Abstract
Background/Objectives: Nutrigenomics explores how dietary components influence genome function, especially via epigenetic mechanisms like DNA methylation. A key challenge is identifying healthy food-derived molecules capable of counteracting epigenetic damage from harmful dietary elements. Pistachio nuts (Pistacia vera L.), particularly the Bronte variety from Sicily, are rich in antioxidant polyphenols. In this study we used a methylomic approach to assess the nutrigenomic potential of a hydrophilic extract from Bronte pistachio (BPHE) in a model of human intestinal epithelium, as well as its capacity to modulate arsenic (As)-induced epigenotoxicity. Methods: BPHE was obtained via ethanol/water Soxhlet extraction. CaCo-2 cells were treated with BPHE, alone and after exposure to sodium arsenite. The methylation pattern of the genomic DNA was assessed by methylation-sensitive arbitrarily primed PCR and the methylomic signature was defined by Next-generation bisulfite sequencing. Results: BPHE alone did not alter DNA methylation pattern but, at the highest dose, modulated the changes induced by As. The identification of differentially methylated gene promoters in cell treatment vs. untreated controls revealed that BPHE and As primarily induced hyper-methylation, with a synergistic effect when combined. In particular, all the treatments increased methylation levels of gene categories such as pseudogenes, key genes of specific pathways, genes for zinc-finger proteins, homeobox proteins, kinases, antisense RNA, and miRNA. Notably, in co-treatment with As, BPHE promoted hypo-methylation of genes involved in tumor suppression, detoxification, mitochondrial function, and cell division. Conclusions: These findings suggest that Bronte pistachio polyphenols may epigenetically steer gene expression toward a protective profile, reducing risks of genomic instability and disease. This supports their potential as nutraceuticals to counter harmful epigenetic effects of toxic food components like arsenic.
1. Introduction
Nutrigenomics is a rapidly developing branch of research that explores how dietary components influence genome function, with a particular focus on gene expression regulated by epigenetic mechanisms such as DNA methylation and histone modifications [1]. Within the framework of nutrient–DNA interactions, nutrigenomics considers food-derived molecules as key modulators of gene expression, often investigated through genome-wide approaches. In contrast, nutrigenetics focuses on individual genetic variations (e.g., allelic polymorphisms) and their influence on how the body metabolizes and responds to nutrients [2].
Most of the previous nutrigenomic studies have focused on human nutrition and the effects of nutrients on disease etiology [3], mainly in relation to a specific alimentary regimen. The Mediterranean diet (MD) has been the object of these approaches [4]. The human health benefits associated with a reduction in the risk of developing non-communicable diseases, such as type 2 diabetes, cardiovascular diseases, some neurodegenerative diseases, and cancers, are well known [5]. Previous nutrigenomic studies demonstrated that indicaxanthin, a phytochemical highly concentrated in the edible fruits of the Mediterranean cactus Opuntia ficus-indica and with known anti-proliferative activity [6], when administrated at dietary doses to human intestinal epithelial cells, is able to specifically reduce DNA hyper-methylation and restore the expression of key silenced cancer-related genes [7], as well as affect the autophagy process [8].
Over the past decade, a wealth of experimental data have significantly reshaped our understanding of the role of nuts in healthy dietary patterns, including the MD. This renewed emphasis on nuts stems from their potential positive impact on serum lipid profiles when incorporated into our diet [9]. Among the 11 species within the Pistacia genus, pistachio (Pistacia vera L.) stands out as the exclusive producer of an edible nut with substantial market value. While less studied than other nuts like almonds and walnuts, an accumulating body of evidence indicates that pistachio nuts offer not only significant nutritional benefits but also hold great promise as a nutraceutical resource. They boast a rich reservoir of polyphenolic components and have earned a place among the top 50 food products recognized for their exceptional antioxidant potential [10]. Indeed, the well-established health-protective effects attributed to pistachio consumption, combined with its favorable lipid profile, can be attributed to the high concentration of bioactive phytochemicals.
Pistachio cultivation thrives across the Mediterranean region and has gained immense popularity in the United States. In fact, California now stands as the world’s leading pistachio producer. Conversely, pistachio production in Italy remains quite limited, with a primary focus in Sicily, renowned for its exceptional Bronte variety, cultivated in the Etna region. The remarkable quality of this pistachio variety is attributed not only to its distinctive sensory characteristics but also to its significantly higher content of antioxidant polyphenols when compared to other pistachio varieties with more substantial commercial presence [11].
Furthermore, some of our previous research has demonstrated the strong antioxidant potential of a hydroalcoholic extract derived from Bronte pistachio, rich in polyphenols and proanthocyanidins [12]. This extract has exhibited notable antioxidant capabilities in various experimental models and has demonstrated anti-inflammatory effects by modulating the NF-κB activation pathway. Specifically, it has effectively inhibited the inflammatory response triggered by LPS (lipopolysaccharide) or inflammatory cytokines in both macrophages and intestinal epithelial cells [13,14].
A new challenge of modern research in nutrigenomics is to find diet-contained molecules that not only have healthy effects at the metabolic level but are also able to modulate the epigenetic damage induced by other non-beneficial food components consumed through diet. In our hypothesis, their simultaneous or subsequent presence in the same metabolic district of the organism can provide the first molecule with the epigenetic possibility of modulating the damage induced by the second, causing an ameliorative final effect. Dietary antioxidants, particularly polyphenols, are among these beneficial molecules with nutrigenomic modulation capabilities; by reducing ROS levels, they help protect DNA from oxidative damage and support the normalization of DNA methylation [2]. This behavior has already been demonstrated by our group using stilbenoids [15]. This usually occurs in the digestive tract when a subject observes a varied diet. Unfortunately, this is an under-studied aspect considering that we are all subjected to continuous insults by the pollutants contained in food, but also by natural poisons. For example, extremely small concentrations of arsenic in drinking water have been linked to many human cancers [16,17,18], given that they play a role in amplifying the genotoxicity of other mutagenic carcinogens, including ultraviolet radiations [19].
This work intends to investigate, with a methylomic approach, the nutrigenomic property of a Bronte pistachio hydrophilic extract (BPHE) and its mechanisms of action at the epigenetic level in differentiated CaCo-2 cells as a model of the human intestinal epithelium. In fact, it is known that the methylation status of a DNA segment or gene is a putative prodromic condition of its function/expression and represents a basal platform for further functional studies. In addition, we aimed at evaluating the in vitro BPHE ability to modulate and reduce the (epi)genotoxic effect of arsenic (As) at DNA methylation level. Given the absence of studies dealing with combined treatments, this study will advance knowledge on such treatment approach.
2. Materials and Methods
2.1. Cell Model
The CaCo-2 cell line (ATCC code: HTB-37, Palo Alto, CA, USA), derived from human colon adenocarcinoma, has largely been used as a study model for colon cancer processes and development and also for drug and intestinal disease studies due to their ability to mimic various aspects of intestinal physiology, as well as provide insights into the pathology of the intestinal epithelium. Moreover, CaCo-2 cells, cultured as a monolayer, have the ability to spontaneously undergo differentiation into epithelium-like cells. Once reaching confluence and establishing contact inhibition, cell-to-cell contact and communication generate a physical cue for the cells to differentiate, taking 21 days to reach full differentiation [20]. Differentiated CaCo-2 cells constitute a polarized monolayer characterized by domes, with microvilli on the apical side and tight junctions between adjacent cells, and they represent a model of the human intestinal epithelium for studying the paracellular movement of substances across a monolayer [15].
In the present study, the CaCo-2 cell line was cultured as previously described [21], and the Caco-2 cell monolayer was employed as an in vitro intestinal system to investigate its interaction with potential bioactive compounds derived from the diet.
2.2. Plant Material
Pistacia vera L. nuts of the Bronte variety were supplied by Pistacchio dell’Etna Srl, located in Bronte, Italy. The seeds were stored in a dark environment at a temperature of 4 °C before extraction.
2.3. Extraction Protocol
The seeds were shelled, placed in a −80 °C freezer for 48 h, and then the kernels, along with their skin, were powdered using a mortar. Subsequently, 5 g samples were extracted by a 4 h Soxhlet extraction process using a 70:30 (v/v) ethanol:water solution with a 1:70 (w/v) extraction ratio.
Following a clean-up step by centrifugation (10 min at 10,000× g, 4 °C) and filtration through a Millex HV 0.45 μm filter (Millipore, Billerica, MA, USA), the extract was collected and stored at −80 °C for future use and analyzed within a span of 5 months. The extraction process was carried out in triplicate.
2.4. Content of Polyphenolic Compounds and Antioxidant Activity of BPHE
2.4.1. Total Polyphenol Content
The total polyphenol content (TPC) was assessed using the Folin–Ciocalteu method [22], following previously established procedures [23]. Gallic acid (GA) served as the standard for calibration curve. The results were quantified and expressed as milligrams of GA equivalents (GAE) per 100 g of fresh weight (FW). Each measurement was conducted in triplicate.
2.4.2. Total Proanthocyanidin Content
The total proanthocyanidin content (PAC) was assessed using the DMAC colorimetric assay, a modified version of the method described by Porter et al. (1985) [24]. The method involves the conversion of proanthocyanidins to anthocyanidins through acid hydrolysis in the presence of iron ions and heating the reaction mixture. The assay was performed as previously described [12]. To subtract the contribution of natural anthocyanins in the sample, the sample was processed also in ice instead of by warming. Cyanidin chloride (CC) was used as the standard and the results are expressed as milligrams CC equivalents (CCE) per 100 g of FW. All measurements were conducted in triplicate.
2.4.3. Antioxidant Activity
Antioxidant activity (AA) was assessed using the ABTS assay [25]. The radical ABTS+ was generated by mixing a 7 mM ABTS stock solution with 2.45 mM K2S2O8 and allowing the mixture to stand in the dark at room temperature for 16 h before use. The assay procedure followed previously established protocols [23,26]. Trolox was employed as a reference standard, and the antioxidant activity was quantified and expressed as millimoles of Trolox equivalent (TE) per 100 g of FW. The experiments were replicated three times.
2.5. Cell Culture, Treatments, and Positive Controls
Differentiated CaCo-2 cells were treated with BPHE for 3 h mimicking the average time of in vivo intestinal absorption. BPHE was applied at three concentrations—10, 20, and 30 µg GAE/mL of cell culture medium, corresponding approximately to 60, 120, and 180 µM GAE, which are commonly used to evaluate the biological activity of polyphenol-rich extracts in cell models [27]. These concentrations also correspond to about 1.4–4.3 mg of fresh pistachio per mL of culture medium, consistent with the range studied in previous research on the anti-inflammatory effects of Bronte pistachio extract in differentiated CaCo-2 cells exposed to IL-1β [14]. Importantly, these concentrations are below the levels likely to be found in the intestinal lumen after consuming a typical 28 g serving of pistachios, assuming an average gastrointestinal fluid volume of roughly 600 mL. A single extraction batch of BPHE was used for all cell culture treatments.
To simulate an (epi)genotoxic insult, CaCo-2 cells were exposed to sodium arsenite (NaAsO2, 10 µg/L of cell culture medium) for 72 h. This concentration is equivalent to the limit of arsenic in drinking water recommended by the World Health Organization’s guidelines [28]. The 72 h incubation period, as previously employed by Volpes et al. (2023) [15] in a similar study, was necessary to reveal the significant differences in DNA methylation, allowing for at least two cycles of cell replication. Three hours before the end of the As treatment, the cells were co-incubated with each concentration of BPHE.
