Octenyl Succinic Anhydride Starch Alleviates Alcoholic Liver Disease by Modulating Gut Microbiota and Metabolism
Abstract
1. Introduction
2. Materials and Methods
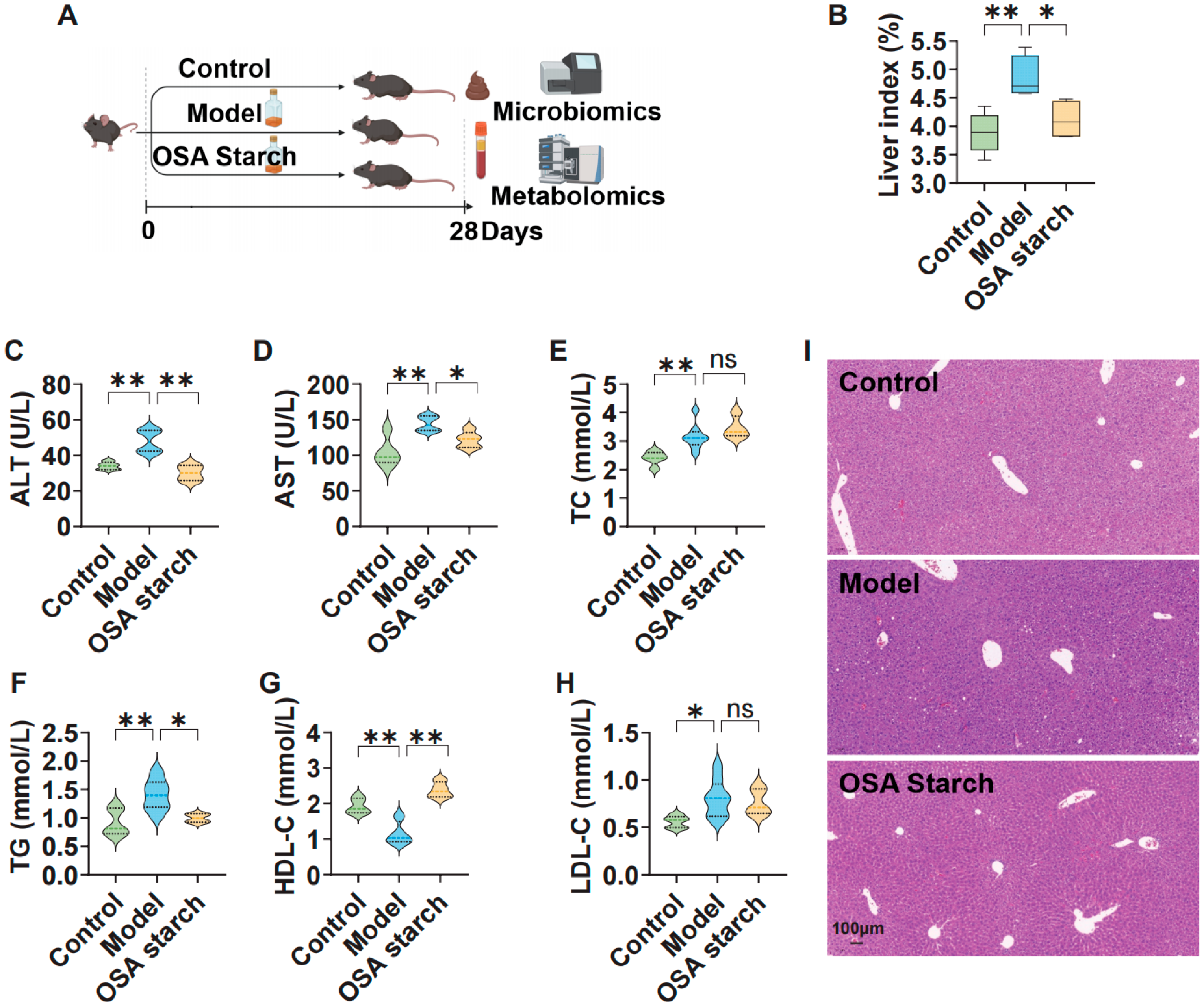
2.1. Establishment of Alcoholic Liver Injury Model and Dietary OSA Starch Intervention
2.2. Serum Biochemical Assays
2.3. Liver Histology via Hematoxylin and Eosin Staining
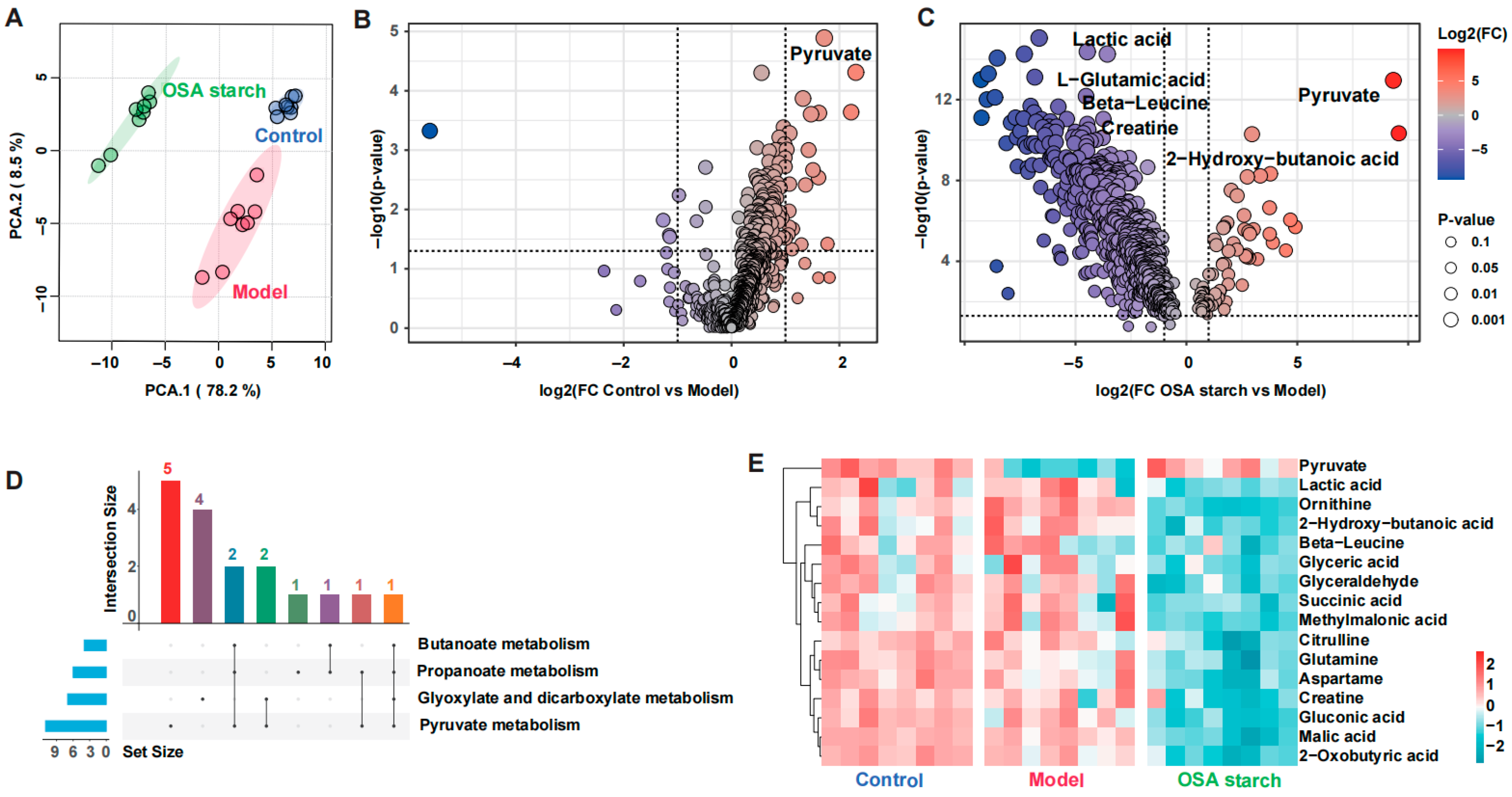
2.4. Untargeted Metabolomics Profiling
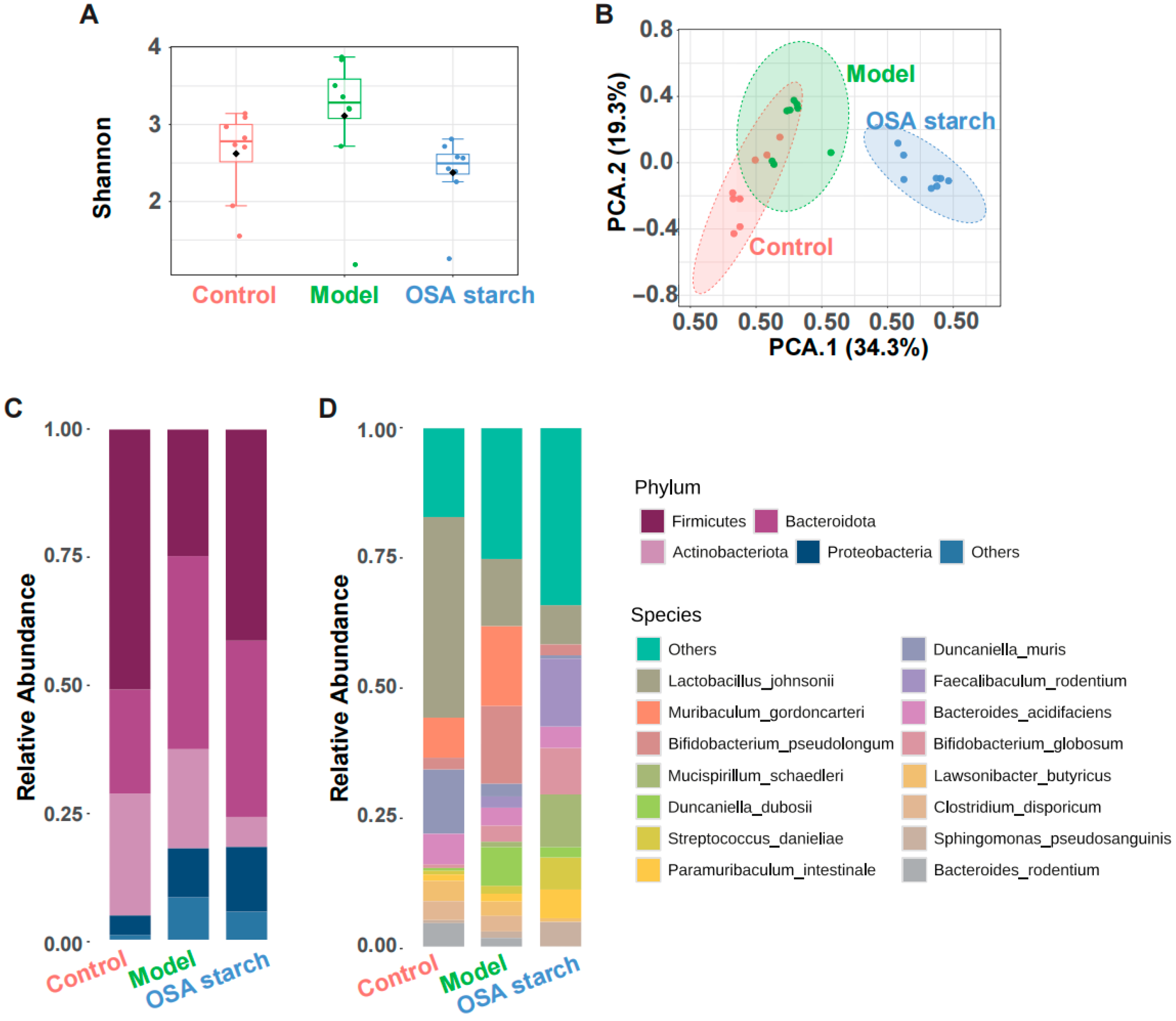
2.5. 16S rRNA Gene Sequencing and Microbial Functional Prediction
2.6. Statistical and Computational Analyses
3. Results and Discussion
3.1. OSA Starch Ameliorates Hepatic Injury and Lipid Dysregulation in ALD Mice
3.2. OSA Starch Intervention Reshapes Gut Microbiota Composition in ALD Mice
3.3. OSA Starch Modulates Key Gut Microbial Taxa and Their Functional Profiles in ALD Mice
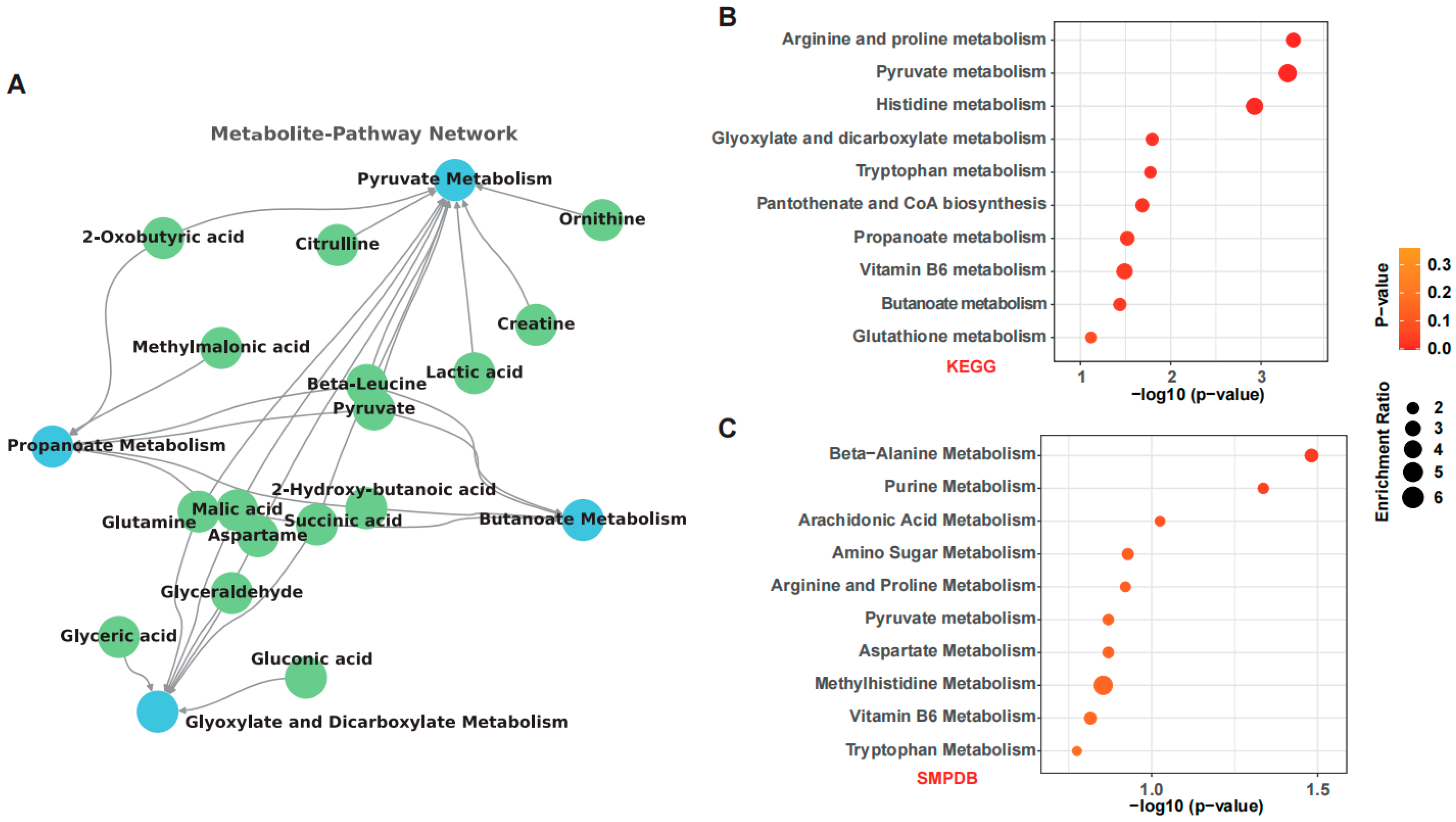
3.4. Key Gut Microbial Metabolic Pathways Modulated by OSA Starch in ALD Mice
3.5. OSA Starch Reshapes Fecal Metabolome and Modulates Key Metabolic Pathways in ALD Mice
3.6. OSA Starch Improves Serum Bile Acid Profiles in ALD Mice
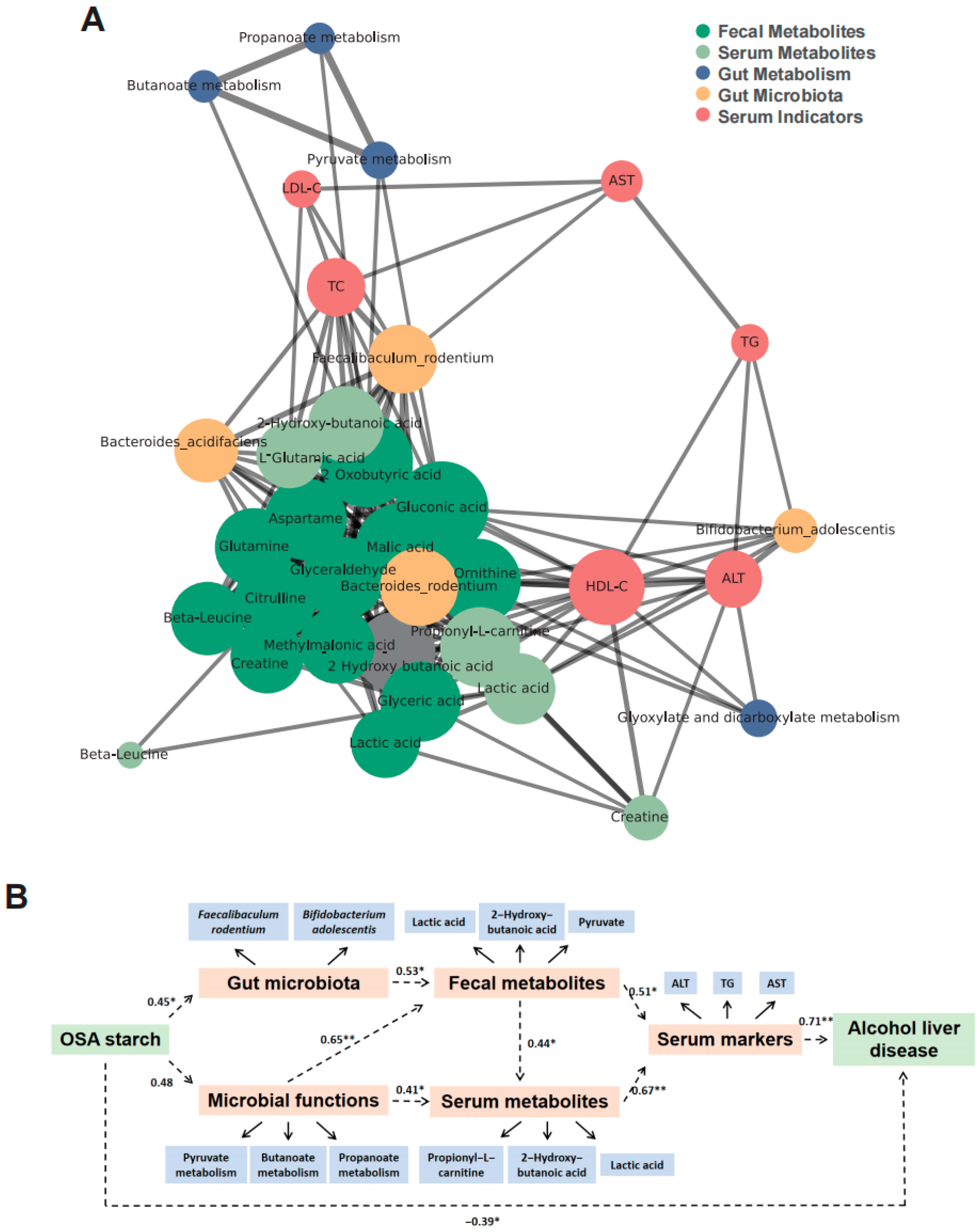
3.7. OSA Starch Modulates Gut Microbiota–Metabolite–Host Axis to Alleviate ALD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALD | Alcoholic liver disease |
| OSA | Octenyl succinic anhydride |
References
- Yan, M.Y.; Man, S.L.; Ma, L.; Guo, L.P.; Huang, L.Q.; Gao, W.Y. Immunological mechanisms in steatotic liver diseases an overview and clinical perspectives. Clin. Mol. Hepatol. 2024, 30, 620–648. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.X.; Jia, W.P. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef]
