Soluble Oncoimmunome Signatures Predict Muscle Mass Response to Enriched Immunonutrition in Cancer Patients: Subanalysis of a Multicenter Randomized Clinical Trial
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Dietary Intervention and Nutritional Evaluation
2.3. Biological Samples
2.4. Soluble Oncoimmunome
2.5. Statistical Analysis
3. Results
3.1. Patients on Enriched ONS Improve Their Weight and Muscle Mass
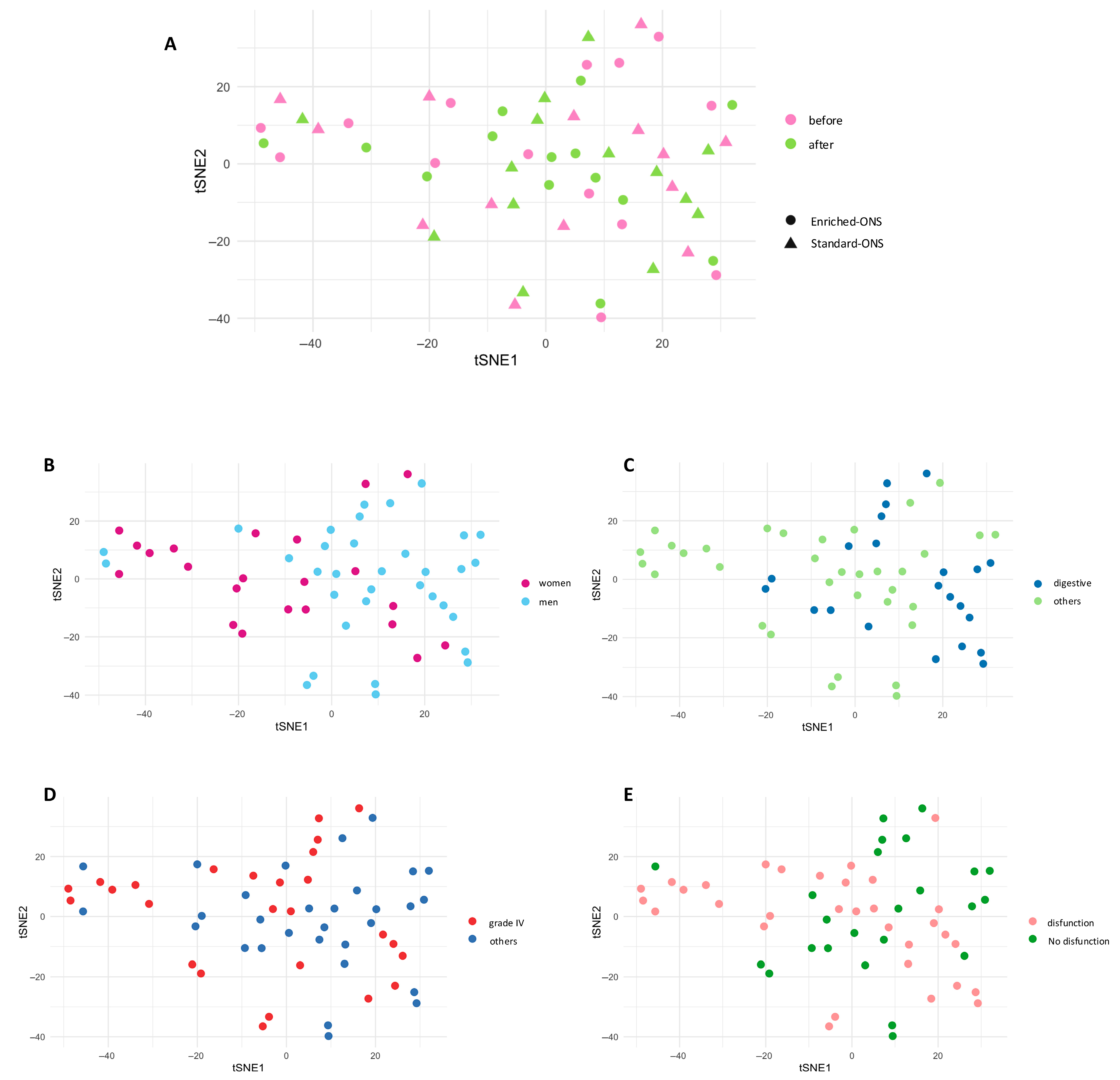
3.2. Each Patient Exhibits a Unique Oncoimmunome Fingerprint
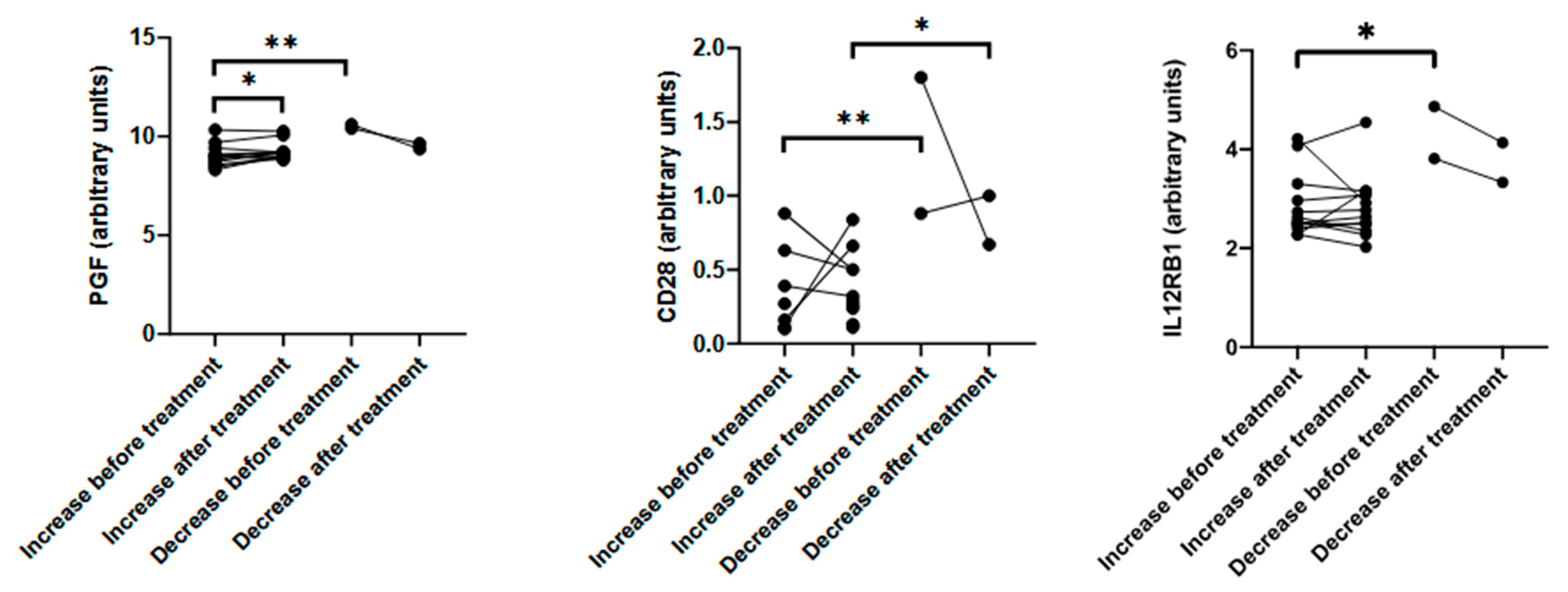
3.3. Enriched ONS Reduces Inflammation and Enhances Myogenic Differentiation
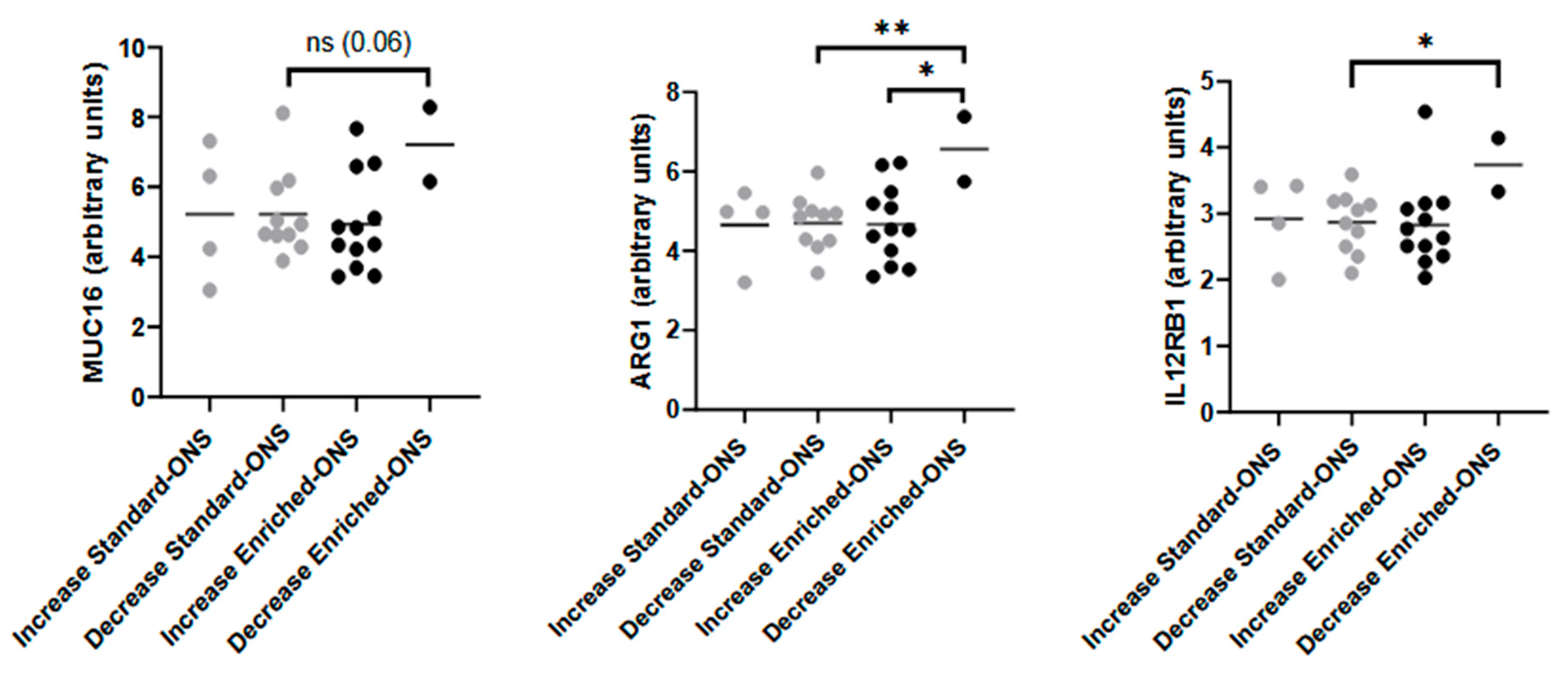
3.4. Baseline Immune Signature Predicts Muscle Mass Response
4. Discussion
5. Strengths and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Planas, M.; Álvarez-Hernández, J.; León-Sanz, M.; Celaya-Pérez, S.; Araujo, K.; García de Lorenzo, A. Prevalence of Hospital Malnutrition in Cancer Patients: A Sub-Analysis of the PREDyCES® Study. Support. Care Cancer 2016, 24, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Bozzetti, F. Screening the Nutritional Status in Oncology: A Preliminary Report on 1,000 Outpatients. Support. Care Cancer 2009, 17, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, T.; Terracina, K.P.; Raza, A.; Matsubara, H.; Takabe, K. Cancer Cachexia, Mechanism and Treatment. World J. Gastrointest. Oncol. 2015, 7, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Milla, S.P.; Luna, P.P.G.; Casariego, A.V.; González, F.V.; Folgueras, T.M.; Jáuregui, O.I.; Rey, S.G.; Fernández, A.C.; Plaza, B.L.; Quintana, T.C.; et al. Adherence and Impact of an Oral Nutritional Supplement Enriched in Leucine, EVOO, EPA and DHA, and Beta-Glucans on the Coverage of Energy and Protein Requirements in Patients with Cancer and Malnutrition: Alisenoc Study. Nutrition 2024, 120, 112355. [Google Scholar] [CrossRef] [PubMed]
- Ostadrahimi, A.; Ziaei, J.E.; Esfahani, A.; Jafarabadi, M.A.; Movassaghpourakbari, A.; Farrin, N. Effect of Beta Glucan on White Blood Cell Counts and Serum Levels of IL-4 and IL-12 in Women with Breast Cancer Undergoing Chemotherapy: A Randomized Double-Blind Placebo-Controlled Clinical Trial. Asian Pac. J. Cancer Prev. 2014, 15, 5733–5739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, G.; Jin, X.; Wang, J.; Yu, S. Pre-Operative Immunonutrition Enhances