As epigenotoxic positive control, the cells were exposed to 5-azacytidine (5-AzaC, 10 μM), a widely known molecule that acts preferentially on the complex DNMTs-S-adenosine methionine (SAM) affecting genomic DNA methylation status, for 48 h (with fresh addition at 24 h due to the low half-life of this molecule) [21]. Three hours before the end of the 5-AzaC treatment, the cells were co-incubated with each concentration of BPHE. BPHE, NaAsO2, and 5-AzaC were dissolved in cell culture medium at the desired concentration. Untreated cells were used as the negative control and are referred to as CTRL. The plan for all of the performed treatments is summarized in Table 1.

Table 1.
Cell treatments plan. BPHE: Bronte pistachio hydrophilic Extract; 5-AzaC: 5-azacytidine; GAE: gallic acid equivalents. The incubation times of the co-treatment with BPHE at various concentrations is indicated in parentheses; BPHE was added during the final hours of the overall treatment period, which lasted 48 or 72 h.
2.6. Genomic DNA Isolation
Isolation of genomic DNA from cells was carried out with the PureLink Genomic DNA Kit (Invitrogen, Renfrewshire, UK) as previously described [15] and DNAzol (Invitrogen®) following the manufacturer’s recommendations. The obtained DNA was quantified using a NanoDrop® ND-1000 (Thermo Fisher Scientific, Wilmington, NC, USA).
2.7. MeSAP-PCR-Based Assessment of Genomic DNA Methylation
To assess the genome-wide changes in DNA methylation status possibly induced by each (co-)treatment, methylation-sensitive arbitrarily primed PCR (MeSAP-PCR) assays were performed, as previously described [29], on genomic DNA extracted from all cell culture conditions (Table 1). Briefly, this assay consists of a PCR amplification using an arbitrary primer (5’-AACTGAAGCAGTGGCCTCGCG-3’), in which genomic DNA, previously endonuclease-digested, was used as a template. In particular, genomic DNA was firstly digested by RsaI restriction endonuclease (single-digested DNA, SD DNA) and then half the amount of the SD DNA was further digested by HpaII methylation-sensitive restriction endonuclease (double-digested DNA, DD DNA). The DNA fingerprinting, obtained by polyacrylamide gel (6%) electrophoresis of the PCR products, was subjected to a densitometric scanning and analyzed using SigmaGel software 1.0 (Jandel Scientific, San Rafael, CA, USA). The SD and DD scans were overlapped to compare any differences between them in terms of disappeared/appeared or intensified/attenuated bands. The greater the number of variations in the band pattern between the SD and DD DNA, the greater the degree of demethylation of the genomic DNA.
2.8. Omic Analyses
2.8.1. Illumina Sequencing and Bioinformatics Processing
The TruSeq Methyl Capture EPIC Library Prep Kit (Illumina, San Diego, CA, USA) was used for library preparation following the manufacturer’s instructions. Genomic DNA samples were quantified by Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). Final libraries were checked with both a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) and Agilent Bioanalyzer DNA assay (Santa Clara, CA, USA). Libraries were then prepared for sequencing and sequenced in the paired-end 150 bp mode on a NovaSeq6000 (Illumina, San Diego, CA, USA).
Processing of raw data for both format conversion and de-multiplexing was performed with the Bcl2Fastq 2.20 version of the Illumina pipeline. Raw paired-end sequencing reads were checked for quality using fastQC 0.11.8 (Brabham Institute, Cambridge, UK), and pre-processing was performed with bbduk v. 38.87 (JGI BBtools). Specifically, the pre-processing involved trimming adapters (k = 13, ktrim = r, mink = 11, tpe, tbo) and removing low-quality reads (maq = 10).
2.8.2. Methylation Analysis from Bisulfite-Sequencing
Methylation signals were obtained from duplicate analysis of next-generation sequencing (NGS) reads using Bismark v.0.22.3 [30]. Bowtie2 v. 2.3.5 [31] was used to map reads to the Bismark-prepared human genome (Homo sapiens GRCh38 primary assembly from ensembl.org). Alignments in the BAM format were sorted with samtools v. 1.10–3 [32]. Methylation signals were analyzed using the R packages methylKit [33], genomation [34], and Granges [35] with gene coordinates taken from the GenCode EBI ftp repository (human release 38). Promoters were defined as regions of 1000 bp before the gene transcription start. Transcription start sites (TSSs) were defined as small regions of 100 bp around the transcription start. Differential analyses between the aforementioned genomic regions (promoters and TSSs) in cells subjected to different treatments were performed using the calculateDiffMeth function of the MethylKit v. 0.99.2, retaining regions showing an adjusted p-value ≤ 0.01 and a difference in methylation ≥25%. All statistical analyses were also performed using Wilcoxon signed-rank test as previously reported [36]. Subsequently, the validation of the methylation data was checked using the bisulfite-specific pyrosequencing.
2.8.3. Intersection Analysis
Differentially methylated regions in the different treatments were compared through intersection analyses using the Venn command of the R package gplots v. 3.2.0 and with other R-base functions such as “table” and “grep”. Among the various possible identifiers for genes, gene symbols were chosen as default identifiers for the analysis.
2.8.4. Functional Analysis
Functional inference of differentially methylated regions were performed by an over-representation analysis using the R package clusterProfiler v3.13.0 [37] using pre-formed gene sets obtained from the Molecular Signature Database (MsigDb v. 7.4, Broad Institute, Cambridge, MA, USA) [38,39].
2.9. Analysis of Variation in Methylation in Some Structural/Functional Categories of Genes
From the general tables of genes showing a significant variation in the methylation status of their promoters, those belonging to some particularly important categories of structural and functional terms for the cells were extracted. The gene categories chosen are the following: pseudogenes, genes for zinc-finger proteins, genes for antisense RNA, genes for homeobox proteins, genes for kinases, and, lastly, genes for key crucial pathways of the cells, designated by us as “pathway-key genes (PKGs)” and synthetically defined as all genes that are crucial in fundamental pathways of the cell, such as basal metabolism, DNA repair, antioxidant activity, and support for fundamental genetic mechanisms. With these categories of specific targets, a complex overlapped histogram with two vertical axes and several colors were built to gain an integrated visual scenario regarding which and how structural/functional categories of genes vary their methylation under treatments and co-treatments. Moreover, all PKGs were compiled in a table with their proper official acronym, name, and specification of function, according to the literature.
3. Results
3.1. Content of Polyphenols and Radical Scavenging Activity of BPHE
The total polyphenol content (TPC) of BPHE was assessed using the Folin–Ciocalteu assay, while the radical scavenging action was estimated by the ABTS assay, a commonly used method for evaluating the antioxidant activity (AA) of plant-derived samples.
Since condensed tannins are particularly abundant in the hydrophilic extracts of pistachio and are responsible for the anti-inflammatory properties associated with these extracts, the total proanthocyanidin content (PAC) within the polyphenolic fraction was estimated using the DMAC colorimetric assay.
The TPC, PAC, and AA values of the BPHE are reported in Table 2.

Table 2.
Total polyphenol content (TPC), total proanthocyanidin content (PAC), and antioxidant activity (AA) of BPHE. Values are expressed as the mean ± SD of three experiments carried out in triplicate. GAE: gallic acid equivalents; CCE: cyanidin chloride equivalents; TE: Trolox equivalents; FW: fresh weight.
3.2. PCR-Based Assessment of BPHE-Induced Genomic DNA Methylation
MeSAP-PCR was performed to assess the changes in the genomic DNA methylation status possibly induced by each co-treatment compared to untreated CaCo-2 cells. The PCR carried out in this assay uses an arbitrary primer with a 3’ tail “CGCG”, which directs the amplification toward the CpG-rich sequences, such as those located in gene promoters. The DNA fingerprinting obtained from each condition of cells (co-)treatment was compared with the band pattern obtained from untreated cells (CTRL) to determine the number of variations, in terms of disappeared/appeared or intensified/attenuated bands. A higher number of band pattern variations in treated cells than in untreated ones denotes a genome-wide demethylation; on the contrary, a lower number of bands in treated cells denotes a more methylated status of the genomic DNA compared to the CTRL. The graph in Figure 1 shows the difference in the band pattern variations between untreated cells and each cell culture condition.
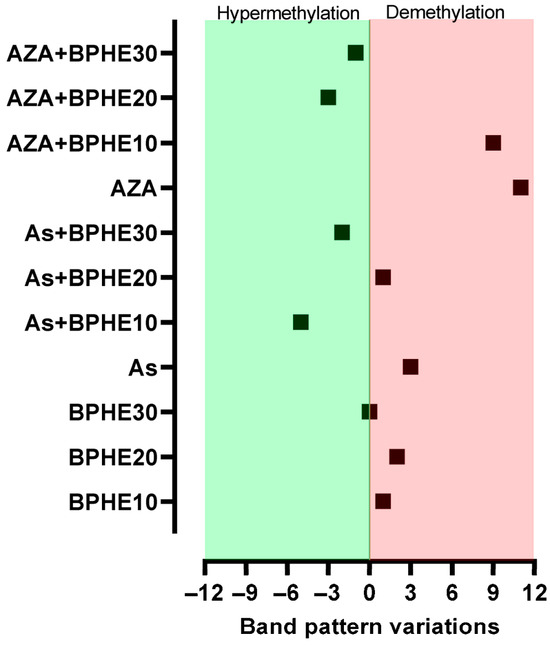
Figure 1.
Number of band pattern variations detected by MeSAP-PCR under the cell culture conditions reported in Table 1. The value for each sample represents the difference between the number of band pattern variations in each (co)-treatment and that of untreated cells.
From treatments with the three concentrations of BPHE (i.e., 10, 20, or 30 µg GAE/mL of cell culture medium), no significant variation in the band pattern of MeSAP-PCR was observed (Figure 1). Thus, no epigenomic action by BPHE was observed in the differentiated CaCo-2 cell line.
Treatment with BPHE Is Able to Decrease As-Induced DNA Demethylation in CaCo-2 Cells
Treatment with As caused the appearance/disappearance of bands in the DNA fingerprinting of SD and DD DNA relative to this treatment (Figure 2), a band pattern variation more consistent compared to those present in the untreated cells. Thus, the genomic DNA of As-treated cells were substantially demethylated compared to the CTRL condition. By inducing DNA demethylation, As confirmed its epigenotoxic insult in the CaCo-2 cells line [15]. However, when BPHE was added in the combined treatments (As + BPHE10, 20, and 30), it was able to restore a DNA methylation status similar to that of untreated cells or even to induce a hyper-methylation status (Figure 1). Thus, BPHE showed a modulation effect against the epigenetic damage induced by As in CaCo-2 cells.
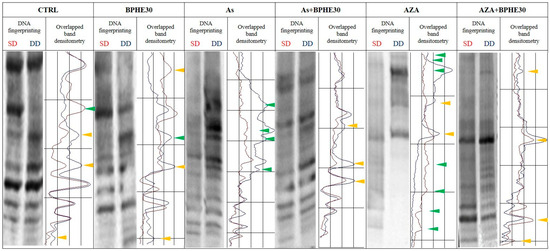
Figure 2.