- Bajaj, J.S. Alcohol, liver disease and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 235–246. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, R.S.; Mu, Y.; Song, Y.; Hao, N.; Wei, Y.B.; Wang, Q.B.; Mackay, C.R. Propionate Ameliorates Alcohol-Induced Liver Injury in Mice via the Gut-Liver Axis: Focus on the Improvement of Intestinal Permeability. J. Agric. Food Chem. 2022, 70, 6084–6096. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ren, Q.X.; Zhou, Z.L.; Li, X.; Ren, D.L.; Ji, Z.W.; Mao, J. Structural elucidation of a highly branched α-D-glucan from Huangjiu and its hepatoprotective activity via gut microbiome regulation and intestinal barrier repairment. Carbohydr. Polym. 2024, 324, 121423. [Google Scholar] [CrossRef]
- Li, F.; McClain, C.; Feng, W. Microbiome dysbiosis and alcoholic liver disease. Liver Res. 2019, 3, 218–226. [Google Scholar] [CrossRef]
- Bluemel, S.; Wang, L.R.; Kuelbs, C.; Moncera, K.; Torralba, M.; Singh, H.; Fouts, D.E.; Schnabl, B. Intestinal and hepatic microbiota changes associated with chronic ethanol administration in mice. Gut Microbes 2020, 11, 265–275. [Google Scholar] [CrossRef]
- Wu, H.Y.; Chen, J.J.; Guo, S.Y.; Deng, J.H.; Zhou, Z.M.; Zhang, X.; Qi, T.T.; Yu, F.; Yang, Q. Advances in the acting mechanism and treatment of gut microbiota in metabolic dysfunction-associated steatotic liver disease. Gut Microbes 2025, 17, 2500099. [Google Scholar] [CrossRef]
- Chang, R.R.; Jin, Z.Y.; Tian, Y.Q. Insights into the structural, morphological, and thermal property changes in simulated digestion and fermentability of four resistant starches from high amylose maize starch. Food Hydrocoll. 2023, 142, 108770. [Google Scholar] [CrossRef]
- Chang, R.R.; Wang, F.; Huang, J.T.; Jin, Z.Y.; Tian, Y.Q. Recrystallized Resistant Starch: Structural Changes in the Stomach, Duodenum, and Ileum and the Impact on Blood Glucose and Intestinal Microbiome in Mice. J. Agric. Food Chem. 2023, 71, 12080–12093. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, C.; Li, H.X.E.; Wu, J.H.; Zhang, D.C.; Li, Y.; Yang, L.; Zhang, N.; Wang, X.Y. Structure properties of Canna edulis RS3 (double enzyme hydrolysis) and RS4 (OS-starch and cross-linked starch): Influence on fermentation products and human gut microbiota. Int. J. Biol. Macromol. 2024, 265, 130700. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.M.; Li, J.Y.; Yang, F.W.; Guo, Y.H.; Yao, W.R.; Qian, H.; Cheng, Y.L. Role of the gut microbiota in dietary patterns rich in torularhodin OSA colon-targeted delivery. Food Funct. 2022, 13, 11034–11048. [Google Scholar] [CrossRef]
- Devarakonda, S.L.S.; Superdock, D.K.; Ren, J.N.F.; Johnson, L.M.; Loinard-González, A.P.; Poole, A.C. Gut microbial features and dietary fiber intake predict gut microbiota response to resistant starch supplementation. Gut Microbes 2024, 16, 2367301. [Google Scholar] [CrossRef]
- Huicui, L.; Min, Z.; Qingyu, M.; Baoming, T.; Chenxi, N.; Zhifei, C.; Juxiu, L. Health beneficial effects of resistant starch on diabetes and obesity via regulation of gut microbiota: A review. Food Funct. 2020, 11, 5749–5767. [Google Scholar] [CrossRef]
- Sun, Z.H.; Chen, Z.W.; Xu, B.; Shi, Y.C. Distribution of octenylsuccinate substituents within a single granule of modified waxy maize starch determined by Raman microspectroscopy. Carbohydr. Polym. 2019, 216, 282–286. [Google Scholar] [CrossRef]
- Chen, J.P.; Maierheba, K.; Zhang, Y.; Cheng, H.; Lin, B.B.; Yue, P.; Wang, L.H.; Liu, F.Z.; Shi, J.W.; Wan, Z.X.; et al. Octenyl Succinic Anhydride-Modified Starch Attenuates Body Weight Gain and Changes Intestinal Environment of High-Fat Diet-Fed Mice. Foods 2022, 11, 2980. [Google Scholar] [CrossRef] [PubMed]
- Li, H.T.; Zhang, L.; Li, J.; Wu, Q.; Qian, L.L.; He, J.S.; Ni, Y.Q.; Kovatcheva-Datchary, P.; Yuan, R.; Liu, S.B.; et al. Resistant starch intake facilitates weight loss in humans by reshaping the gut microbiota. Nat. Metab. 2024, 6, 578–597. [Google Scholar] [CrossRef]