Postoperative Outcomes and Elevates Tumor-Infiltrating Lymphocyte Counts in Colorectal Cancer Patients: A Meta-Analysis of Randomized Controlled Trials. Nutr Cancer 2024, 76, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Cangelosi, G.; Mancin, S.; Pantanetti, P.; Sguanci, M.; Morales Palomares, S.; De Luca, A.; Biondini, F.; Tartaglia, F.; Ferrara, G.; Petrelli, F. Impact of the COVID-19 Pandemic on Lifestyle Behavior and Clinical Care Pathway Management in Type 2 Diabetes: A Retrospective Cross-Sectional Study. Medicina 2024, 60, 1624. [Google Scholar] [CrossRef] [PubMed]
- Cangelosi, G.; Palomares, S.M.; Pantanetti, P.; De Luca, A.; Biondini, F.; Nguyen, C.T.T.; Mancin, S.; Sguanci, M.; Petrelli, F. COVID-19, Nutrients and Lifestyle Eating Behaviors: A Narrative Review. Diseases 2024, 12, 193. [Google Scholar] [CrossRef] [PubMed]
- Franco-Romero, A.; Sandri, M. Role of Autophagy in Muscle Disease. Mol. Asp. Med. 2021, 82, 101041. [Google Scholar] [CrossRef] [PubMed]
- Hua, D.-X.; Ma, K.-S.; Cheng, J.-Y.; Liu, Y.; Sun, J.; He, Q.-Y.; Deng, Y.-P.; Yang, J.; Fu, L.; Zhao, H. Serum TRAIL Predicts Severity and Prognosis in Patients with Community-Acquired Pneumonia: A Prospective Cohort Study. Intern. Emerg. Med. 2022, 17, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Chen, Y.; Liu, D.; Li, F.; Li, X.; Jia, W. High Expression of LAMP3 Is a Novel Biomarker of Poor Prognosis in Patients with Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 17655–17667. [Google Scholar] [CrossRef] [PubMed]
- Nagelkerke, A.; Bussink, J.; Mujcic, H.; Wouters, B.G.; Lehmann, S.; Sweep, F.C.; Span, P.N. Hypoxia Stimulates Migration of Breast Cancer Cells via the PERK/ATF4/LAMP3-Arm of the Unfolded Protein Response. Breast Cancer Res. 2013, 15, R2. [Google Scholar] [CrossRef] [PubMed]
- Mujcic, H. Hypoxia-Induced Metastasis: The Role of the Unfolded Protein Response. Ph.D. Thesis, Maastricht University, Maastricht, The Netherlands, 2013. [Google Scholar]
- Sun, R.; Wang, X.; Zhu, H.; Mei, H.; Wang, W.; Zhang, S.; Huang, J. Prognostic Value of LAMP3 and TP53 Overexpression in Benign and Malignant Gastrointestinal Tissues. Oncotarget 2014, 5, 12398–12409. [Google Scholar] [CrossRef] [PubMed]
- Kanao, H.; Enomoto, T.; Kimura, T.; Fujita, M.; Nakashima, R.; Ueda, Y.; Ueno, Y.; Miyatake, T.; Yoshizaki, T.; Buzard, G.S.; et al. Overexpression of LAMP3/TSC403/DC-LAMP Promotes Metastasis in Uterine Cervical Cancer. Cancer Res. 2005, 65, 8640–8645. [Google Scholar] [CrossRef] [PubMed]