Representative MeSAP-DNA fingerprinting and relative scanning densitometry, indicating genomic DNA methylation of untreated CaCo-2 cells (CTRL) or cells treated with BPHE 30 µg GAE/mL (BPHE30), NaAsO2 10 µg/L (As), NaAsO2 10 µg/L plus BPHE 30 µg GAE/mL (As + BPHE30), 5-AzaC 10 µM (AZA), and 5-AzaC 10 µM plus BPHE 30 µg GAE/mL (AZA + BPHE30). Band pattern variation, in terms of intensification/attenuation (yellow triangles) and appearance/disappearance (green triangles), was evaluated by densitometer scanning of single-digested DNA (SD, profile in red) compared with double-digested DNA (DD, profile in blue).
A similar protective behavior of BPHE, at 20 and 30 µg GAE/mL doses, has been shown toward the significant epigenetic insult by 5-AzaC, used as the demethylating positive control in the combined treatments AZA + BPHE20 and AZA + BPHE30 (Figure 1). Indeed, 5-AzaC-treated cells (AZA) showed nine band pattern variations, most of which involved the appearance/disappearance of bands (Figure 2), showing genome-wide demethylation with respect to the CTRL cells. In the combined treatments, BPHE was able to counteract 5-AzaC-induced DNA demethylation, showing a reduction in band pattern variations with respect to the AZA treatment.
3.3. Differential Promoter Methylation Analysis of Treated Cells from Omic Data
In order to exactly locate the position and identity of differential methylation sites (genes) in response to the treatments, we sequenced the DNA of the samples, as well as their counterparts, upon treatment with sodium bisulfite using NGS. Specifically, we investigated the promoter sequences and the TSSs searching for hyper- or hypo-methylation signals, indicating a reprogramming of chromatin accessibility in response to treatments.
Table 3 summarizes the results of the study by reporting the number of genes detected as differentially methylated (hyper-methylated or hypo-methylated) in the promoter regions in the different (co-)treatments with respect to the untreated control cells.

Table 3.
Number of genes resulting as differentially methylated (hyper- or hypo-methylated) in the promoter regions in the different cell (co-)treatments with respect to the untreated control cells (see Supplementary File S1 for the complete report).
The same results for the number of genes detected as differentially methylated in TSSs are reported in Supplementary File S2.
Our results for the differential methylation of gene promoters between the treated and untreated cells reveal that the most pronounced effect was observed for the treatment with BPHE 30 µg GAE/mL (BPHE30, 745 genes), followed by 5-AzaC (AZA, 705 genes) and sodium arsenite (As, 465 genes) treatments. The co-treatment with BPHE led to a supplement effect under both AZA (823 genes) and As (894 genes) conditions. Overall, we found higher methylation than demethylation effects, stronger in As (377 hyper- vs. 88 hypo-methylated genes) and BPHE30 (614 hyper- vs. 131 hypo-methylated genes) than in AZA (414 hyper- vs. 291 hypo-methylated genes). As expected, 5-AzaC was the strongest demethylating agent; however, when AZA + BPHE30 co-incubation is considered, the number of genes demethylated by 5-AzaC was reduced (from 291 in AZA to 147 in AZA + BPHE30). Treatment with As caused a moderate-to-low variation in the number of genes with different methylation compared to the other treatments, with the majority showing hyper-methylation. However, when As + BPHE30 co-treatment is considered, the number of hyper-methylated genes nearly doubles (from 377 in As to 746 in As + BPHE30), and the number of hypo-methylated genes also increases (from 88 in As to 148 in As + BPHE30) (Table 3).
In Figure 3, the Venn diagram presents the number of genes with differential methylation upon treatment with 5-AzaC, NaAsO2, or BPHE 30 µg GAE/mL versus the untreated control. As shown, this intersection analysis allowed for isolation of the specific contributions of the different treatments (see Supplementary File S3 for a complete list of affected genes). We found that the three treatments induced peculiar variations in the methylation levels of the gene promoters, with a slightly higher overlap between BPHE30 and As than between BPHE30 and AZA.
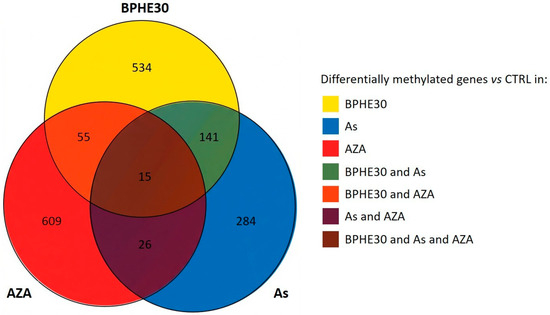
Figure 3.
Venn diagram representing the number of genes with evidence of differential methylation under BPHE30, As, or AZA treatment conditions vs. the untreated control.
Despite the modest overlap among the three treatments, functional analyses of genes peculiar to the different treatments revealed a common theme related to histone 3 tri-methylation at K27, with potential impacts on cell differentiation and proliferation [40].
The addition of BPHE 30 μg GAE/mL to NaAsO2 or 5-AzaC in the co-treatments (As + BPHE30 and AZA + BPHE, respectively) was further challenged against BPHE alone in order to decouple their effects. The residual pistachio effect was found to be higher (more than double) in As than in AZA and resulted in a large hyper-methylation (four times higher than hypo-methylation) (Figure 4, see Supplementary File S4 for a complete list of affected genes).
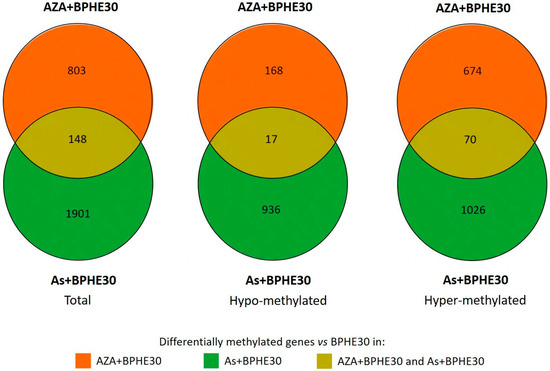
Figure 4.
Venn diagrams representing the number of genes with evidence of differential methylation (total, hypo-methylated, or hyper-methylated) in As + BPHE30 and AZA + BPHE30 co-treatments vs. BPHE30 single treatment.
3.4. Effects of Treatments and Co-Treatments on the Methylation of Structural/Functional Categories of Genes
We wanted to test, in detail, the variation in the number of differentially methylated promoters, with respect to the untreated control, within some categories of genes in the As and AZA treatments and in their respective co-treatments with BPHE 30 µg GAE/mL (As + BPHE30 and AZA + BPHE30). In particular, pseudogenes; genes for zinc-finger proteins, antisense RNA, homeobox proteins, microRNA, and kinases; and key genes of some pathways (called pathway-key genes, PKGs) were analyzed. The results are shown in Figure 5 and in Table 4, Table 5, Table 6 and Table 7. As shown, all treatments tended to increase methylation levels. Furthermore, most of the methylation variation, regardless of treatment and type of variation induced, is observed in pseudogenes.
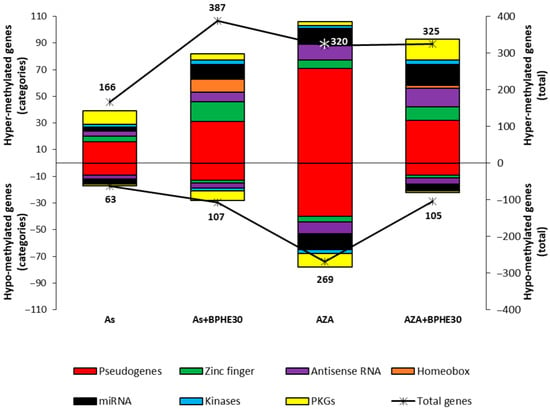
Figure 5.
Effects of the treatment with NaAsO2 (As) or 5-AzaC (AZA) and of the relative co-treatments with BPHE 30 µg GAE/mL (As + BPHE30 and AZA + BPHE30, respectively) on the methylation status of promoter regions of the structural/functional categories of genes (pseudogenes; genes for zinc-finger proteins, antisense RNA, homeobox proteins, miRNA, and kinases; and pathway-key genes (PKGs)). The chart presents the results as two overlapping graphs. The colored histograms correspond to the left vertical axis, indicating the number of differentially methylated (hyper- or hypo-methylated) genes within each specific structural/functional gene category. The black lines marked with asterisks correspond to the right vertical axis and represent the total number of hyper- or hypo-methylated genes for each individual treatment or co-treatment condition.

Table 4.
Pathway-key genes (PKGs) differentially methylated by 5-AzaC treatment (AZA) in CaCo-2 cells.

Table 5.
Pathway-key genes (PKGs) differentially methylated in AZA + BPHE30 co-treatment in CaCo-2 cells.

Table 6.
Pathway-key genes (PKGs) differentially methylated by NaAsO2 treatment (As) in CaCo-2 cells.

Table 7.
Pathway-key genes (PKGs) differentially methylated in As + BPHE30 co-treatment in CaCo-2 cells.
This analysis further confirms that 5-AzaC treatment (AZA) causes the most evident increase in demethylation, with 269 hypo-methylated genes recorded including an appreciable quantity of PKGs (Table 4).
When the cells were co-treated with 5-AzaC together with BPHE 30 µg GAE/mL (AZA + BPHE30), the important variations in both the hyper- and hypo-methylating sense already described for the treatment with 5-AzaC alone were reduced. It is noteworthy that the demethylated PKGs from the AZA treatment, as reported in Table 4, were not found among the list of those from the AZA + BPHE30 co-treatment. The PKGs for which the methylation was changed with the AZA + BPHE30 co-treatment are reported in Table 5.
Treatment with sodium arsenite (As) produced an appreciable increase in hyper-methylated PKGs and only one hypo-methylated PKG (Table 6).
Interestingly, when cells are co-treated with NaAsO2 together with BPHE 30 µg GAE/mL (As + BPHE30), the variation in differentially methylated genes assumes important numbers recording the maximum of hyper-methylated genes (387). In particular, there was an increase in the number of hyper-methylated genes for zinc-finger proteins, antisense RNA, homeobox proteins, and microRNA. Furthermore, this co-treatment varied the methylation of some PKGs (Table 7).
An integrated comparison was also made between two treatments or co-treatments in order to highlight some methylation variations in noteworthy genes. In particular, Figure 6 shows the number of variations in differentially methylated genes within structural/functional categories by comparing between the treatments As and As + BPHE30 and also between the co-treatments AZA + BPHE30 and As + BHPE30.
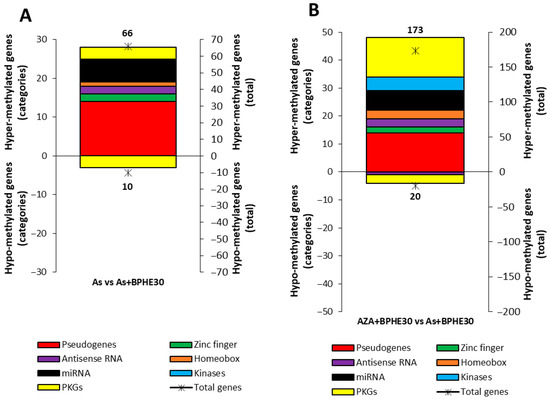
Figure 6.