- Ni, Y.; Qian, L.; Siliceo, S.L.; Long, X.; Nychas, E.; Liu, Y.; Ismaiah, M.J.; Leung, H.; Zhang, L.; Gao, Q.; et al. Resistant starch decreases intrahepatic triglycerides in patients with NAFLD via gut microbiome alterations. Cell Metab. 2023, 35, 1530–1547. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, C.; Ma, R.R.; Pan, X.H.; Shen, W.Y.; Tian, Y.Q. Esterified starches enhance short-term satiety in mice structural and physicochemical alterations. Food Funct. 2024, 15, 12058–12068. [Google Scholar] [CrossRef]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Kwon, Y.-J.; Lee, E.; Lee, H.S.; Youn, Y.H.; Baik, S.J.; Shin, H.J.; Chae, H.W. Optimal cutoffs of fatty liver index and hepatic steatosis index in diagnosing pediatric MASLD. Clin. Gastroenterol. Hepatol. 2025, 25, 00530. [Google Scholar] [CrossRef]
- Abeysekera, K.W.A.; Fernandes, G.S.; Hammerton, G.; Portal, A.J.; Gordon, F.H.; Heron, J.; Hickman, M. Prevalence of steatosis and fibrosis in young adults in the UK: A population-based study. Lancet Gastroenterol. 2020, 5, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.Y.; Zhou, L.L.; Zhang, H.; Yang, Y.R.; Jiang, L.; Wu, D.Q.; Shu, H.; Zhang, H.J.; Xie, L.X.; Zhou, K.C.; et al. Dietary fiber alleviates alcoholic liver injury via Bacteroides acidifaciens and subsequent ammonia detoxification. Cell Host Microbe 2024, 32, 1331–1346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Liu, X.J.; Jin, X.; Mao, X.D.; Xu, X.L.; Zhang, X.; Shang, K.; Xu, Y.; Zhang, Y.H.; Meng, G.F.; et al. Liver-specific inactivation of Cideb improves metabolic profiles and ameliorates steatohepatitis and fibrosis in animal models for MASH. Pharmacol. Res. 2025, 214, 107664. [Google Scholar] [CrossRef]
