Pseudomonas | 0.05 | |
| Pseudopropionibacterium | 0.05 | |
| Ruminiclostridium | 0.05 | |
| communication—total (adequate > low) | Dielma | 0.05 |
| Lachnospiraceae NK4A136 | 0.05 | |
| Oscillibacter | 0.05 | |
| Paraprevotella | 0.05 | |
| Phascolarctobacterium | 0.05 | |
| Ruminiclostridium | 0.05 | |
| communication—total (adequate > high) | Phascolarctobacterium | 0.05 |
| communication—total (high > low) | Anaerococcus | 0.05 |
| CAG.352 | 0.01 | |
| Eisenbergiella | 0.01 | |
| Enorma | 0.05 | |
| Enterococcus | 0.05 | |
| Flavonifractor | 0.05 | |
| Gemella | 0.05 | |
| Hungatella | 0.05 | |
| Tyzzerella | 0.05 | |
| communication—total (high > adequate) | Granullicatella | 0.05 |
| Streptococcus | 0.05 | |
| daily living skill personal (low > high) | Collinsella | 0.05 |
| daily living skill personal (adequate > low) | Clostidium sensu stricto | 0.05 |
| Veillonella | 0.01 | |
| Dolosigranulum | 0.05 | |
| Lachnospiraceae FCS020 | 0.05 | |
| Sarcina | 0.05 | |
| daily living skill personal (high > low) | CAG.352 | 0.05 |
| Eubacterium brachy | 0.05 | |
| Eubacterium coprostanoligenes | 0.05 | |
| Dialister | 0.05 | |
| Flavonifractor | 0.05 | |
| Incertae sedis | 0.05 | |
| Intestinibacter | 0.05 | |
| Romboutsia | 0.05 | |
| Veillonella | 0.05 | |
| daily living skill personal (high > adequate) | Anaerococcus | 0.05 |
| CAG.352 | 0.05 | |
| Colidextribacter | 0.05 | |
| GCA.900066575 | 0.05 | |
| daily living skill domestic (low > adequate) | Ruminiclostridium | 0.05 |
| daily living skill domestic (low > high) | Anaerostipes | 0.05 |
| Butyricicoccus | 0.05 | |
| Lachnospiraceae UCG.003 | 0.05 | |
| Oxalobacter | 0.05 | |
| Pseudochrobacter | 0.05 | |
| Pseudomonas | 0.05 | |
| daily living skill domestic (adequate > low) | Butyricimonas | 0.05 |
| Clostridium sensu stricto | 0.01 | |
| Coprococcus | 0.05 | |
| DTU089 | 0.05 | |
| Intestinibacter | 0.05 | |
| Veillonella | 0.05 | |
| daily living skill domestic (high > low) | Dialister | 0.05 |
| Enterococcus | 0.05 | |
| Escherichia/Shigella | 0.05 | |
| Hungatella | 0.05 | |
| Incertae sedis | 0.05 | |
| Intestinibacter | 0.05 | |
| Papillibacter | 0.05 | |
| Tyzzerella | 0.01 | |
| daily living skills—community skills (low > adequate) | Ruminococcus gnavus | 0.05 |
| daily living skills—community skills (low > high) | Granullicatella | 0.05 |
| Parvimonas | 0.05 | |
| daily living skill community (adequate > low) | Barnesiella | 0.05 |
| Eubacterium siraeum | 0.05 | |
| Coprococcus | 0.05 | |
| Dialister | 0.01 | |
| Lachnospiraceae FCS020 | 0.01 | |
| NK4A214 | 0.05 | |
| Odoribacter | 0.05 | |
| Oscillibacter | 0.01 | |
| Parabacteroides | 0.05 | |
| Ruminiclostridium | 0.01 | |
| Sarcina | 0.05 | |
| Turicibacter | 0.05 | |
| UCG.003 | 0.05 | |
| daily living skill community (adequate > high) | Agathobacter | 0.05 |
| Allistipes | 0.01 | |
| Barnesiella | 0.05 | |
| Ruminococcus gauvreaui | 0.05 | |
| Haemophilus | 0.05 | |
| Parabacteroides | 0.05 | |
| UCG.003 | 0.05 | |
| daily living skills—community skills (high > low) | Escherichia/Shigella | 0.05 |
| Hungatella | 0.05 | |