Integrated methylation analysis of promotor regions of the structural/functional categories of genes (pseudogenes; genes for zinc-finger proteins, antisense RNA, homeobox proteins, miRNA, and kinases; and pathway-key genes (PKGs)) between the (A) As treatment and As + BPHE30 co-treatment and the (B) AZA + BPHE30 and As + BPHE30 co-treatments. The charts present the results as two overlapping graphs. The colored histograms correspond to the left vertical axis, indicating the number of differentially methylated (hyper- or hypo-methylated) genes within each specific structural/functional gene category. The black asterisks correspond to the right vertical axis and represent the total number of hyper- or hypo-methylated genes for each condition.
The PKGs detected by an integrated comparison between the As and As + BPHE30 treatments are reported in Table 8.

Table 8.
Pathway-key genes (PKGs) differentially methylated in the integrated comparison between the As treatment and As + BPHE30 co-treatment in CaCo-2 cells.
The PKGs detected by an integrated comparison between the AZA + BPHE30 and As + BPHE30 co-treatments are reported in Table 9.

Table 9.
Pathway-key genes (PKGs) differentially methylated in the integrated comparison between AZA + BPHE30 and As + BPHE30 co-treatments in CaCo-2 cells.
4. Discussion
Nutrigenomics, a modern branch of genetics and genomics, investigates how dietary components influence gene expression through epigenetic mechanisms. Understanding whether bioactive food molecules can exert beneficial effects—complementing conventional therapies—may offer added value in enhancing therapeutic outcomes with minimal additional effort. This becomes particularly relevant when such molecules exhibit genotoxicity-modulating properties, potentially counteracting the damage caused by harmful dietary components. In such cases, the same molecule may provide dual benefits by both supporting genome stability and mitigating genotoxic insults. Among various dietary elements, we focused on the Bronte pistachio (Pistacia vera L. var. Bronte), which represents a peculiarity of the Mediterranean diet, a plant that grows in the foothills region of the Etna volcano (Southern Italy). In a previous study, we carried out a comprehensive qualitative and quantitative chemical characterization of the phenolic compounds and fatty acids of this variety of Pistacia vera L. [11]. To assess the bioactive molecules content and the antioxidant properties of a hydrophilic extract obtained from Bronte pistachio (BPHE), we preliminarily evaluated its total polyphenol content (TPC) and its radical scavenging activity using an ABTS assay. Considering the known bioactivity of the proanthocyanidin fraction of hydrophilic pistachio extracts in experimental models [13,14], we also estimated the total proanthocyanidin content (PAC). The results reveal extremely high levels of antioxidant polyphenols in our BPHE (Table 2). Soxhlet extraction with an ethanol/water mixture (70:30) extracted polyphenols in quantities four times greater than those obtained using the maceration method with a methanol/water mixture (70:30), as reported in the literature [12], and this method also resulted in an extract with superior antioxidant activity. BPHE demonstrates superior radical scavenging activity, as indicated by a higher ABTS value, compared to the extract obtained through maceration with methanol/water. This suggests that Soxhlet extraction not only improves the yield of polyphenols but also maintains, or potentially enhances, their antioxidant efficacy. We based our evaluation on total phenolic content (TPC) and proanthocyanidins (PACs), which are widely recognized in the literature as major contributors to epigenetic modulation [109,110]. These compounds influence chromatin accessibility and the activity of epigenetic enzymes, such as DNMTs and HDACs. Thus, we evaluated the nutrigenomic effect and the ability to modulate genotoxic damage by BPHE, and we did so according to two parallel methods applied to human enterocytes (differentiated CaCo-2 cells). Notably the concentrations of BPHE used here support the physiological relevance of our findings.
At first, we used an epigenetic approach in order to evaluate the DNA methylation status of cells treated with three different concentrations of BPHE compared to molecules with a known epigenotoxic effect on DNA methylation; subsequently, we determined the methylome of the same cells, choosing the higher dose of BPHE, with an omic approach. In particular, we performed co-treatments with BPHE and arsenic, normally contained in drinking water, to investigate whether the pistachio extract also has the ability to modulate genotoxicity.
In order to evaluate the effect of BPHE, alone or in combination with genotoxic molecules (As or 5-AzaC), on DNA methylation, we preliminary used MeSAP-PCR. This assay is able to define DNA methylation status at a genome-wide level, although it is directed, in particular, toward the CpG islands thanks to the 3’ tail “CGCG” of the arbitrary primer used in the amplification step. As expected, this assay revealed that BPHE (10, 20, and 30 µg GAE/mL of cell culture medium) did not have an effect on the DNA methylation of differentiated CaCo-2 cells, maintaining a condition similar to that of untreated cells; BPHE 30 µg GAE/mL, in particular, resulted in exactly the same methylation condition of the untreated cells (Figure 1). It is known that As induces genomic instability [111], and genomic instability has also been reported to be associated with genomic demethylation of the DNA of unstable cells [112]. Here, the treatment with NaAsO2 resulted in a certain degree of demethylation of genomic DNA compared to untreated cells (Figure 1), also configuring As as an epigenotoxic agent, as already demonstrated by us [15]. In the combined treatments, when BPHE was added at the end of the incubation period with As, the DNA demethylation As induced was recovered. Furthermore, BPHE induced hyper-methylation at 10 and 30 µg GAE/mL doses (Figure 1). Thus, BPHE demonstrated the ability to modulate the epigenotoxic damage caused by As in CaCo-2 cells. The known demethylating agent 5-AzaC, used as the epigenotoxic positive control, induced in differentiated CaCo-2 cells a significant DNA demethylation, also in combination with BPHE 10 µg GAE/mL (Figure 1). However, higher BPHE doses added in combination with 5-AzaC induced DNA hyper-methylation. In particular, BPHE 30 µg GAE/mL induced a similar effect on DNA hyper-methylation when added after both As and 5-AzaC (Figure 1). These results suggest a protective effect of Bronte pistachio extract on the insults of dangerous molecules commonly present in our diet, such as arsenic.
To better investigate the epigenetic effect of BPHE and its activity toward DNA methylation alterations, we used an omic approach to define the methylomic signature of the differentiated CaCo-2 cell line treated with BPHE, alone or in combination with As or 5-AzaC. For this purpose, we chose only the highest BPHE dose—30 µg GAE/mL—for the cell culture medium, as it was the one for which the MeSAP-PCR showed a DNA methylation status more similar to that of the untreated cells.
As can be seen in Table 3, BPHE changed the methylation status of 745 gene promoters compared to the untreated control cells, most of which were hyper-methylated. Similarly, As mainly showed a hyper-methylating effect on promoter regions of differentiated CaCo-2 cells, although for a smaller number of genes (about 50%) compared to those hyper-methylated by BPHE alone. Previous studies from other laboratories, performed on tumoral CaCo-2 cells, have reported that As has a similar hyper-methylating effect [113]. Since we cannot make a comparison with other data on differentiated CaCo-2 cells, we can only hypothesize that the effect of As on DNA methylation is independent from the condition of genomic instability of CaCo-2 cells. Our data are the first to report on differentiated CaCo-2 cells, considered as normal enterocytes in vitro. Although MeSAP-PCR on As-treated cells has shown a certain degree of DNA demethylation, at the omic level, the As treatment resulted in the genome of differentiated CaCo-2 cells having a higher overall number of hyper-methylated regions than that of the control cells. Indeed, as shown in Figure 3 and Figure 4, the As + BPHE30 co-treatment exhibited a synergistic action: the number of hyper-methylated gene promoters increased compared to that of the individual treatments, indicating that, under these specific conditions and in this experimental system, As and BPHE have similar and additive effects. The discrepancy between the results of the MeSAP-PCR and the methylome evaluation, both for the BPHE30 and As treatments, can be attributed to the inherent differences between the two methodologies in terms of resolution, genomic coverage, and sensitivity to methylation patterns. MeSAP-PCR relies on methylation-sensitive restriction enzymes that recognize specific sequences whose cleavage is inhibited by methylation. Consequently, it provides a snapshot of methylation changes at selected genomic loci, often enriched in repetitive or highly methylated regions (e.g., CpG islands), but it lacks nucleotide-level resolution and may miss methylation changes occurring in non-CpG contexts. In contrast, next-generation bisulfite sequencing enables single-base resolution mapping of methylated cytosines across a broader portion of the genome, offering a much more comprehensive view of DNA methylation patterns. Therefore, the methylome fraction assessed by MeSAP-PCR may not accurately represent the entire methylome, as revealed by omic analysis, but rather a focus of the methylation status of the genomic CpG islands. Considering this, the observed discrepancies between the two methodologies do not necessarily reflect contradictory biological results but rather highlight the methodological and interpretative differences between targeted enzymatic fingerprinting and genome-wide methylation profiling. Integrating both approaches, as performed in the present study, can provide complementary insights into DNA methylation dynamics, particularly when interpreting complex or subtle epigenetic modifications.
Regarding the AZA treatment, if, on the one hand, 5-AzaC is confirmed as a molecule capable of greatly influencing genomic DNA methylation, on the other hand, it shows its specific and atypical behavior. As it is possible to observe, in this normaloid cellular system, 5-AzaC shows a pleiotropic behavior causing both an increase and decrease in the methylation of gene promoters. However, this action is not perfectly balanced, and it can be said that 5-AzaC induces an increase in methylation in a slightly higher number of genes. Probably, also for 5-AzaC, it is also important to consider the normal-like nature of differentiated CaCo-2 cells, particularly with regard to their baseline level of DNA methylation. Interestingly, when the cells were co-treated with 5-AzaC and BPHE, the number of differentially methylated genes increased with a clear prevalence of hyper-methylated ones. It seems that BPHE enforces the slight hyper-methylating action carried out by 5-AzaC alone.
Even more interesting are the insights that can be drawn from methylome analysis focused on specific structural/functional categories of genes. As shown in Figure 5, the upper part of the figure appears more expanded compared to the lower part, indicating that all the treatments cause a greater increase in the number of hyper-methylated genes than demethylated ones. Moreover, by following the red color, it becomes immediately apparent that most of the methylation changes—regardless of the treatment or the type of variation—are concentrated in pseudogenes. There are currently few contributions in the literature regarding the relationship between methylation of pseudogenes and phenotypic outcomes. While it can be hypothesized that pseudogene methylation plays a role in cell genome managing, assigning biological relevance to this finding is challenging, given that many pseudogenes are known to be not expressed. However, as reported by Kovalenko et al. (2021) [114], some pseudogenes, like PTENP1, were transcribed with the aim of forming long noncoding RNAs able to regulate gene expression. In our experiments, we observed a massive modification of the methylation pattern of pseudogenes. This evidence offers fascinating insights into the putative indirect power of dietary molecules that affect not only the known activity of regulating the DNA methylation of specific genes but also the still unknown regulation of gene expression related to pseudogenes. On the other hand, it is important to remember that pseudogenes are non-functional genes and that non-functionality can, in general, be linked to both silencing gene mutations and epigenetic modifications that inhibit expression.