- Blazev, R.; Ashwood, C.; Abrahams, J.L.; Chung, L.H.; Francis, D.; Yang, P.; Watt, K.I.; Qian, H.; Quaife-Ryan, G.A.; Hudson, J.E.; et al. Integrated Glycoproteomics Identifies a Role of N-Glycosylation and Galectin-1 on Myogenesis and Muscle Development. Mol. Cell. Proteom. 2021, 20, 100030. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Liu, X.; Ren, X.; Zhang, Q.; Zhang, T.; Qian, Q.; Liu, W.; Jiang, C. Galectin-1-Induced Skeletal Muscle Cell Differentiation of Mesenchymal Stem Cells Seeded on an Acellular Dermal Matrix Improves Injured Anal Sphincter. Discov. Med. 2016, 21, 331–340. [Google Scholar] [PubMed]
- Vilen, Z.; Joeh, E.; Critcher, M.; Parker, C.G.; Huang, M.L. Proximity Tagging Identifies the Glycan-Mediated Glycoprotein Interactors of Galectin-1 in Muscle Stem Cells. ACS Chem. Biol. 2021, 16, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Aithal, A.; Rauth, S.; Kshirsagar, P.; Shah, A.; Lakshmanan, I.; Junker, W.M.; Jain, M.; Ponnusamy, M.P.; Batra, S.K. MUC16 as a Novel Target for Cancer Therapy. Expert Opin. Ther. Targets 2018, 22, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Lu, Q.; Lou, T.; Cao, G.; Wang, S.; Zhang, Z. MUC16 Affects the Biological Functions of Ovarian Cancer Cells and Induces an Antitumor Immune Response by Activating Dendritic Cells. Ann. Transl. Med. 2020, 8, 1494. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Yu, Y.; Li, Z.; Ren, Y.; Li, Z.; Ye, Q.; Liu, P.; Ji, C.; Qian, L.; Xiong, Y. Arginase: An Emerging and Promising Therapeutic Target for Cancer Treatment. Biomed. Pharmacother. 2022, 149, 112840. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiang, H.; Toyoshima, Y.; Shen, W.; Shichi, S.; Nakamoto, H.; Kimura, S.; Sugiyama, K.; Homma, S.; Miyagi, Y.; et al. Arginase-1 Inhibition Reduces Migration Ability and Metastatic Colonization of Colon Cancer Cells. Cancer Metab. 2023, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.T. IL12Rβ1: The Cytokine Receptor That We Used to Know. Cytokine 2015, 71, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Horsley, V.; Pavlath, G.K. Prostaglandin F2α Stimulates Growth of Skeletal Muscle Cells via an NFATC2-Dependent Pathway. J. Cell Biol. 2003, 161, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Frauwirth, K.A.; Riley, J.L.; Harris, M.H.; Parry, R.V.; Rathmell, J.C.; Plas, D.R.; Elstrom, R.L.; June, C.H.; Thompson, C.B. The CD28 Signaling Pathway Regulates Glucose Metabolism. Immunity 2002, 16, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.K.; Wong, C.K.; Leung, T.F.; Lam, C.W.K. Elevation of Plasma Soluble T Cell Costimulatory Molecules CTLA-4, CD28 and CD80 in Children with Allergic Asthma. Int. Arch. Allergy Immunol. 2005, 137, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.K.; Wong, C.K.; Leung, T.F.; Lam, C.W.K. Plasma Concentrations of Soluble CTLA-4, CD28, CD80 and CD86 Costimulatory Molecules Reflect Disease Severity of Acute Asthma in Children. Pediatr. Pulmonol. 2006, 41, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Lit, L.C.W.; Tam, L.S.; Li, E.K.; Lam, C.W.K. Aberrant Production of Soluble Costimulatory Molecules CTLA-4, CD28, CD80 and CD86 in Patients with Systemic Lupus Erythematosus. Rheumatology 2005, 44, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Yi, L.; Tao, H.; Huang, J.; Jin, Z.; Xiao, Y.; Feng, C.; Sun, J. Clinical Immunology Enhancement of Soluble CD28 Levels in the Serum of Graves’ Disease. Cent. Eur. J. Immunol. 2014, 39, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Aprilia, A.; Handono, K.; Sujuti, H.; Sabarudin, A.; Winaris, N. SCD163, SCD28, SCD80, and SCTLA-4 as Soluble Marker Candidates for Detecting Immunosenescence. Immun. Ageing 2024, 21, 9. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Lee, S.; Yoo, J.; Baek, K. The Protective Effect of IL-12/23 Neutralizing Antibody in Sarcopenia Associated with Dextran Sulfate Sodium-induced Experimental Colitis. J. Cachexia Sarcopenia Muscle 2023, 14, 1096–1106. [Google Scholar] [CrossRef] [PubMed]

| Enriched ONS (A) | Standard ONS (B) | |
|---|---|---|
| Energy (kcal) | 200 | 200 |
Proteins, g/TE%, (ingredients)
| 10 g/20% [whey protein (63%), caseinate, and vegetal protein]
| 10 g/20% [caseinate and whey protein (25%)]
|
| Carbohydrates: g/TE%, (ingredients)
| 20 g/39.7% (dextrin and maltodextrin)
| 18.2 g/46% (maltodextrin)
|
| Fat g/TE% (ingredients) | 8.6 g/38.5% (EVOO, canola, MCT, and fish oil) | 5 g/(31%) (canola and high-oleic sunflower oil) |
|
|
|
| Fiber g/TE% (ingredients) | 1.8 g/1.8% (FOS, acacia fiber and beta glucans) 100% soluble | 1.5 g/1.5%(FOS and oat fiber) 60% soluble–40% insoluble |
| Osmolarity (mOsm/L) | 420 | 390 |