| Incertae sedis | 0.05 | |
| Senegalimassilia | 0.05 | |
| Tyzzerella | 0.05 | |
| daily living skills—community skills (high > adequate) | Ruminococcus gnavus | 0.05 |
| daily living skills—total (low > high) | Anaerostipes | 0.05 |
| Butyricicoccus | 0.05 | |
| Lachnospiraceae UCG.003 | 0.05 | |
| Oxalobacter | 0.05 | |
| Pseudochrobactrum | 0.05 | |
| Pseudomonas | 0.05 | |
| daily living skill total (adequate > low) | Butyricimonas | 0.05 |
| Clostidium sensu stricto | 0.05 | |
| Oscillibacter | 0.05 | |
| Ruminiclostridium | 0.05 | |
| Staphylococcus | 0.05 | |
| daily living skill total (adequate > high) | Anaerostipes | 0.05 |
| daily living skills—total (high > low) | Dialister | 0.05 |
| Enterococcus | 0.05 | |
| Escherichia/Shigella | 0.05 | |
| Flavonifractor | 0.05 | |
| Hungatella | 0.05 | |
| Papillibacter | 0.05 | |
| Tyzzerella | 0.01 | |
| socialization—interpersonal skills (low > adequate) | Ruminiclostridium | 0.05 |
| socialization—interpersonal skills (low > high) | Anaerostipes | 0.05 |
| Barnesiella | 0.05 | |
| Butyricicoccus | 0.05 | |
| Collinsella | 0.05 | |
| socialization—interpersonal skills (adequate > low) | Ruminococcus torques | 0.05 |
| Coprococcus | 0.05 | |
| Dialister | 0.05 | |
| Lachnospiraceae FCS020 | 0.05 | |
| Odoribacter | 0.05 | |
| Subdoligranulum | 0.01 | |
| socialization—interpersonal skills (adequate > high) | Barnesiella | 0.05 |
| Butyricicoccus | 0.05 | |
| Lachnospiraceae NK4A136 | 0.05 | |
| Monoglobus | 0.05 | |
| socialization—interpersonal skills (high > low) | Ruminococcus gnavus | 0.05 |
| F0032 | 0.05 | |
| Lachnospiraceae UCG.003 | 0.05 | |
| Oxalobacter | 0.05 | |
| Veillonella | 0.01 | |
| socialization—interpersonal skills (high > adequate) | Clostidium innocuum | 0.05 |
| Ruminococcus gnavus | 0.05 | |
| socialization—play and leisure skills (adequate > low) | Barnesiella | 0.05 |
| Odoribacter | 0.05 | |
| Ruminiclostridium | 0.05 | |
| UCG.003 | 0.05 | |
| Veillonella | 0.05 | |
| socialization—play and leisure skills (high > low) | CAG.352 | 0.01 |
| Dialister | 0.05 | |
| Enorma | 0.05 | |
| Pseudochrobactrum | 0.01 | |
| Pseudomonas | 0.01 | |
| socialization—coping skills (adequate > low) | Actinomyces | 0.05 |
| Eubacterium brachy | 0.01 | |
| F0332 | 0.05 | |
| Hungatella | 0.05 | |
| Ruminococcus torques | 0.05 | |
| TM7x | 0.01 | |
| Veillonella | 0.05 | |
| Weissella | 0.01 | |
| socialization—coping skills (high > low) | Eubacterium brachy | 0.01 |
| Defluviitaleaceae UCG.011 | 0.001 | |
| Dialister | 0.05 | |
| Dorea | 0.05 | |
| Enorma | 0.001 | |
| Enterobacter | 0.05 | |
| F0332 | 0.001 | |
| Flavonifractor | 0.05 | |
| Oscillibacter | 0.05 | |
| Parasutterella | 0.01 | |
| Pseudopropionibacterium | 0.001 | |
| Ruminiclostridium | 0.05 | |
| TM7x | 0.01 | |
| Varibaculum | 0.05 | |
| socialization—coping skills (high > adequate) | Parasutterella | 0.05 |
| socialization—total (low > high) | Enorma | 0.05 |
| socialization—total (adequate > low) | Clostridium sensu stricto | 0.05 |
| Dialister | 0.05 | |
| Enterobacter | 0.05 | |
| Enterococcus | 0.05 | |