- Hersberger, M.; von Eckardstein, A. Low high-density lipoprotein cholesterol—Physiological background, clinical importance and drug treatment. Drugs 2003, 63, 1907–1945. [Google Scholar] [CrossRef]
- Tsiavos, A.; Antza, C.; Trakatelli, C.; Kotsis, V. The Microbial Perspective: A Systematic Literature Review on Hypertension and Gut Microbiota. Nutrients 2024, 16, 3698. [Google Scholar] [CrossRef]
- Wang, R.; Tang, R.Q.; Li, B.; Ma, X.; Schnabl, B.; Tilg, H. Gut microbiome, liver immunology, and liver diseases. Cell. Mol. Immunol. 2021, 18, 4–17. [Google Scholar] [CrossRef]
- Shi, Y.J.; Zou, Y.X.; Xiong, Y.H.; Zhang, S.Y.; Song, M.M.; An, X.F.; Liu, C.; Zhang, W.X.; Chen, S.Y. Host Gasdermin D restrains systemic endotoxemia by capturing Proteobacteria in the colon of high-fat diet-feeding mice. Gut Microbes 2021, 13, 1946369. [Google Scholar] [CrossRef]
- DeMinicis, S.; Rychlicki, C.; Agostinelli, L.; Saccomanno, S.; Candelaresi, C.; Trozzi, L.; Mingarelli, E.; Facinelli, B.; Magi, G.; Palmieri, C.; et al. Dysbiosis Contributes to Fibrogenesis in the Course of Chronic Liver Injury in Mice. Hepatology 2014, 59, 1738–1749. [Google Scholar] [CrossRef]
- Dobranowski, P.A.; Stintzi, A. Resistant starch, microbiome, and precision modulation. Gut Microbes 2021, 13, 1926842. [Google Scholar] [CrossRef]
- Olagunju, A.; Martinez, J.; Kenny, D.; Gideon, P.; Mookadam, F.; Unzek, S. Virulent endocarditis due to Haemophilus parainfluenzae: A systematic review of the literature. World J. Cardiol. 2022, 14, 546–556. [Google Scholar] [CrossRef]
- Leibovitzh, H.; Nayeri, S.; Borowski, K.; Hernandez-Rocha, C.; Lee, S.H.; Turpin, W.; Stempak, J.M.; Sandhu, I.; Milgrom, R.; Smith, M.; et al. Inflammatory Bowel Disease Associated with Primary Sclerosing Cholangitis is Associated with an Altered Gut Microbiome and Bile Acid Profile. J. Crohn’s Colitis 2024, 18, 1957–1966. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Gueimonde, M.; Duncan, S.H.; Flint, H.J.; de los Reyes-Gavilan, C.G. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015, 362, 176. [Google Scholar] [CrossRef]