| Flavor | Natural vanilla | Natural vanilla |
| Sweetener | Sucralose | Sucralose |
| Standard ONS | Unit | Before | After |
| Energy intake | kcal | 1309.75 (372.18) | 2147.48 (347.28) *** |
| Protein | g | 55.78 (16.76) | 96.25 (13.19) *** |
| Leucine | g | 0.7 (0.6) | 1.3 (0.7) |
| Carbohydrate | g | 126.78 (42.72) | 221.06 (40.32) *** |
| Fat | g | 61.34 (23.82) | 92.12 (23.07) * |
| SFA | g | 18.1 (7.62) | 25.22 (6.27) * |
| MUFA | g | 28.43 (11.54) | 43.57 (12.72) ** |
| PUFA | g | 6.94 (3.15) | 13.88 (4.39) *** |
| EPA and DHA | g | 0.5 (0.3) | 0.7 (0.4) |
| Fiber | g | 13.56 (8.18) | 19.25 (9.28) *** |
| Energy per body weight | kcal/kg | 24.68 (8.65) | 39.82 (9.57) *** |
| Protein per body weight | g/kg | 1.07 (0.45) | 1.78 (0.36) *** |
| Enriched ONS | Units | Before | After |
| Energy intake | kcal | 1459.58 (276.37) | 2222.01 (470.05) *** |
| Protein | g | 61.03 (13.71) | 101.91 (20.03) *** |
| Leucine | g | 0.6 (0.4) | 3.3 (0.9) ** |
| Carbohydrate | g | 137.76 (35.77) | 210.91 (51.14) *** |
| Fat | g | 71.18 (18.98) | 102.98 (24.84) ** |
| SFA | g | 22.42 (8.34) | 29.68 (8.50) * |
| MUFA | g | 34.14 (10.34) | 49.69 (13.36) * |
| PUFA | g | 7.57 (2.14) | 15.97 (3.65) *** |
| EPA and DHA | g | 0.4 (0.2) | 1.4 (0.3) ** |
| Fiber | g | 11.79 (4.64) | 20.92 (6.76) *** |
| Energy per body weight | kcal/kg | 25.78 (8.62) | 37.16 (8.73) ** |
| Protein per body weight | g/kg | 1.09 (0.45) | 1.70 (0.37) *** |
| Standard (n = 14) | Enriched (n = 14) | |||
|---|---|---|---|---|
| Before | After | Before | After | |
| Body weight (kg) | 61.7 ± 13.1 | 61.4 ± 12.3 | 67.3 ± 17.1 | 68.7 ± 6.1 * |
| Muscle mass (kg) | 19.6 ± 7.1 | 18.7 ± 6.2 | 18.5 ± 8.0 | 20.7 ± 8.5 * |
| BMI (kg/m2) | 23.1 ± 4.1 | 23.0 ± 3.6 | 24.5 ± 5.6 | 25.1 ± 6.1 |
| Enriched ONS (A) (n = 14) | Standard ONS (B) (n = 14) | |||
|---|---|---|---|---|
| Before | After | Before | After | |
| IL8 | 8.46 ± 1.52 | 8.23 ± 1.51 | 8.64 ± 1.98 | 7.6 ± 1.05 |
| TNFRSF9 | 6.11 ± 0.96 | 5.98 ± 0.75 | 5.81 ± 0.71 | 5.92 ± 0.81 |
| TIE2 | 6.96 ± 0.35 | 7.05 ± 0.29 | 7.08 ± 0.24 | 7.09 ± 0.27 |
| MCP_3 | 3.95 ± 1.6 | 3.76 ± 0.78 | 3.71 ± 0.95 | 3.53 ± 1.22 |
| CD40_L | 7.93 ± 1.71 | 8.49 ± 0.77 | 8.18 ± 1.31 | 7.69 ± 1.63 |
| IL_1alpha | 0.75 ± 0.53 | 1.18 ± 1.01 | 0.85 ± 0.4 | 0.69 ± 0.36 |
| CD244 | 6.63 ± 0.47 | 6.62 ± 0.32 | 6.43 ± 0.34 | 6.56 ± 0.64 |
| EGF | 9.35 ± 1.52 | 9.74 ± 0.69 | 9.3 ± 1.31 | 9 ± 1.33 |