| Lachnospira | 0.05 | |
| Subdoligranulum | 0.05 | |
| socialization—total (high > low) | Bilophila | 0.01 |
| F0332 | 0.05 | |
| Oscillibacter | 0.05 | |
| Parasutterella | 0.05 | |
| socialization—total (high > adequate) | Bilophila | 0.01 |
| Olsenella | 0.01 | |
| using large muscles (adequate > low) | Romboutsia | 0.05 |
| using large muscles (high > low) | Bifidobacterium | 0.05 |
| using large muscles (high > adequate) | Flavonifractor | 0.05 |
| Staphylococcus | 0.05 | |
| TM7x | 0.05 | |
| using small muscles (adequate > low) | Clostridium sensu stricto | 0.05 |
| Coprococcus | 0.05 | |
| Dolosigranulum | 0.05 | |
| Lachnospiraceae FCS020 | 0.05 | |
| Romboutsia | 0.05 | |
| Veillonella | 0.01 | |
| using small muscles (adequate > high) | Monoglobus | 0.05 |
| using small muscles (high > low) | Butyricimonas | 0.05 |
| Clostridium sensu stricto | 0.05 | |
| Eubacterium coprostanoligenes | 0.05 | |
| Ruminococcus torques | 0.05 | |
| Incertae sedis | 0.05 | |
| Intestinibacter | 0.05 | |
| Parasutterella | 0.05 | |
| Romboutsia | 0.05 | |
| Ruminiclostridium | 0.05 | |
| Sarcina | 0.01 | |
| UCG.005 | 0.05 | |
| Veillonella | 0.05 | |
| using small muscles (high > adequate) | Butyricimonas | 0.05 |
| Sarcina | 0.05 | |
| motor skills—total (adequate > low) | Eubacterium siraeum | 0.01 |
| Eubacterium ventriosum | 0.05 | |
| Ruminococcus gauvreauii | 0.01 | |
| Ruminococcus torques | 0.01 | |
| Coprococcus | 0.05 | |
| Frisingicoccus | 0.05 | |
| Lachnospiraceae FCS020 | 0.05 | |
| Monoglobus | 0.05 | |
| motor skills—total (high > low) | Clostridium sensu stricto | 0.05 |
| Eubacterium coprostanoligenes | 0.05 | |
| Intestinibacter | 0.05 | |
| Lachnospiraceae FCS020 | 0.05 | |
| motor skills—total (high > adequate) | Clostridium sensu stricto | 0.01 |
| Intestinibacter | 0.01 | |
| Species | ||
| age (adolescents > school children) | Clostridum perfingens | 0.01 |
| Lactobacillus paracasei | 0.01 | |
| Lactobacillus sakei | 0.05 | |
| Pediococcus pentosaceus | 0.01 | |
| diagnosis: childhood autism | Streptococcus mutans | 0.05 |
| sex: female | Lactobacillus fermentum | 0.01 |
| Streptococcus sobrinus | 0.01 | |
| high-protein diet | Lactobacillus curvatus | 0.05 |
| FGID | Leuconostoc mesenteroides | 0.05 |
| diarrhoea | Leuconostoc lactis | 0.05 |
| Leuconostoc citreum | 0.05 | |
| no diarrhoea | Enterobacter cloacae | 0.05 |
| Lactobacillus oligofermentans | 0.05 | |
| Lactobacillus brevis | 0.05 | |
| bloating | Eubacterium hallii | 0.05 |
| no bloating | Lactobacillus delbrueckii | 0.05 |
| Pediococcus pentosaceus | 0.05 | |
| constipation | Eubacterium hallii | 0.05 |
| no constipation | Lactococcus lactis | 0.05 |
| communication—expressive skills (high > low) | Lactococcus garvieae | 0.05 |
| communication—receptive skills (adequate > low) | Leuconostoc citreum | 0.05 |
| Leuconostoc lactis | 0.05 | |
| communication—receptive skills (high > low) | Lactococcus garvieae | 0.01 |
| communication—writing skills (low > high) | Clostridium tertium | 0.05 |
| communication—writing skills (adequate > low) | Enterococcus casseliflavus | 0.001 |