- Zagato, E.; Pozzi, C.; Bertocchi, A.; Schioppa, T.; Saccheri, F.; Guglietta, S.; Fosso, B.; Melocchi, L.; Nizzoli, G.; Troisi, J.; et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat. Microbiol. 2020, 5, 511–523. [Google Scholar] [CrossRef]
- Kawano, Y.; Edwards, M.; Huang, Y.M.; Bilate, A.M.; Araujo, L.P.; Tanoue, T.; Atarashi, K.; Ladinsky, M.S.; Reiner, S.L.; Wang, H.H.; et al. Microbiota imbalance induced by dietary sugar disrupts immune-mediated protection from metabolic syndrome. Cell 2022, 185, 3501–3519. [Google Scholar] [CrossRef]
- Ogura, Y.; Suzuki, S.; Shirakawa, T.; Masuda, M.; Nakamura, H.; Iijima, K.; Yoshikawa, N. antigen and antibody in children with IgA nephropathy and Henoch-Schonlein nephritis. Am. J. Kidney Dis. 2000, 36, 47–52. [Google Scholar] [CrossRef]
- Geng, X.; Zhuang, M.M.; Tian, W.N.; Shang, H.Y.; Gong, Z.Y.; Lv, Y.F.; Li, J.R. Green Radish Polysaccharide Prevents Alcoholic Liver Injury by Interfering with Intestinal Bacteria and Short-Chain Fatty Acids in Mice. Foods 2024, 13, 3733. [Google Scholar] [CrossRef] [PubMed]
- Yiew, N.K.H.; Vazquez, J.H.; Martino, M.R.; Kennon-McGill, S.; Price, J.R.; Allard, F.D.; Yee, E.U.; Layman, A.J.; James, L.P.; Mccommis, K.S.; et al. Hepatic pyruvate and alanine metabolism are critical and complementary for maintenance of antioxidant capacity and resistance to oxidative insult. Mol. Metab. 2023, 77, 101808. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.T.; Tran, L.; Beaven, S.; Tontonoz, P.; Reue, K.; Dipple, K.M.; Liao, J.C. Resistance to Diet-Induced Obesity in Mice with Synthetic Glyoxylate Shunt. Cell Metab. 2009, 9, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Recharla, N.; Geesala, R.; Shi, X.Z. Gut Microbial Metabolite Butyrate and Its Therapeutic Role in Inflammatory Bowel Disease: A Literature Review. Nutrients 2023, 15, 2275. [Google Scholar] [CrossRef]