The addition of BPHE in the co-incubation with As increases the number of methylated genes for zinc-finger proteins, homeobox proteins, and miRNA, as well as distributes the number of methylated/demethylated PKGs differently. It is important to remember that DNA methylation is closely associated with miRNA expression, and, in turn, miRNA genes regulate DNA methylation by targeting DNMT mRNAs [55]. Moreover, it may be considered that re-expression, out of morphogenesis, of HOX genes is associated with different solid tumors and that genes for homeobox proteins are known as widespread transcriptional regulators. In particular, CpG island hyper-methylation in several HOX genes contributes to the silencing of DNA repair genes with detrimental effects on genomic stability that can lead to oncogenic transformation, including in colorectal cancer [115]. In this complex scenario, our data demonstrate that BPHE greatly modulates all this gene subset and provides insight into the integration rate of its effects on the entire epigenome.
Regarding the co-treatment with 5-AzaC or As together with BPHE, the most evident data show that the addition of BPHE significantly increases the number of methylated PKGs. Attempting to draw conclusions from this intricate network of methylation and demethylation can be arduous, especially regarding the nutrigenomic role of BPHE. Nevertheless, looking at Table 4, Table 5, Table 6 and Table 7, which report the PKGs for each treatment or co-treatment, some conclusions emerge. It can be noted that 5-AzaC alone demethylated genes of the (i) RAS-family, (ii) histone deacetylase, (iii) Src family kinases (indirectly), and (iv) a protein involved in cell migration, adhesion, and in cytoskeleton management (Table 4). These genes, the expression of which is probably unblocked by 5-AzaC-induced demethylation, can increase genomic instability and could promote a tumoral transformation. Co-treatment AZA + BPHE30, which hyper-methylates three genes implicated in DNA repair, the pentose phosphate pathway, and xenobiotic detoxification, was, notably, also able to putatively block the expression of genes involved in autophagy, genotoxic-stress inducement, genomic instability, tumor pathogenesis, and dysfunction of peroxisomes, as well as promoting hypo-methylation of a gene coding for a cell-cycle regulator (Table 7). In parallel, while As alone hyper-methylated and verisimilarly blocked genes implicated in MEOS detoxification, redox management, autophagy, and assembly of repair DNA polymerase (Table 5), the co-presence of As and BPHE, interestingly, hypo-methylated genes coding a tumor-suppressor gene, cytochrome c oxidase, aldehyde dehydrogenase with known tumor-suppressive properties, and kinesin required for spindle assembly and chromosome movement (Table 6). From both the co-treatments, it can be deduced that the presence of BPHE could epigenetically modulate the gene network expression of differentiated CaCo-2 cells toward a better landscape, decreasing all conditions that are prodromal to genomic instability, cancer, and, in general, to a genotoxic condition. Clearly, further functional gene expression studies have to be performed to move from a hypothesis to the certainty that these methylomic changes are reflected in changes in gene expression.
The omic nature of our data also offers the mathematic possibility of conducting an integrated comparison between the two classes of data coming from the treatments or co-treatments to highlight some methylation variations in noteworthy genes. It is possible to overlap the cohorts of data regarding the As treatment and AZA + BPHE30 co-treatment versus the As + BHPE30 co-treatment. Figure 6 shows the number variations in differentially methylated genes within specific structural/functional categories via a comparison between the As treatment and As + BPHE30 and also between the AZA + BPHE30 and As + BHPE30 co-treatments. In Figure 6A, in particular, the comparison of As vs. As + BPHE30 highlights the behavior of BPHE: almost the entire distribution is positioned in the upper part of the histogram, showing that BHPE mainly had a methylating effect. Table 8, in addition, reports that the PKGs involved in changing the methylation in this integrated analysis were all genes not particularly important for enterocyte cell functions. Similarly, Figure 6B, for the comparison of AZA + BPHE30 versus As + BHPE30, highlights the different behaviors of 5-AzaC and As. In addition to the large number of hyper-methylated pseudogenes, it is worth noting that there were a large number, similar to pseudogenes, of hyper-methylated PKGs, greater in number than the corresponding hypo-methylated ones. These results confirm the methylating power of As and underline the ineffectiveness of 5-AzaC in counteracting this effect. Table 8 and Table 9 provide further details regarding the gene activities involved in these integrated analyses; it should be noted that a DNA repair gene and a gene for nucleotide synthesis are hyper-methylated, while genes for cell adhesion and histone deacetylase are hypo-methylated.
Intestinal epithelial cells are naturally exposed to continuous and repetitive stress, often of genotoxic origin, and it is possible that compounds derived from pistachio could exert a compensatory effect that supports cellular health. The present findings suggest that BPHE exerts a protective epigenetic effect against genotoxic insult on differentiated CaCo-2 cells. Although these preliminary in vitro findings are promising, further investigations are needed to confirm this protective effect in vivo and, ideally, in human models. Similar effects could also be explored for other dietary bioactives, particularly those characteristic of the Mediterranean diet. Such research may contribute to a better understanding of the diet’s established protective role against gastrointestinal diseases, including cancer.
5. Conclusions
In this study Bronte pistachio nuts were used to obtain a hydrophilic extract (BPHE) particularly rich in polyphenols with a strong antioxidant and radical scavenging activity. Our BPHE was tested in vitro on a model of human intestinal epithelium (differentiated CaCo-2 cells) in order to assess its ability to modulate DNA at the epigenetic level and identify the methylomic signature of genomic DNA after an epigenotoxic insult induced by arsenic.
Both BPHE and As induce a predominantly hyper-methylating effect on gene promoter regions, and, interestingly, their combination exerts a synergistic increase in DNA hyper-methylation, highlighting that BPHE is able to modulate the methylation changes induced by As. A gene category-specific analysis revealed that most of the methylation changes occurred in pseudogenes and that the co-treatment with BPHE and As increased the number of methylated genes for zinc-finger proteins, homeobox proteins, miRNA, and antisense RNA. Regarding PKGs, while As alone hyper-methylates genes implicated in MEOS detoxification, redox managing, autophagy, and assembly of repair DNA polymerase, the co-presence of As and BPHE hypo-methylated a subset of critical genes involved in tumor suppression, detoxification, mitochondrial function, and cell division. These findings suggest that BPHE exerts a protective epigenetic influence on the gene network expression of differentiated CaCo-2 cells, restoring a more balanced methylation landscape and potentially reducing the risk of genomic instability and related pathologies, including cancer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nu17162678/s1. File S1: differential_analysis_promoter; File S2: differential_analysis_TSS; File S3: venn_AZA_As_BPHE30; File S4: venn_BPHE30_effect_on_As_and_AZA; Table S1: Number of genes resulting as differentially methylated (hyper- or hypo-methylated) in transcription start sites (TSSs) with the different cell (co-)treatments with respect to the untreated control cells (see Supplementary File S2 for the complete report).
Author Contributions
Conceptualization, F.C.; methodology, I.C., F.N., M.R., C.G. and F.C.; software, M.R.; validation, I.C., F.N., M.R. and F.C.; formal analysis, I.C., F.N. and M.R.; investigation, I.C., F.N., A.P. and C.G.; resources, F.M.; data curation, M.R.; writing—original draft preparation, I.C., F.N., M.R., C.G. and F.C.; writing—review and editing, P.S.C., S.V., L.G., G.S., M.R., C.G. and F.C.; visualization, M.R.; supervision, C.G. and F.C.; funding acquisition, F.M. and F.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was also partially funded by (i) proceeds from a research assignment given to F.C. and C.G. by a private Italian company in the agri-food sector; (ii) grants from the University of Palermo (FFR 2024–2025 to F.N., C.G., and F.C.); (iii) National Biodiversity Future Center (identification code: CN00000033, CUP B73C22000790001) on “Biodiversity”, financed under the National Recovery and Resilience Plan (NRRP), Mission 4, Component 2, Investment 1.4, “Strengthening of Research Structures and Creation of R&D ‘National Champions’ on Some Key Enabling Technologies”—Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of the Italian Ministry of University and Research, funded by the European Union—NextGenerationEU; and award number project code: CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, CUP B73C22000790001, Project title “National Biodiversity Future Center—NBFC” (FC).