| ANGPT1 | 7.06 ± 0.86 | 7.33 ± 0.16 | 7.11 ± 0.88 | 7.18 ± 0.26 |
| IL7 | 5.06 ± 1.37 | 5.32 ± 1.08 | 5.38 ± 1.09 | 5.44 ± 0.99 |
| PGF | 9.25 ± 0.75 | 9.28 ± 0.43 | 8.92 ± 0.52 | 8.94 ± 0.46 |
| IL6 | 4.83 ± 1.7 | 4.36 ± 1.76 | 5.35 ± 2 | 4.54 ± 1.78 |
| ADGRG1 | 2.05 ± 1.31 | 2.1 ± 1.2 | 2.02 ± 1.4 | 2.04 ± 1.01 |
| MCP_1 | 12.63 ± 0.86 | 12.82 ± 0.44 | 12.63 ± 0.72 | 12.47 ± 0.73 |
| CRTAM | 5.84 ± 1 | 5.65 ± 0.75 | 5.57 ± 0.8 | 5.43 ± 0.73 |
| CXCL11 | 9.3 ± 1.27 | 9.59 ± 1.23 | 9.61 ± 1.32 | 9.34 ± 1.21 |
| MCP_4 | 10.79 ± 0.97 | 11.08 ± 0.57 | 10.63 ± 0.82 | 10.62 ± 0.78 |
| TRAIL | 8.5 ± 0.44 | 8.51 ± 0.4 * | 8.72 ± 0.29 | 8.78 ± 0.22 |
| FGF2 | 0.94 ± 0.75 | 1.12 ± 0.43 | 0.99 ± 1.11 | 1.03 ± 1.24 |
| CXCL9 | 8.28 ± 1.28 | 8.26 ± 0.83 | 8.18 ± 1.08 | 8.09 ± 1 |
| CD8A | 9.63 ± 0.64 | 9.48 ± 0.54 | 9.37 ± 0.63 | 9.31 ± 0.93 |
| CAIX | 5.2 ± 1.12 | 4.86 ± 0.84 | 5.18 ± 0.81 | 5.39 ± 0.97 |
| MUC_16 | 5.67 ± 1.47 | 5.26 ± 1.56 | 5.49 ± 1.2 | 5.23 ± 1.38 |
| ADA | 6.11 ± 0.58 | 5.93 ± 0.54 | 6 ± 0.63 | 6.06 ± 0.53 |
| CD4 | 4.09 ± 0.53 | 4.05 ± 0.41 | 3.98 ± 0.39 | 3.93 ± 0.36 |
| NOS3 | 2.71 ± 0.83 | 2.61 ± 0.45 | 2.61 ± 0.66 | 2.53 ± 0.9 |
| IL2 | 0.57 ± 0.79 | 0.72 ± 0.56 | 0.94 ± 0.39 | 0.63 ± 0.38 |
| Gal_9 | 9.09 ± 0.56 | 9.08 ± 0.43 | 8.98 ± 0.44 | 9.03 ± 0.33 |
| VEGFR_2 | 8.55 ± 0.46 | 8.63 ± 0.29 | 8.6 ± 0.27 | 8.56 ± 0.25 |
| CD40 | 11.83 ± 0.73 | 11.84 ± 0.5 | 11.58 ± 0.58 | 11.62 ± 0.77 |
| IL18 | 9.5 ± 0.64 | 9.55 ± 0.86 | 9.63 ± 0.89 | 9.75 ± 0.8 |
| GZMH | 4.74 ± 1.25 | 4.41 ± 0.67 | 4.52 ± 1.31 | 4.32 ± 1.01 |
| KIR3DL1 | 2.23 ± 1.64 | 2.15 ± 1.58 | 2.27 ± 0.98 | 2.35 ± 1.13 |
| LAPTGF_beta_1 | 8.74 ± 0.66 | 8.82 ± 0.45 | 8.8 ± 0.67 | 8.77 ± 0.6 |
| CXCL1 | 9.39 ± 0.88 | 9.58 ± 0.75 | 9.73 ± 0.75 | 9.31 ± 0.94 |
| TNFSF14 | 6.33 ± 1.34 | 6.29 ± 1.07 | 6.62 ± 1.34 | 6.08 ± 1.04 |
| IL33 | 0.44 ± 0.24 | 0.44 ± 0.34 | 0.39 ± 0.2 | 0.34 ± 0.22 |
| TWEAK | 9.13 ± 0.62 | 9.21 ± 0.3 | 9.09 ± 0.43 | 9.06 ± 0.45 |
| PDGFsubunitB | 10.56 ± 0.56 | 10.82 ± 0.11 | 10.64 ± 0.47 | 10.67 ± 0.28 |
| PDCD1 | 8.1 ± 1.3 | 8.3 ± 1.11 | 8.06 ± 1.3 | 7.86 ± 1.04 |
| FASLG | 7.33 ± 1.16 | 7.32 ± 0.7 | 7.56 ± 0.67 | 7.37 ± 0.97 |
| CD28 | 0.31 ± 0.57 | 0.37 ± 0.35 | 0.33 ± 0.38 | 0.35 ± 0.46 |
| CCL19 | 11.67 ± 0.92 | 11.44 ± 0.8 | 11.48 ± 0.59 | 11.36 ± 0.61 |
| MCP_2 | 8.91 ± 0.95 | 9 ± 0.51 | 8.64 ± 0.56 | 8.61 ± 0.6 |