- Cani, P.D. Is colonic propionate delivery a novel solution to improve metabolism and inflammation in overweight or obese subjects? Gut 2019, 68, 1352–1353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.; Deng, Y.K.; Zou, Y.F.; Han, B.L.; Pu, J.Z.; Rao, J.Q.; Huang, D.; Luo, H.B. Linking Microbial Functional Gene Abundance and Daqu Extracellular Enzyme Activity: Implications for Carbon Metabolism during Fermentation. Foods 2022, 11, 3623. [Google Scholar] [CrossRef] [PubMed]
- Deja, S.; Fletcher, J.A.; Kim, C.W.; Kucejova, B.; Fu, X.R.; Mizerska, M.; Villegas, M.; Pudelko-Malik, N.; Browder, N.; Inigo-Vollmer, M.; et al. Hepatic malonyl-CoA synthesis restrains gluconeogenesis by suppressing fat oxidation, pyruvate carboxylation, and amino acid availability. Cell Metab. 2024, 36, 1088–1104. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Li, G.L.; Zheng, Q.X.; Gu, X.Y.; Shi, Q.M.; Su, Y.S.; Chu, Q.F.; Yuan, X.; Bao, Z.Y.; Lu, J.; et al. Tryptophan metabolism in health and disease. Cell Metab. 2023, 35, 1304–1326. [Google Scholar] [CrossRef]
- Harris, P.S.; McGinnis, C.D.; Michel, C.R.; Marentette, J.O.; Reisdorph, R.; Roede, J.R.; Fritz, K.S. Click chemistry-based thiol redox proteomics reveals significant cysteine reduction induced by chronic ethanol consumption. Redox Biol. 2023, 64, 102792. [Google Scholar] [CrossRef]
- Puhakka, E.; Ahmed, H.; Haikonen, R.; Leclercq, S.; Hanhineva, K.; Maccioni, L.; Amadieu, C.; Lehtonen, M.; Maennistoe, V.; Rysae, J.; et al. Serum Metabolite Profile in Progressive Versus Nonprogressive Alcohol-Related Liver Disease: A Cross-Sectional Metabolomics Study. Liver Int. 2025, 45, 70128. [Google Scholar] [CrossRef]