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors are grateful to Prof. Antonino Lauria for his support in obtaining the hydrophilic extract of Bronte pistachio. Special thanks also to Fondazione Umberto Veronesi.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 5-AzaC | 5-Azacytidine |
| AA | Antioxidant activity |
| As | Arsenic |
| BPHE | Bronte pistachio hydrophilic extract |
| CC | Cyanidin chloride |
| CCE | Cyanidin chloride equivalents |
| DD | Double-digested DNA |
| FW | Fresh weight |
| GA | Gallic acid |
| GAE | Gallic acid equivalents |
| MD | Mediterranean diet |
| MeSAP-PCR | Methylation-sensitive arbitrarily primed PCR |
| NGS | Next-generation sequencing |
| PAC | Total proanthocyanidin content |
| PKGs | Pathway-key genes |
| SD | Single-digested DNA |
| TE | Trolox equivalent |
| TPC | Total polyphenol content |
| TSSs | Transcription start sites |
References
- Bordoni, L.; Gabbianelli, R. Primers on Nutrigenetics and Nutri(Epi)Genomics: Origins and Development of Precision Nutrition. Biochimie 2019, 160, 156–171. [Google Scholar] [CrossRef]
- Caradonna, F.; Consiglio, O.; Luparello, C.; Gentile, C. Science and Healthy Meals in the World: Nutritional Epigenomics and Nutrigenetics of the Mediterranean Diet. Nutrients 2020, 12, 1748. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Palika, R.; Ismail, A.; Pullakhandam, R.; Reddy, G.B. Nutrigenomics: Opportunities & Challenges for Public Health Nutrition. Indian J. Med. Res. 2018, 148, 632–641. [Google Scholar] [CrossRef]
- Caradonna, F.; Cruciata, I.; Luparello, C. Nutrigenetics, Nutrigenomics and Phenotypic Outcomes of Dietary Low-Dose Alcohol Consumption in the Suppression and Induction of Cancer Development: Evidence from in Vitro Studies. Crit. Rev. Food Sci. Nutr. 2022, 62, 2122–2139. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sánchez, M.L.; García-Vigara, A.; Hidalgo-Mora, J.J.; García-Pérez, M.-Á.; Tarín, J.; Cano, A. Mediterranean Diet and Health: A Systematic Review of Epidemiological Studies and Intervention Trials. Maturitas 2020, 136, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Naselli, F.; Tesoriere, L.; Caradonna, F.; Bellavia, D.; Attanzio, A.; Gentile, C.; Livrea, M.A. Anti-Proliferative and pro-Apoptotic Activity of Whole Extract and Isolated Indicaxanthin from Opuntia Ficus-Indica Associated with Re-Activation of the Onco-Suppressor p16INK4a Gene in Human Colorectal Carcinoma (Caco-2) Cells. Biochem. Biophys. Res. Commun. 2014, 450, 652–658. [Google Scholar] [CrossRef]
- Naselli, F.; Belshaw, N.J.; Gentile, C.; Tutone, M.; Tesoriere, L.; Livrea, M.A.; Caradonna, F. Phytochemical Indicaxanthin Inhibits Colon Cancer Cell Growth and Affects the DNA Methylation Status by Influencing Epigenetically Modifying Enzyme Expression and Activity. Lifestyle Genom. 2015, 8, 114–127. [Google Scholar] [CrossRef]
- Ragusa, M.A.; Naselli, F.; Cruciata, I.; Volpes, S.; Schimmenti, C.; Serio, G.; Mauro, M.; Librizzi, M.; Luparello, C.; Chiarelli, R.; et al. Indicaxanthin Induces Autophagy in Intestinal Epithelial Cancer Cells by Epigenetic Mechanisms Involving DNA Methylation. Nutrients 2023, 15, 3495. [Google Scholar] [CrossRef]
- Abbasifard, M.; Jamialahmadi, T.; Reiner, Ž.; Eid, A.H.; Sahebkar, A. The Effect of Nuts Consumption on Circulating Oxidized Low-density Lipoproteins: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Phytother. Res. 2023, 37, 1678–1687. [Google Scholar] [CrossRef] [PubMed]
- Mandalari, G.; Barreca, D.; Gervasi, T.; Roussell, M.A.; Klein, B.; Feeney, M.J.; Carughi, A. Pistachio Nuts (Pistacia vera L.): Production, Nutrients, Bioactives and Novel Health Effects. Plants 2021, 11, 18. [Google Scholar] [CrossRef]
- Mannino, G.; Gentile, C.; Maffei, M.E. Chemical Partitioning and DNA Fingerprinting of Some Pistachio (Pistacia vera L.) Varieties of Different Geographical Origin. Phytochemistry 2019, 160, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Gentile, C.; Tesoriere, L.; Butera, D.; Fazzari, M.; Monastero, M.; Allegra, M.; Livrea, M.A. Antioxidant Activity of Sicilian Pistachio (Pistacia vera L. Var. Bronte) Nut Extract and Its Bioactive Components. J. Agric. Food Chem. 2007, 55, 643–648. [Google Scholar] [CrossRef]
- Gentile, C.; Allegra, M.; Angileri, F.; Pintaudi, A.M.; Livrea, M.A.; Tesoriere, L. Polymeric Proanthocyanidins from Sicilian Pistachio (Pistacia vera L.) Nut Extract Inhibit Lipopolysaccharide-Induced Inflammatory Response in RAW 264.7 Cells. Eur. J. Nutr. 2012, 51, 353–363. [Google Scholar] [CrossRef]
- Gentile, C.; Perrone, A.; Attanzio, A.; Tesoriere, L.; Livrea, M.A. Sicilian Pistachio (Pistacia vera L.) Nut Inhibits Expression and Release of Inflammatory Mediators and Reverts the Increase of Paracellular Permeability in IL-1β-Exposed Human Intestinal Epithelial Cells. Eur. J. Nutr. 2015, 54, 811–821. [Google Scholar] [CrossRef]
- Volpes, S.; Cruciata, I.; Ceraulo, F.; Schimmenti, C.; Naselli, F.; Pinna, C.; Mauro, M.; Picone, P.; Dallavalle, S.; Nuzzo, D.; et al. Nutritional Epigenomic and DNA-Damage Modulation Effect of Natural Stilbenoids. Sci. Rep. 2023, 13, 658. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Patlolla, A.K.; Centeno, J.A. Invited Reviews: Carcinogenic and Systemic Health Effects Associated with Arsenic Exposure—A Critical Review. Toxicol. Pathol. 2003, 31, 575–588. [Google Scholar] [CrossRef]
- Tapio, S.; Grosche, B. Arsenic in the Aetiology of Cancer. Mutat. Res. Mutat. Res. 2006, 612, 215–246. [Google Scholar] [CrossRef]
- Wang, C.-H.; Hsiao, C.K.; Chen, C.-L.; Hsu, L.-I.; Chiou, H.-Y.; Chen, S.-Y.; Hsueh, Y.-M.; Wu, M.-M.; Chen, C.-J. A Review of the Epidemiologic Literature on the Role of Environmental Arsenic Exposure and Cardiovascular Diseases. Toxicol. Appl. Pharmacol. 2007, 222, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Guzman, E.; Langowski, J.L.; Owen-Schaub, L. Mad Dogs, Englishmen and Apoptosis: The Role of Cell Death in UV-Induced Skin Cancer. Apoptosis 2003, 8, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Fekete, E.E.; Wang, A.; Creskey, M.; Cummings, S.E.; Lavoie, J.R.; Ning, Z.; Li, J.; Figeys, D.; Chen, R.; Zhang, X. Multilevel Proteomic Profiling of Colorectal Adenocarcinoma Caco-2 Cell Differentiation to Characterize an Intestinal Epithelial Model. J. Proteome Res. 2024, 23, 2561–2575. [Google Scholar] [CrossRef]
- Caradonna, F.; Cruciata, I.; Schifano, I.; La Rosa, C.; Naselli, F.; Chiarelli, R.; Perrone, A.; Gentile, C. Methylation of Cytokines Gene Promoters in IL-1β-Treated Human Intestinal Epithelial Cells. Inflamm. Res. 2018, 67, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Rossi, J.A. Colorimetry of Total Phenolics with Phosphomolybdic-Phosphotungstic Acid Reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar] [CrossRef]
- Mannino, G.; Perrone, A.; Campobenedetto, C.; Schittone, A.; Margherita Bertea, C.; Gentile, C. Phytochemical Profile and Antioxidative Properties of Plinia Trunciflora Fruits: A New Source of Nutraceuticals. Food Chem. 2020, 307, 125515. [Google Scholar] [CrossRef] [PubMed]
- Porter, L.J.; Hrstich, L.N.; Chan, B.G. The Conversion of Procyanidins and Prodelphinidins to Cyanidin and Delphinidin. Phytochemistry 1985, 25, 223–230. [Google Scholar] [CrossRef]
- Fellegrini, N.; Ke, R.; Yang, M.; Rice-Evans, C. [34] Screening of Dietary Carotenoids and Carotenoid-Rich Fruit Extracts for Antioxidant Activities Applying 2,2’-Azinobis(3-Ethylenebenzothiazoline-6-Sulfonic Acid Radical Cation Decolorization Assay. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1999; Volume 299, pp. 379–389. ISBN 978-0-12-182200-2. [Google Scholar]
- Mannino, G.; Serio, G.; Asteggiano, A.; Gatti, N.; Bertea, C.M.; Medana, C.; Gentile, C. Bioactive Compounds and Antioxidant Properties with Involved Mechanisms of Eugenia Involucrata DC Fruits. Antioxidants 2022, 11, 1769. [Google Scholar] [CrossRef]
- Amatori, S.; Mazzoni, L.; Alvarez-Suarez, J.M.; Giampieri, F.; Gasparrini, M.; Forbes-Hernandez, T.Y.; Afrin, S.; Errico Provenzano, A.; Persico, G.; Mezzetti, B.; et al. Polyphenol-Rich Strawberry Extract (PRSE) Shows in Vitro and in Vivo Biological Activity against Invasive Breast Cancer Cells. Sci. Rep. 2016, 6, 30917. [Google Scholar] [CrossRef]
- Frisbie, S.H.; Mitchell, E.J. Arsenic in Drinking Water: An Analysis of Global Drinking Water Regulations and Recommendations for Updates to Protect Public Health. PLoS ONE 2022, 17, e0263505. [Google Scholar] [CrossRef]
- Naselli, F.; Catanzaro, I.; Bellavia, D.; Perez, A.; Sposito, L.; Caradonna, F. Role and Importance of Polymorphisms with Respect to DNA Methylation for the Expression of CYP2E1 Enzyme. Gene 2014, 536, 29–39. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A Flexible Aligner and Methylation Caller for Bisulfite-Seq Applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. methylKit: A Comprehensive R Package for the Analysis of Genome-Wide DNA Methylation Profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef]
- Akalin, A.; Franke, V.; Vlahoviček, K.; Mason, C.E.; Schübeler, D. Genomation: A Toolkit to Summarize, Annotate and Visualize Genomic Intervals. Bioinformatics 2015, 31, 1127–1129. [Google Scholar] [CrossRef]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Ragusa, M.A.; Gentile, C.; Nicosia, A.; Costa, S.; Volpes, S.; Greco, L.; Naselli, F.; Caradonna, F. Epigenetic Remodeling of Regulatory Regions by Indicaxanthin Suggests a Shift in Cell Identity Programs in Colorectal Cancer Cells. Int. J. Mol. Sci. 2025, 26, 6072. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Shahhosseini, A.; Bourova-Flin, E.; Derakhshan, S.; Aminishakib, P.; Goudarzi, A. High Levels of Histone H3 K27 Acetylation and Tri-Methylation Are Associated with Shorter Survival in Oral Squamous Cell Carcinoma Patients. BioMedicine 2023, 13, 22–38. [Google Scholar] [CrossRef]