| CCL4 | 7.68 ± 0.78 | 7.62 ± 1.02 | 7.71 ± 1.03 | 7.11 ± 0.68 |
| IL15 | 5.65 ± 0.47 | 5.66 ± 0.36 | 5.89 ± 0.62 | 5.77 ± 0.45 |
| Gal_1 | 8.05 ± 0.4 | 8.14 ± 0.29 * | 7.85 ± 0.31 | 7.86 ± 0.3 |
| PD_L1 | 7.15 ± 0.61 | 7.1 ± 0.44 | 7.15 ± 0.62 | 7.14 ± 0.53 |
| CD27 | 9.36 ± 0.69 | 9.33 ± 0.57 | 9.23 ± 0.56 | 9.16 ± 0.51 |
| CXCL5 | 9.72 ± 1.09 | 10.09 ± 0.64 | 9.76 ± 0.69 | 9.53 ± 0.95 |
| IL5 | 1.27 ± 0.85 | 1.2 ± 0.75 | 1.1 ± 0.76 | 1.17 ± 0.54 |
| HGF | 10.08 ± 0.8 | 10.09 ± 0.61 | 10.32 ± 0.86 | 10.01 ± 0.59 |
| GZMA | 7.12 ± 0.79 | 6.9 ± 0.49 | 6.92 ± 0.65 | 6.79 ± 0.65 |
| HO_1 | 12.76 ± 0.5 | 12.64 ± 0.35 | 12.84 ± 0.38 | 12.84 ± 0.3 |
| CX3CL1 | 4.59 ± 0.74 | 4.56 ± 0.53 | 4.55 ± 0.45 | 4.54 ± 0.46 |
| CXCL10 | 9.54 ± 1.06 | 9.63 ± 0.84 | 9.28 ± 0.95 | 9.2 ± 0.89 |
| CD70 | 5.09 ± 0.82 | 5.05 ± 0.48 | 4.93 ± 0.74 | 5.04 ± 0.65 |
| IL10 | 3.74 ± 0.99 | 3.42 ± 0.71 | 3.71 ± 1.13 | 3.29 ± 0.81 |
| TNFRSF12A | 8.89 ± 0.77 | 8.92 ± 0.53 | 8.55 ± 0.78 | 8.59 ± 0.55 |
| CCL23 | 12.6 ± 0.61 | 12.48 ± 0.6 | 12.39 ± 0.65 | 12.3 ± 0.6 |
| CD5 | 5.73 ± 0.72 | 5.64 ± 0.49 | 5.65 ± 0.55 | 5.54 ± 0.43 |
| CCL3 | 8.33 ± 1.57 | 7.93 ± 1.58 | 8.14 ± 1.73 | 7.33 ± 0.82 |
| MMP7 | 12.61 ± 1.04 | 12.89 ± 0.39 | 12.27 ± 1.49 | 12.55 ± 0.68 |
| ARG1 | 4.8 ± 1.17 | 4.94 ± 1.18 | 5.19 ± 1.1 | 4.69 ± 0.75 |
| NCR1 | 4.49 ± 0.81 | 4.61 ± 0.69 | 4.41 ± 0.68 | 4.47 ± 0.65 |
| DCN | 4.63 ± 0.49 | 4.6 ± 0.28 | 4.49 ± 0.37 | 4.55 ± 0.39 |
| TNFRSF21 | 10.75 ± 0.28 | 10.8 ± 0.21 | 10.73 ± 0.26 | 10.74 ± 0.22 |
| TNFRSF4 | 8.22 ± 0.83 | 8.13 ± 0.6 | 7.9 ± 0.62 | 7.98 ± 0.53 |
| MIC_A/B | 5.94 ± 2.15 | 6 ± 2.01 | 6.13 ± 2.02 | 6.05 ± 1.9 |
| CCL17 | 10.72 ± 1.19 | 11.25 ± 0.72 | 10.8 ± 1.09 | 10.79 ± 0.84 |
| ANGPT2 | 6.14 ± 0.74 | 6.05 ± 0.66 | 5.87 ± 0.73 | 5.83 ± 0.41 |
| PTN | 1.69 ± 0.62 | 1.78 ± 0.57 | 1.41 ± 0.65 | 1.8 ± 0.59 |
| CXCL12 | 1.55 ± 0.5 | 1.54 ± 0.33 | 1.51 ± 0.3 | 1.49 ± 0.51 |
| IFN_gamma | 7.98 ± 1.24 | 8.15 ± 0.97 | 8.9 ± 1.99 | 8.71 ± 1.61 |
| LAMP3 | 4.54 ± 0.65 | 4.63 ± 0.72 * | 4.98 ± 0.74 | 5.24 ± 0.82 |
| CASP_8 | 6.64 ± 0.95 | 6.57 ± 1.17 | 6.46 ± 1.05 | 6.39 ± 1.35 |
| ICOSLG | 6.19 ± 0.61 | 6.45 ± 0.77 | 6.01 ± 0.38 | 6.04 ± 0.36 |
| MMP12 | 8.8 ± 0.83 | 8.45 ± 0.65 | 8.69 ± 1.45 | 8.26 ± 0.58 |
| CXCL13 | 9.67 ± 1.04 | 9.43 ± 0.7 | 9.55 ± 0.93 | 9.61 ± 0.89 |
| PD_L2 | 3.25 ± 0.6 | 3.2 ± 0.54 | 3.14 ± 0.55 | 3.2 ± 0.57 |