- Zhu, X.P.; Huang, Q.X.; Ma, S.; Chen, L.Y.; Wu, Q.; Wu, L.; Ma, H.; Li, X.M.; Li, Q.; Aleteng, Q.; et al. Presence of sarcopenia identifies a special group of lean NAFLD in middle-aged and older people. Hepatol. Int. 2023, 17, 313–325. [Google Scholar] [CrossRef]
- Tamai, Y.; Chen, Z.; Wu, Y.; Okabe, J.; Kobayashi, Y.; Chiba, H.; Hui, S.P.; Eguchi, A.; Iwasa, M.; Ito, M.; et al. Branched-chain amino acids and l-carnitine attenuate lipotoxic hepatocellular damage in rat cirrhotic liver. Biomed. Pharmacother. 2021, 135, 111181. [Google Scholar] [CrossRef]
- Song, X.; Cui, W.; Meng, F.; Xia, Q.; Li, X.; Hou, M.; Jia, L.; Zhang, J. Glucopyranose from Pleurotus geesteranus prevent alcoholic liver diseases by regulating Nrf2/HO-1-TLR4/NF-κB signalling pathways and gut microbiota. Food Funct. 2022, 13, 2441–2455. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liu, T.; Ma, R.; Pan, X.; Tian, Y. Octenyl Succinic Anhydride Starch Alleviates Alcoholic Liver Disease by Modulating Gut Microbiota and Metabolism. Nutrients 2025, 17, 2779. https://doi.org/10.3390/nu17172779
Liu C, Liu T, Ma R, Pan X, Tian Y. Octenyl Succinic Anhydride Starch Alleviates Alcoholic Liver Disease by Modulating Gut Microbiota and Metabolism. Nutrients. 2025; 17(17):2779. https://doi.org/10.3390/nu17172779
Chicago/Turabian StyleLiu, Chang, Tangqian Liu, Rongrong Ma, Xiaohua Pan, and Yaoqi Tian. 2025. "Octenyl Succinic Anhydride Starch Alleviates Alcoholic Liver Disease by Modulating Gut Microbiota and Metabolism" Nutrients 17, no. 17: 2779. https://doi.org/10.3390/nu17172779
APA StyleLiu, C., Liu, T., Ma, R., Pan, X., & Tian, Y. (2025). Octenyl Succinic Anhydride Starch Alleviates Alcoholic Liver Disease by Modulating Gut Microbiota and Metabolism. Nutrients, 17(17), 2779. https://doi.org/10.3390/nu17172779