| Enterococcus faecalis | 0.001 | |
| Lactobacillus fermentum | 0.05 | |
| Pseudomonas spp. | 0.001 | |
| Streptococcus sobrinus | 0.05 | |
| communication—writing skills (adequate > high) | Bifidobacterium bifidum | 0.05 |
| communication—total (low > high) | Clostridium tertium | 0.05 |
| Enterococcus casseliflavus | 0.05 | |
| Enterococcus faecalis | 0.05 | |
| Haemophilus pittmaniae | 0.05 | |
| Pseudomonas spp. | 0.05 | |
| communication—total (high > low) | Bifidobacterium animalis | 0.05 |
| Lactococcus garvieae | 0.01 | |
| communication—total (high > adequate) | Streptococcus salivarius | 0.01 |
| daily living skill personal (adequate > low) | Odoribacter splanchnicus | 0.05 |
| daily living skill personal (high > low) | Lactococcus garvieae | 0.05 |
| daily living skill domestic (low > adequate) | Staphylococcus epidermidis | 0.01 |
| Streptococcus gallolyticus | 0.05 | |
| Streptococcus sobrinus | 0.05 | |
| daily living skill domestic (low > high) | Clostridium tertium | 0.05 |
| Enterococcus casseliflavus | 0.05 | |
| Enterococcus faecalis | 0.05 | |
| Lactococcus garvieae | 0.05 | |
| Pseudomonas spp. | 0.05 | |
| daily living skill domestic (adequate > low) | Lactobacillus fermentum | 0.05 |
| Odoribacter splanchnicus | 0.01 | |
| daily living skill community (adequate > low) | Lactobacillus fermentum | 0.01 |
| Lactobacillus salivarius | 0.05 | |
| Odoribacter splanchnicus | 0.01 | |
| Streptococcus sobrinus | 0.01 | |
| daily living skill total (low > high) | Clostridium tertium | 0.05 |
| Enterococcus casseliflavus | 0.05 | |
| Enterococcus faecalis | 0.05 | |
| Lactococcus garvieae | 0.05 | |
| Pseudomonas spp. | 0.05 | |
| socialization—interpersonal skills (low > adequate) | Lactobacillus fermentum | 0.05 |
| Streptococcus sobrinus | 0.05 | |
| socialization—interpersonal skills (adequate > low) | Streptococcus mutans | 0.05 |
| socialization—interpersonal skills (high > low) | Clostridium tertium | 0.05 |
| Lactococcus garvieae | 0.05 | |
| Staphylococcus haemolyticus | 0.05 | |
| socialization—play and leisure skills (adequate > low) | Lactobacillus fermentum | 0.05 |
| Lactococcus garvieae | 0.05 | |
| Odoribacter splanchnicus | 0.05 | |
| Streptococcus sobrinus | 0.05 | |
| socialization—play and leisure skills (high > low) | Enterococcus casseliflavus | 0.01 |
| Enterococcus faecalis | 0.01 | |
| Pseudomonas spp. | 0.01 | |
| socialization—coping skills (adequate > low) | Leuconostoc citreum | 0.01 |
| Leuconostoc lactis | 0.01 | |
| socialization—coping skills (high > low) | Haemophilus pittmaniae | 0.001 |
| Lactococcus garvieae | 0.001 | |
| Leuconostoc citreum | 0.001 | |
| Streptococcus salivarius | 0.05 | |
| socialization—total (low > adequate) | Lactococcus garvieae | 0.05 |
| using large muscles (adequate > low) | Streptococcus mutans | 0.05 |
| using small muscles (high > low) | Bifidobacterium bifidum | 0.01 |
| Lactobacillus salivarius | 0.05 | |
| Lactococcus garvieae | 0.05 | |
| Streptococcus anginosus | 0.05 | |
| motor skills—total (adequate > low) | Eubacterium hallii | 0.05 |
