2D Prediction of the Nutritional Composition of Dishes from Food Images: Deep Learning Algorithm Selection and Data Curation Beyond the Nutrition5k Project
Highlights
- Eight deep learning architectures were benchmarked for the direct 2D prediction of dish mass and nutrient content from food images, combining four feature extractors (ResNet 50 and 101, InceptionV3, ViT-B-16) with two regression heads.
- Model performance was evaluated across four curated ground-truth datasets derived from Nutrition5k, incorporating both U.S. and Italian food composition data and ingredient-level corrections.
- ResNet101 and ViT-B-16 architectures consistently outperformed the benchmark InceptionV3, suggesting their suitability for real-time, image-based nutrient estimation in digital health tools.
- An analysis of consistently mispredicted dishes across most algorithms revealed issues related to image quality and labeling, emphasizing the importance of automated quality control in scalable app development.
- Beyond dishes requiring ingredient mass corrections, commonly mispredicted groups included complex salads, chicken-based and egg-based dishes, and Western-style breakfasts.
- Instead of frame filtering on a subset of the test set including mispredicted dishes, ingredient mass correction substantially improved prediction metrics, highlighting the need for curated inputs in future mobile dietary assessment apps.
Abstract
1. Introduction
2. Materials and Methods
2.1. Analytical Framework and Ground Truth Datasets for Analysis
2.2. Deep Learning for Food Image Recognition
2.2.1. Splitting Data into Training, Validation, and Test Sets
2.2.2. Selecting the Architecture and Pre-Training Weights for the Automatic Feature Extraction
2.2.3. Extracting and Preprocessing Frames
2.2.4. Setting the Architecture for Problem Solving
2.2.5. Setting the Optimization of the Loss Function for Feature Extraction and Problem Solving