| VEGFA | 10.37 ± 0.98 | 10.37 ± 0.88 | 10.58 ± 1.02 | 10.35 ± 0.73 |
| IL4 | 1.2 ± 0.54 | 0.88 ± 0.58 | 1.13 ± 0.37 | 0.81 ± 0.58 |
| LAG3 | 4.93 ± 0.77 | 5.19 ± 0.51 | 4.98 ± 0.8 | 4.96 ± 0.76 |
| IL12RB1 | 3.07 ± 0.84 | 2.96 ± 0.7 | 2.86 ± 0.53 | 2.88 ± 0.5 |
| IL13 | 0.31 ± 1.15 | 0.18 ± 0.57 | 0.66 ± 0.65 | 0.41 ± 0.68 |
| CCL20 | 8.52 ± 1.35 | 7.89 ± 1.23 | 8.51 ± 1.95 | 8.25 ± 1.75 |
| TNF | 4.95 ± 1.35 | 4.71 ± 1.61 | 4.85 ± 1.31 | 4.2 ± 0.79 |
| KLRD1 | 7.69 ± 0.85 | 7.61 ± 0.68 | 7.22 ± 0.87 | 7.25 ± 0.86 |
| GZMB | 5.28 ± 0.96 | 4.88 ± 0.65 | 5.14 ± 1.1 | 4.89 ± 0.78 |
| CD83 | 2.87 ± 0.76 | 2.88 ± 0.57 | 2.93 ± 0.54 | 2.92 ± 0.46 |
| IL12 | 5.37 ± 1.69 | 5.73 ± 1.07 | 5.38 ± 1.24 | 5.34 ± 1.31 |
| CSF_1 | 10.06 ± 0.32 | 10.01 ± 0.19 | 10.04 ± 0.31 | 9.98 ± 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuesta-Sancho, S.; Gomez, J.J.L.; García-Luna, P.P.; Primo, D.; Martínez-Ortega, A.J.; Izaola, O.; Casañas, T.; Calleja, A.; Bernardo, D.; de Luis, D. Soluble Oncoimmunome Signatures Predict Muscle Mass Response to Enriched Immunonutrition in Cancer Patients: Subanalysis of a Multicenter Randomized Clinical Trial. Nutrients 2025, 17, 2421. https://doi.org/10.3390/nu17152421
Cuesta-Sancho S, Gomez JJL, García-Luna PP, Primo D, Martínez-Ortega AJ, Izaola O, Casañas T, Calleja A, Bernardo D, de Luis D. Soluble Oncoimmunome Signatures Predict Muscle Mass Response to Enriched Immunonutrition in Cancer Patients: Subanalysis of a Multicenter Randomized Clinical Trial. Nutrients. 2025; 17(15):2421. https://doi.org/10.3390/nu17152421
Chicago/Turabian StyleCuesta-Sancho, Sara, Juan José López Gomez, Pedro Pablo García-Luna, David Primo, Antonio J. Martínez-Ortega, Olatz Izaola, Tamara Casañas, Alicia Calleja, David Bernardo, and Daniel de Luis. 2025. "Soluble Oncoimmunome Signatures Predict Muscle Mass Response to Enriched Immunonutrition in Cancer Patients: Subanalysis of a Multicenter Randomized Clinical Trial" Nutrients 17, no. 15: 2421. https://doi.org/10.3390/nu17152421
APA StyleCuesta-Sancho, S., Gomez, J. J. L., García-Luna, P. P., Primo, D., Martínez-Ortega, A. J., Izaola, O., Casañas, T., Calleja, A., Bernardo, D., & de Luis, D. (2025). Soluble Oncoimmunome Signatures Predict Muscle Mass Response to Enriched Immunonutrition in Cancer Patients: Subanalysis of a Multicenter Randomized Clinical Trial. Nutrients, 17(15), 2421. https://doi.org/10.3390/nu17152421