| motor skills—total (high > adequate) | Bifidobacterium bifidum | 0.05 |
| Feature | Bacteria | Correlation (r) |
|---|---|---|
| Order | ||
| age | Micrococcales | −0.47 |
| fat saturated (mean)/fat total (mean) | Peptostreptococcales/Tissierellales | −0.46 |
| complex carbohydrates (mean) | Clostridiales | 0.5 |
| Peptostreptococcales/Tissierellales | 0.47 | |
| daily living skills—personal skills (v-score) | Peptostreptococcales/Tissierellales | 0.5 |
| using small muscles (v-score) | Clostridiales | 0.53 |
| Peptostreptococcales/Tissierellales | 0.57 | |
| Genus | ||
| age | Rothia | −0.47 |
| fat saturated (mean)/fat total (mean) | Eubacterium coprostanoligenes | −0.46 |
| Eubacterium hallii | −0.59 | |
| Dorea | −0.6 | |
| Leucostonoc | 0.49 | |
| fat unsaturated (mean)/fat total (mean) | Dielma | 0.55 |
| fat (mean)/weight (kg) | Dielma | 0.47 |
| simple carbohydrates (mean)/carbohydrates total (mean) | Epulopiscium | 0.53 |
| Flavonifractor | −0.5 | |
| Weissella | 0.51 | |
| complex carbohydrates (mean) | Romboutsia | 0.52 |
| kcal from carbohydrates/total carbohydrates (mean) | Epulopiscium | 0.5 |
| Klebsiella | 0.66 | |
| Weissella | 0.51 | |
| kcal from carbohydrates/total kcal | Epulopiscium | 0.5 |
| Klebsiella | 0.66 | |
| protein (mean)/weight (kg) | Dielma | 0.52 |
| communication—receptive skills (v-score) | Flavonifractor | 0.49 |
| communication—writing skills (v-score) | Anaerostipes | −0.51 |
| daily living skills—personal skills (v-score) | Romboutsia | 0.5 |
| daily living skills—domestic skills (v-score) | Intestinibacter | 0.5 |
| socialization—coping skills (v-score) | F0032 | 0.5 |
| using small muscles (v-score) | Romboutsia | 0.55 |
| Species | ||
| fat saturated (mean)/fat total (mean) | Enterobacter cloacae | 0.68 |
| Leuconostoc citreum | 0.53 | |
| Leuconostoc lactis | 0.56 | |
| Staphylococcus epidermidis | 0.46 | |
| Streptococcus gallolyticus | 0.66 | |
| kcal from carbohydrates/total carbohydrates (mean) | Clostridium perfingens | 0.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewandowska-Pietruszka, Z.; Figlerowicz, M.; Mazur-Melewska, K. Gut Microbiota and Autism Spectrum Disorders: Neurodevelopmental, Behavioral, and Gastrointestinal Interactions. Nutrients 2025, 17, 2781. https://doi.org/10.3390/nu17172781
Lewandowska-Pietruszka Z, Figlerowicz M, Mazur-Melewska K. Gut Microbiota and Autism Spectrum Disorders: Neurodevelopmental, Behavioral, and Gastrointestinal Interactions. Nutrients. 2025; 17(17):2781. https://doi.org/10.3390/nu17172781
Chicago/Turabian StyleLewandowska-Pietruszka, Zuzanna, Magdalena Figlerowicz, and Katarzyna Mazur-Melewska. 2025. "Gut Microbiota and Autism Spectrum Disorders: Neurodevelopmental, Behavioral, and Gastrointestinal Interactions" Nutrients 17, no. 17: 2781. https://doi.org/10.3390/nu17172781
APA StyleLewandowska-Pietruszka, Z., Figlerowicz, M., & Mazur-Melewska, K. (2025). Gut Microbiota and Autism Spectrum Disorders: Neurodevelopmental, Behavioral, and Gastrointestinal Interactions. Nutrients, 17(17), 2781. https://doi.org/10.3390/nu17172781