- Muranova, L.K.; Vostrikova, V.M.; Shatov, V.M.; Sluchanko, N.N.; Gusev, N.B. Interaction of the C-Terminal Immunoglobulin-like Domains (Ig 22–24) of Filamin C with Human Small Heat Shock Proteins. Biochimie 2024, 219, 146–154. [Google Scholar] [CrossRef]
- Cao, D.-L.; Ma, L.-J.; Jiang, B.-C.; Gu, Q.; Gao, Y.-J. Cytochrome P450 26A1 Contributes to the Maintenance of Neuropathic Pain. Neurosci. Bull. 2024, 40, 293–309. [Google Scholar] [CrossRef] [PubMed]
- Sternisha, S.M.; Miller, B.G. Molecular and Cellular Regulation of Human Glucokinase. Arch. Biochem. Biophys. 2019, 663, 199–213. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J. WARP: A Unique Extracellular Matrix Component of Cartilage, Muscle, and Endothelial Cell Basement Membranes. Anat. Rec. 2020, 303, 1619–1623. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Lee, H.Y. Rab25 and RCP in Cancer Progression. Arch. Pharm. Res. 2019, 42, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Pihlström, S.; Richardt, S.; Määttä, K.; Pekkinen, M.; Olkkonen, V.M.; Mäkitie, O.; Mäkitie, R.E. SGMS2 in Primary Osteoporosis with Facial Nerve Palsy. Front. Endocrinol. 2023, 14, 1224318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Teng, Z.; Wang, Z.; Zhu, P.; Wang, Z.; Liu, F.; Liu, X. Human Umbilical Cord Mesenchymal Stem Cells (hUC-MSCs) Alleviate Paclitaxel-Induced Spermatogenesis Defects and Maintain Male Fertility. Biol. Res. 2023, 56, 47. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Tan, Z.; Li, Q.; Fan, W.; Chen, G.; Bin, Y.; Zhou, Y.; Yi, J.; Luo, X.; Tan, J.; et al. Combined Analysis of Expression Profiles in a Mouse Model and Patients Identified BHMT2 as a New Regulator of Lipid Metabolism in Metabolic-Associated Fatty Liver Disease. Front. Cell Dev. Biol. 2021, 9, 741710. [Google Scholar] [CrossRef]
- Catalanotto, M.; Vaz, J.M.; Abshire, C.; Youngblood, R.; Chu, M.; Levine, H.; Jolly, M.K.; Dragoi, A.-M. Dual Role of CASP8AP2/FLASH in Regulating Epithelial-to-Mesenchymal Transition Plasticity (EMP). Transl. Oncol. 2024, 39, 101837. [Google Scholar] [CrossRef]
- Tolleson, C.; Claassen, D. The Function of Tyrosine Hydroxylase in the Normal and Parkinsonian Brain. CNS Neurol. Disord—Drug Targets 2012, 11, 381–386. [Google Scholar] [CrossRef]
- Ju, H.; Lim, B.; Kim, M.; Kim, Y.S.; Kim, W.H.; Ihm, C.; Noh, S.-M.; Han, D.S.; Yu, H.-J.; Choi, B.Y.; et al. A Regulatory Polymorphism at Position -309 in PTPRCAP Is Associated with Susceptibility to Diffuse-Type Gastric Cancer and Gene Expression. Neoplasia 2009, 11, 1340–1347. [Google Scholar] [CrossRef]
- Janssen, W.J.M.; Grobarova, V.; Leleux, J.; Jongeneel, L.; Van Gijn, M.; Van Montfrans, J.M.; Boes, M. Proline-Serine-Threonine Phosphatase Interacting Protein 1 (PSTPIP1) Controls Immune Synapse Stability in Human T Cells. J. Allergy Clin. Immunol. 2018, 142, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Noh, E.J.; Lim, D.-S.; Lee, J.-S. A Novel Role for Methyl CpG-Binding Domain Protein 3, a Component of the Histone Deacetylase Complex, in Regulation of Cell Cycle Progression and Cell Death. Biochem. Biophys. Res. Commun. 2009, 378, 332–337. [Google Scholar] [CrossRef]
- Žižić, M.; Atlagić, K.; Karaman, M.; Živić, M.; Stanić, M.; Maksimović, V.; Zakrzewska, J. Uptake of Vanadium and Its Intracellular Metabolism by Coprinellus truncorum Mycelial Biomass. J. Trace Elem. Med. Biol. 2024, 83, 127381. [Google Scholar] [CrossRef]
- Yang, M.; Hu, X.; Tang, B.; Deng, F. Exploring the Interplay between Methylation Patterns and Non-Coding RNAs in Non-Small Cell Lung Cancer: Implications for Pathogenesis and Therapeutic Targets. Heliyon 2024, 10, e24811. [Google Scholar] [CrossRef]
- Zhang, S.; Huangfu, H.; Zhao, Q.; Li, Y.; Wu, L. Downregulation of Long Noncoding RNA HCP5/miR-216a-5p/ZEB1 Axis Inhibits the Malignant Biological Function of Laryngeal Squamous Cell Carcinoma Cells. Front. Immunol. 2022, 13, 1022677. [Google Scholar] [CrossRef]
- Xiao, Y.; Wei, R.; Yuan, Z.; Lan, X.; Kuang, J.; Hu, D.; Song, Y.; Luo, J. Rutin Suppresses FNDC1 Expression in Bone Marrow Mesenchymal Stem Cells to Inhibit Postmenopausal Osteoporosis. Am. J. Transl. Res. 2019, 11, 6680–6690. [Google Scholar] [PubMed]
- Nagamani, S.C.S.; Erez, A.; Lee, B. Argininosuccinate Lyase Deficiency. In GeneReviews; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Yi, C.; Cai, C.; Cheng, Z.; Zhao, Y.; Yang, X.; Wu, Y.; Wang, X.; Jin, Z.; Xiang, Y.; Jin, M.; et al. Genome-Wide CRISPR-Cas9 Screening Identifies the CYTH2 Host Gene as a Potential Therapeutic Target of Influenza Viral Infection. Cell Rep. 2022, 38, 110559. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Calderone, V.; Ciofi-Baffoni, S.; Giachetti, A.; Jaiswal, D.; Mikolajczyk, M.; Piccioli, M.; Winkelmann, J. Molecular View of an Electron Transfer Process Essential for Iron–Sulfur Protein Biogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 7136–7141. [Google Scholar] [CrossRef]
- Cui, M.; Yamano, K.; Yamamoto, K.; Yamamoto-Imoto, H.; Minami, S.; Yamamoto, T.; Matsui, S.; Kaminishi, T.; Shima, T.; Ogura, M.; et al. HKDC1, a Target of TFEB, Is Essential to Maintain Both Mitochondrial and Lysosomal Homeostasis, Preventing Cellular Senescence. Proc. Natl. Acad. Sci. USA 2024, 121, e2306454120. [Google Scholar] [CrossRef]
- Wu, Y.; Lee, Y.; Li, W.; Hsu, W.; Lin, H.; Chang, L.; Huang, A.; Jhan, J.; Wu, W.; Li, C.; et al. High Transaldolase 1 Expression Predicts Poor Survival of Patients with Upper Tract Urothelial Carcinoma. Pathol. Int. 2021, 71, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ye, R.D. Serum Amyloid A1: Structure, Function and Gene Polymorphism. Gene 2016, 583, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Uyama, T.; Inoue, M.; Okamoto, Y.; Shinohara, N.; Tai, T.; Tsuboi, K.; Inoue, T.; Tokumura, A.; Ueda, N. Involvement of Phospholipase A/Acyltransferase-1 in N-Acylphosphatidylethanolamine Generation. Biochim. Biophys. Acta BBA—Mol. Cell Biol. Lipids 2013, 1831, 1690–1701. [Google Scholar] [CrossRef]
- Nagata, M.; Arakawa, S.; Yamaguchi, H.; Torii, S.; Endo, H.; Tsujioka, M.; Honda, S.; Nishida, Y.; Konishi, A.; Shimizu, S. Dram1 Regulates DNA Damage-Induced Alternative Autophagy. Cell Stress 2018, 2, 55–65. [Google Scholar] [CrossRef]
- Lin, T.; Zhang, L.; Wu, M.; Jiang, D.; Li, Z.; Yang, Z. Repair of Hypoxanthine in DNA Revealed by DNA Glycosylases and Endonucleases from Hyperthermophilic Archaea. Front. Microbiol. 2021, 12, 736915. [Google Scholar] [CrossRef]
- Amai, K.; Fukami, T.; Ichida, H.; Watanabe, A.; Nakano, M.; Watanabe, K.; Nakajima, M. Quantitative Analysis of mRNA Expression Levels of Aldo-Keto Reductase and Short-Chain Dehydrogenase/Reductase Isoforms in Human Livers. Drug Metab. Pharmacokinet. 2020, 35, 539–547. [Google Scholar] [CrossRef]
- Hong, B.; Wu, Y.; Li, W.; Wang, X.; Wen, Y.; Jiang, S.; Dimitrov, D.S.; Ying, T. In-Depth Analysis of Human Neonatal and Adult IgM Antibody Repertoires. Front. Immunol. 2018, 9, 128. [Google Scholar] [CrossRef]
- Shuptrine, C.W.; Perez, V.M.; Selitsky, S.R.; Schreiber, T.H.; Fromm, G. Shining a LIGHT on Myeloid Cell Targeted Immunotherapy. Eur. J. Cancer 2023, 187, 147–160. [Google Scholar] [CrossRef]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA Circ-CBFB Promotes Proliferation and Inhibits Apoptosis in Chronic Lymphocytic Leukemia through Regulating miR-607/FZD3/Wnt/β-Catenin Pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Chamorro, J.G.; Castagnino, J.P.; Aidar, O.; Musella, R.M.; Frías, A.; Visca, M.; Nogueras, M.; Costa, L.; Perez, A.; Caradonna, F.; et al. Effect of Gene–Gene and Gene–Environment Interactions Associated with Antituberculosis Drug-Induced Hepatotoxicity. Pharmacogenet. Genom. 2017, 27, 363–371. [Google Scholar] [CrossRef]
- Saghaeian Jazi, M.; Samaei, N.M.; Mowla, S.J.; Arefnezhad, B.; Kouhsar, M. SOX2OT Knockdown Derived Changes in Mitotic Regulatory Gene Network of Cancer Cells. Cancer Cell Int. 2018, 18, 129. [Google Scholar] [CrossRef] [PubMed]
- Christians, E.S.; Ishiwata, T.; Benjamin, I.J. Small Heat Shock Proteins in Redox Metabolism: Implications for Cardiovascular Diseases. Int. J. Biochem. Cell Biol. 2012, 44, 1632–1645. [Google Scholar] [CrossRef]
- Padilla, J.; Lee, J. A Novel Therapeutic Target, BACH1, Regulates Cancer Metabolism. Cells 2021, 10, 634. [Google Scholar] [CrossRef]
- Kalafatakis, I.; Savvaki, M.; Velona, T.; Karagogeos, D. Implication of Contactins in Demyelinating Pathologies. Life 2021, 11, 51. [Google Scholar] [CrossRef]
- Baker, N.E. Founding the Wnt Gene Family: How Wingless Was Found to Be a Positional Signal and Oncogene Homolog. BioEssays 2024, 46, 2300156. [Google Scholar] [CrossRef]
- Cristofani, R.; Piccolella, M.; Crippa, V.; Tedesco, B.; Montagnani Marelli, M.; Poletti, A.; Moretti, R.M. The Role of HSPB8, a Component of the Chaperone-Assisted Selective Autophagy Machinery, in Cancer. Cells 2021, 10, 335. [Google Scholar] [CrossRef]
- Kitazawa, T.; Kaiya, H. Motilin Comparative Study: Structure, Distribution, Receptors, and Gastrointestinal Motility. Front. Endocrinol. 2021, 12, 700884. [Google Scholar] [CrossRef] [PubMed]
- Kappé, G.; Verschuure, P.; Philipsen, R.L.A.; Staalduinen, A.A.; Van De Boogaart, P.; Boelens, W.C.; De Jong, W.W. Characterization of Two Novel Human Small Heat Shock Proteins: Protein Kinase-Related HspB8 and Testis-Specific HspB9. Biochim. Biophys. Acta BBA—Gene Struct. Expr. 2001, 1520, 1–6. [Google Scholar] [CrossRef]
- Li, G.; Cai, Z.; Ma, X.; Sun, M.; Li, J.; Sun, W.; Hua, Y. Screening of serum marker proteins in osteosarcoma and preliminary bioinformatic analysis on POLR3F. Zhonghua Zhong Liu Za Zhi 2011, 33, 836–841. [Google Scholar] [PubMed]
- Berger, J.; Berger, S.; Li, M.; Currie, P.D. Myo18b Is Essential for Sarcomere Assembly in Fast Skeletal Muscle. Hum. Mol. Genet. 2017, 26, ddx025. [Google Scholar] [CrossRef]
- Taub, M.; Springate, J.E.; Cutuli, F. Targeting of Renal Proximal Tubule Na,K-ATPase by Salt-Inducible Kinase. Biochem. Biophys. Res. Commun. 2010, 393, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Skariah, N.; James, O.J.; Swamy, M. Signalling Mechanisms Driving Homeostatic and Inflammatory Effects of Interleukin-15 on Tissue Lymphocytes. Discov. Immunol. 2024, 3, kyae002. [Google Scholar] [CrossRef]
- Shindo, S.; Shioya, A.; Watanabe, M.; Sasaki, T.; Suzuki, H.; Kumagai, T.; Hwang, G.-W.; Nagata, K. Development of an Adenovirus-Mediated Reporter Assay System to Detect a Low Concentration of Retinoic Acid in MCF-7 Cells. J. Toxicol. Sci. 2022, 47, 249–255. [Google Scholar] [CrossRef]
- Chotiner, J.Y.; Wolgemuth, D.J.; Wang, P.J. Functions of Cyclins and CDKs in Mammalian Gametogenesis. Biol. Reprod. 2019, 101, 591–601. [Google Scholar] [CrossRef]
- Fard, D.; Testa, E.; Panzeri, V.; Rizzolio, S.; Bianchetti, G.; Napolitano, V.; Masciarelli, S.; Fazi, F.; Maulucci, G.; Scicchitano, B.M.; et al. SEMA6C: A Novel Adhesion-Independent FAK and YAP Activator, Required for Cancer Cell Viability and Growth. Cell. Mol. Life Sci. 2023, 80, 111. [Google Scholar] [CrossRef]
- Nawaz, S.; Hussain, S.; Bilal, M.; Syed, N.; Liaqat, K.; Ullah, I.; Akil, A.A.; Fakhro, K.A.; Ahmad, W. A Variant in Sperm-specific Glycolytic Enzyme Enolase 4 (ENO4) Causes Human Male Infertility. J. Gene Med. 2024, 26, e3583. [Google Scholar] [CrossRef]
- Taheri, M.; Shirvani-Farsani, Z.; Harsij, A.; Fathi, M.; Khalilian, S.; Ghafouri-Fard, S.; Baniahmad, A. A Review on the Role of KCNQ1OT1 lncRNA in Human Disorders. Pathol.-Res. Pract. 2024, 255, 155188. [Google Scholar] [CrossRef]
- Li, D.; Peng, W.; Wu, B.; Liu, H.; Zhang, R.; Zhou, R.; Yao, L.; Ye, L. Metallothionein MT1M Suppresses Carcinogenesis of Esophageal Carcinoma Cells through Inhibition of the Epithelial-Mesenchymal Transition and the SOD1/PI3K Axis. Mol. Cells 2021, 44, 267–278. [Google Scholar] [CrossRef]
- Mota, G.A.F.; De Souza, S.L.B.; Vileigas, D.F.; Da Silva, V.L.; Sant’Ana, P.G.; Costa, L.C.D.S.; Padovani, C.R.; Zanatti Bazan, S.G.; Buzalaf, M.A.R.; Santos, L.D.D.; et al. Myocardial Proteome Changes in Aortic Stenosis Rats Subjected to Long-term Aerobic Exercise. J. Cell. Physiol. 2024, 239, e31199. [Google Scholar] [CrossRef]
- Lv, Q.; Shi, J.; Miao, D.; Tan, D.; Zhao, C.; Xiong, Z.; Zhang, X. miR-1182-Mediated ALDH3A2 Inhibition Affects Lipid Metabolism and Progression in ccRCC by Activating the PI3K-AKT Pathway. Transl. Oncol. 2024, 40, 101835. [Google Scholar] [CrossRef]
- Manning, A.L.; Ganem, N.J.; Bakhoum, S.F.; Wagenbach, M.; Wordeman, L.; Compton, D.A. The Kinesin-13 Proteins Kif2a, Kif2b, and Kif2c/MCAK Have Distinct Roles during Mitosis in Human Cells. Mol. Biol. Cell 2007, 18, 2970–2979. [Google Scholar] [CrossRef]
- Kolijn, P.M.; Huijser, E.; Wahadat, M.J.; Van Helden-Meeuwsen, C.G.; Van Daele, P.L.A.; Brkic, Z.; Rijntjes, J.; Hebeda, K.M.; Groenen, P.J.T.A.; Versnel, M.A.; et al. Extranodal Marginal Zone Lymphoma Clonotypes Are Detectable Prior to eMZL Diagnosis in Tissue Biopsies and Peripheral Blood of Sjögren’s Syndrome Patients through Immunogenetics. Front. Oncol. 2023, 13, 1130686. [Google Scholar] [CrossRef]
- Steele, E.J.; Franklin, A.; Lindley, R.A. Somatic Mutation Patterns at Ig and Non-Ig Loci. DNA Repair 2024, 133, 103607. [Google Scholar] [CrossRef]
- Zheng, Z.; Zheng, H.; Yan, W. Fank1 Is a Testis-Specific Gene Encoding a Nuclear Protein Exclusively Expressed during the Transition from the Meiotic to the Haploid Phase of Spermatogenesis. Gene Expr. Patterns 2007, 7, 777–783. [Google Scholar] [CrossRef]
- Hong, S.N.; Park, J.-Y.; Yang, S.-Y.; Lee, C.; Kim, Y.-H.; Joung, J.-G. Reduced Diversity of Intestinal T-Cell Receptor Repertoire in Patients with Crohn’s Disease. Front. Cell. Infect. Microbiol. 2022, 12, 932373. [Google Scholar] [CrossRef]
- Yuan, J.; Jia, J.; Wu, T.; Du, Z.; Chen, Q.; Zhang, J.; Wu, Z.; Yuan, Z.; Zhao, X.; Liu, J.; et al. Long Intergenic Non-Coding RNA DIO3OS Promotes Osteosarcoma Metastasis via Activation of the TGF-β Signaling Pathway: A Potential Diagnostic and Immunotherapeutic Target for Osteosarcoma. Cancer Cell Int. 2023, 23, 215. [Google Scholar] [CrossRef]
- Gnanagobal, H.; Cao, T.; Hossain, A.; Dang, M.; Hall, J.R.; Kumar, S.; Van Cuong, D.; Boyce, D.; Santander, J. Lumpfish (Cyclopterus lumpus) Is Susceptible to Renibacterium Salmoninarum Infection and Induces Cell-Mediated Immunity in the Chronic Stage. Front. Immunol. 2021, 12, 733266. [Google Scholar] [CrossRef]
- Guo, Y.; Miao, Q.; Zhang, Y.; Wang, C.; Liang, M.; Li, X.; Qiu, W.; Shi, G.; Zhai, Q.; Chen, Z. A Novel Missense Creatine Mutant of CaBP4, c.464G>A (p.G155D), Associated with Autosomal Dominant Nocturnal Frontal Lobe Epilepsy (ADNFLE), Reduces the Expression of CaBP4. Transl. Pediatr. 2022, 11, 396–402. [Google Scholar] [CrossRef]
- Adhikari, S.; Üren, A.; Roy, R. Excised Damaged Base Determines the Turnover of Human N-Methylpurine-DNA Glycosylase. DNA Repair 2009, 8, 1201–1206. [Google Scholar] [CrossRef]
- Petrungaro, C.; Zimmermann, K.M.; Küttner, V.; Fischer, M.; Dengjel, J.; Bogeski, I.; Riemer, J. The Ca2+-Dependent Release of the Mia40-Induced MICU1-MICU2 Dimer from MCU Regulates Mitochondrial Ca2+ Uptake. Cell Metab. 2015, 22, 721–733. [Google Scholar] [CrossRef]
- Bou-Nader, C.; Barraud, P.; Pecqueur, L.; Pérez, J.; Velours, C.; Shepard, W.; Fontecave, M.; Tisné, C.; Hamdane, D. Molecular Basis for Transfer RNA Recognition by the Double-Stranded RNA-Binding Domain of Human Dihydrouridine Synthase 2. Nucleic Acids Res. 2019, 47, 3117–3126. [Google Scholar] [CrossRef]
- Saunders, A.S.; Bender, D.E.; Ray, A.L.; Wu, X.; Morris, K.T. Colony-Stimulating Factor 3 Signaling in Colon and Rectal Cancers: Immune Response and CMS Classification in TCGA Data. PLoS ONE 2021, 16, e0247233. [Google Scholar] [CrossRef]
- Xie, J.; Lan, T.; Zheng, D.-L.; Ding, L.-C.; Lu, Y.-G. CDH4 Inhibits Ferroptosis in Oral Squamous Cell Carcinoma Cells. BMC Oral Health 2023, 23, 329. [Google Scholar] [CrossRef]
- Piórkowska, K.; Żukowski, K.; Nowak, J.; Połtowicz, K.; Ropka-Molik, K.; Gurgul, A. Genome-wide RNA -Seq Analysis of Breast Muscles of Two Broiler Chicken Groups Differing in Shear Force. Anim. Genet. 2016, 47, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Rivedal, M.; Mikkelsen, H.; Marti, H.-P.; Liu, L.; Kiryluk, K.; Knoop, T.; Bjørneklett, R.; Haaskjold, Y.L.; Furriol, J.; Leh, S.; et al. Glomerular Transcriptomics Predicts Long Term Outcome and Identifies Therapeutic Strategies for Patients with Assumed Benign IgA Nephropathy. Kidney Int. 2024, 105, 717–730. [Google Scholar] [CrossRef]
- Ortega, M.A.; De Leon-Oliva, D.; Garcia-Montero, C.; Fraile-Martinez, O.; Boaru, D.L.; Del Val Toledo Lobo, M.; García-Tuñón, I.; Royuela, M.; García-Honduvilla, N.; Bujan, J.; et al. Understanding HAT1: A Comprehensive Review of Noncanonical Roles and Connection with Disease. Genes 2023, 14, 915. [Google Scholar] [CrossRef]
- Miller, P.P.; Caza, T.; Larsen, C.P.; Charu, V. EXT1 and NCAM1-Associated Membranous Lupus Nephritis in a Cohort of Patients Undergoing Repeat Kidney Biopsies. Nephrol. Dial. Transplant. 2023, 38, 396–404. [Google Scholar] [CrossRef]
- Nowrasteh, G.; Zand, A.; Raposa, L.B.; Szabó, L.; Tomesz, A.; Molnár, R.; Kiss, I.; Orsós, Z.; Gerencsér, G.; Gyöngyi, Z.; et al. Fruit Extract, Rich in Polyphenols and Flavonoids, Modifies the Expression of DNMT and HDAC Genes Involved in Epigenetic Processes. Nutrients 2023, 15, 1867. [Google Scholar] [CrossRef]
- Del Saz-Lara, A.; López De Las Hazas, M.-C.; Visioli, F.; Dávalos, A. Nutri-Epigenetic Effects of Phenolic Compounds from Extra Virgin Olive Oil: A Systematic Review. Adv. Nutr. 2022, 13, 2039–2060. [Google Scholar] [CrossRef]
- Sciandrello, G.; Mauro, M.; Catanzaro, I.; Saverini, M.; Caradonna, F.; Barbata, G. Long-lasting Genomic Instability Following Arsenite Exposure in Mammalian Cells: The Role of Reactive Oxygen Species. Environ. Mol. Mutagen. 2011, 52, 562–568. [Google Scholar] [CrossRef]
- Mauro, M.; Caradonna, F.; Klein, C.B. Dysregulation of DNA Methylation Induced by Past Arsenic Treatment Causes Persistent Genomic Instability in Mammalian Cells. Environ. Mol. Mutagen. 2016, 57, 137–150. [Google Scholar] [CrossRef]
- Davis, C.D.; Uthus, E.O.; Finley, J.W. Dietary Selenium and Arsenic Affect DNA Methylation In Vitro in Caco-2 Cells and In Vivo in Rat Liver and Colon. J. Nutr. 2000, 130, 2903–2909. [Google Scholar] [CrossRef]
- Kovalenko, T.F.; Morozova, K.V.; Pavlyukov, M.S.; Anufrieva, K.S.; Bobrov, M.Y.; Gamisoniya, A.M.; Ozolinya, L.A.; Dobrokhotova, Y.E.; Shakhparonov, M.I.; Patrushev, L.I. Methylation of the PTENP1 Pseudogene as Potential Epigenetic Marker of Age-Related Changes in Human Endometrium. PLoS ONE 2021, 16, e0243093. [Google Scholar] [CrossRef]
- Rodrigues, M.F.S.D.; Esteves, C.M.; Xavier, F.C.A.; Nunes, F.D. Methylation Status of Homeobox Genes in Common Human Cancers. Genomics 2016, 108, 185–193. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).