2.2.6. Setting the Performance Metrics for Problem Solving
2.2.7. Predicting Mass, Energy, and Macronutrient Content with and Without Ingredient–Mass Correction and Italian Nutritional Value Calculation
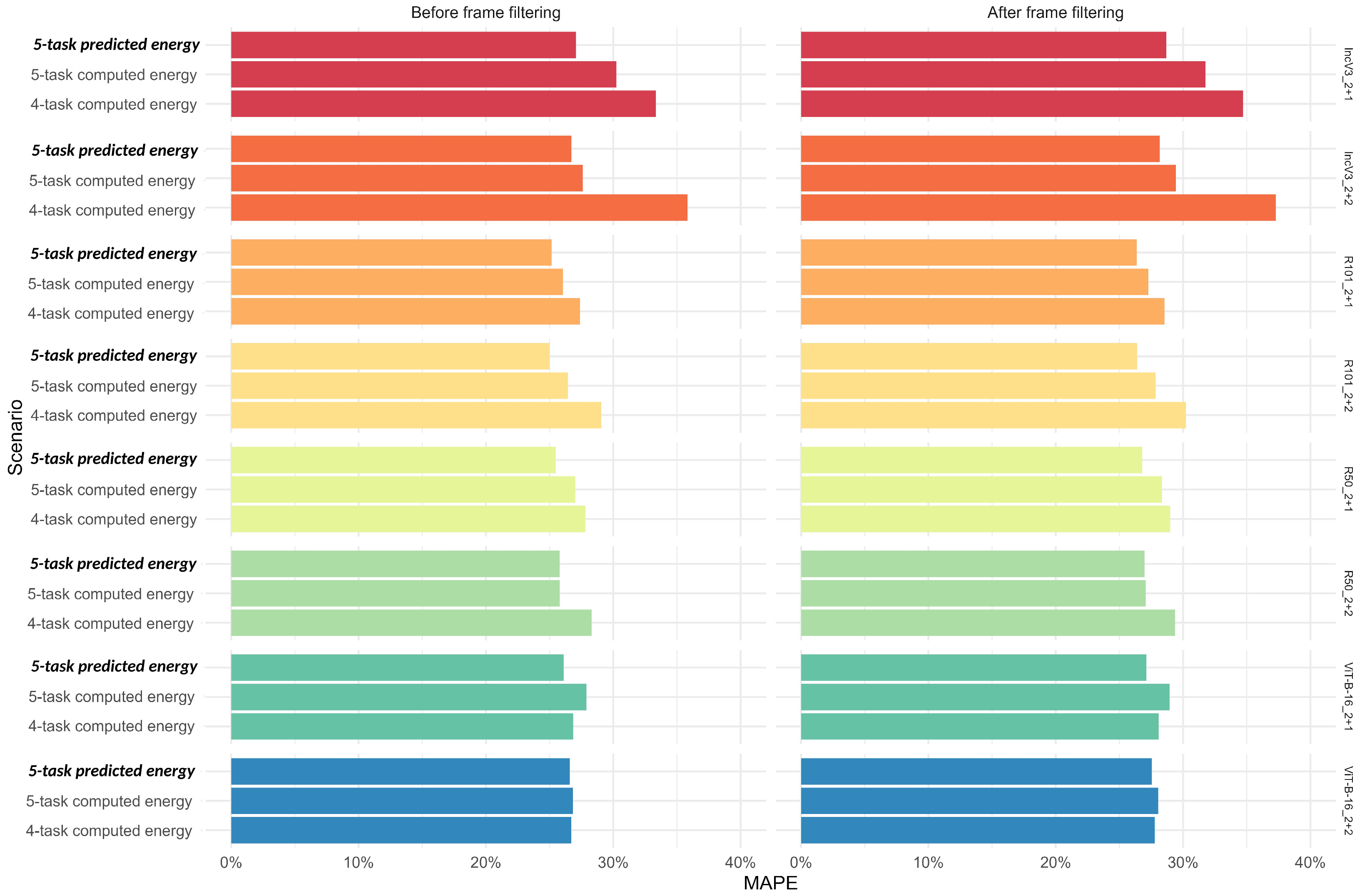
2.2.8. Sensitivity Analysis: Comparison of Predicted Versus Calculated Energy Content in 4-Task and 5-Task Scenarios
2.3. Statistical Analysis and Visual Inspection of Frames
3. Results
3.1. Descriptive Statistics on Observed and Predicted Values from the Test Set with and Without Ingredient–Mass Correction and Italian Nutritional Value Calculation
3.2. Comparison Between Observed and Predicted Values from the Test Set with and Without Ingredient–Mass Correction and Italian Nutritional Value Calculation
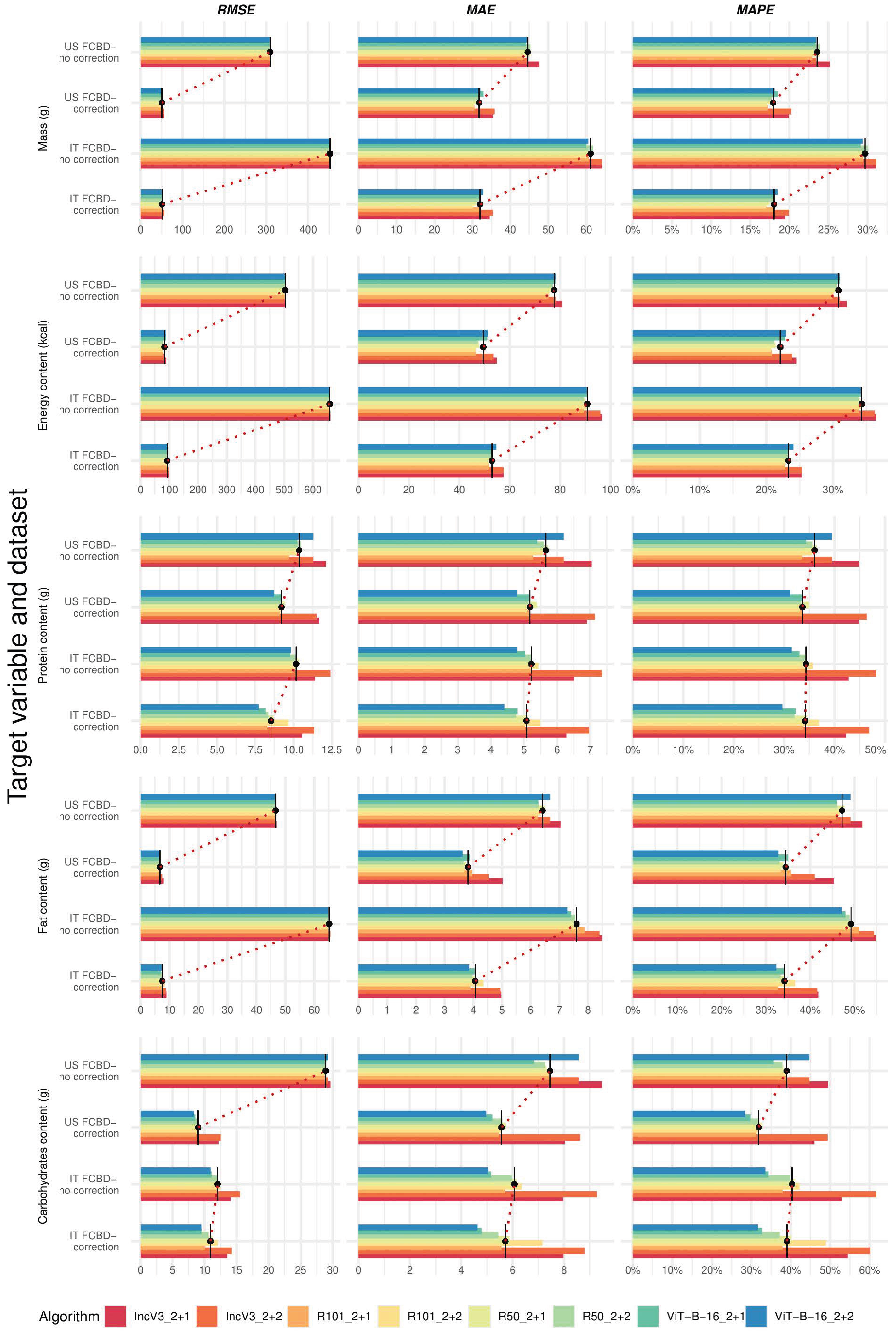
3.3. Comparison Between Metrics with and Without Ingredient–Mass Correction and Italian Nutritional Value Calculation
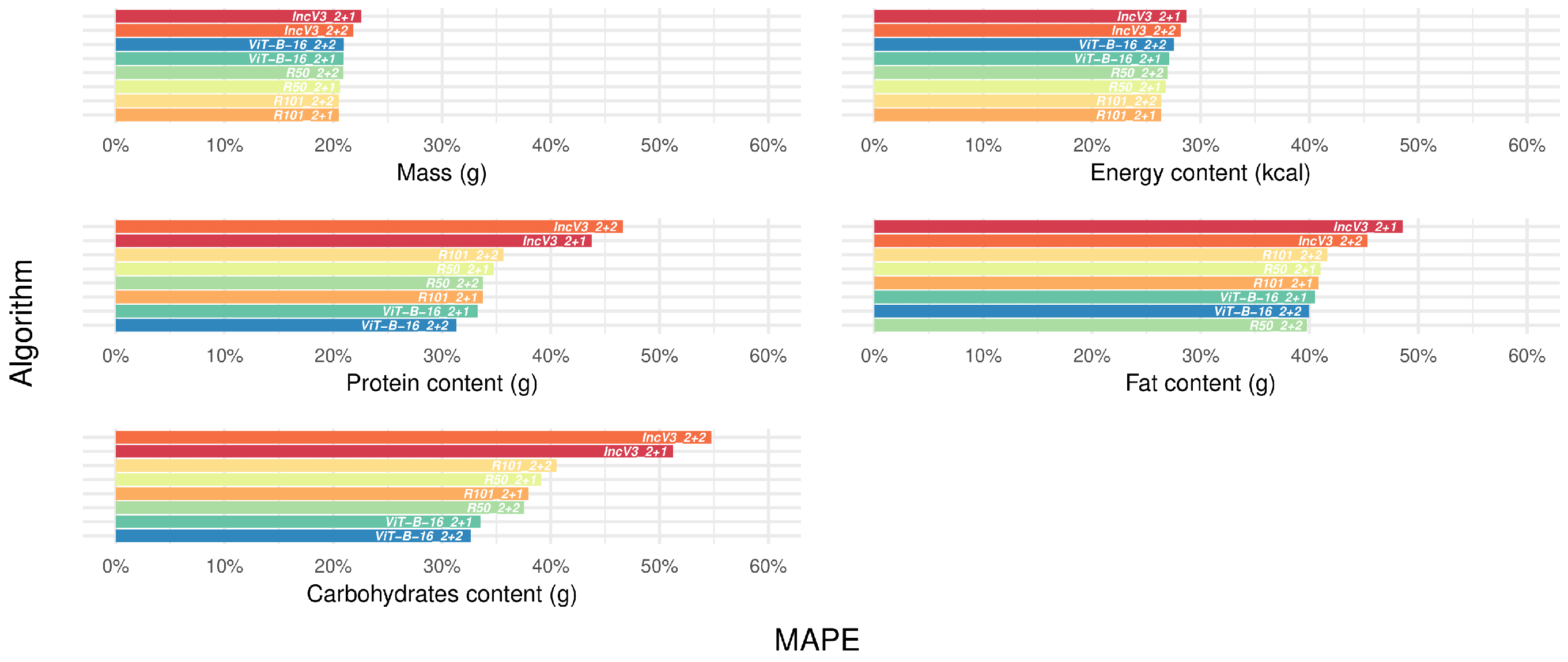
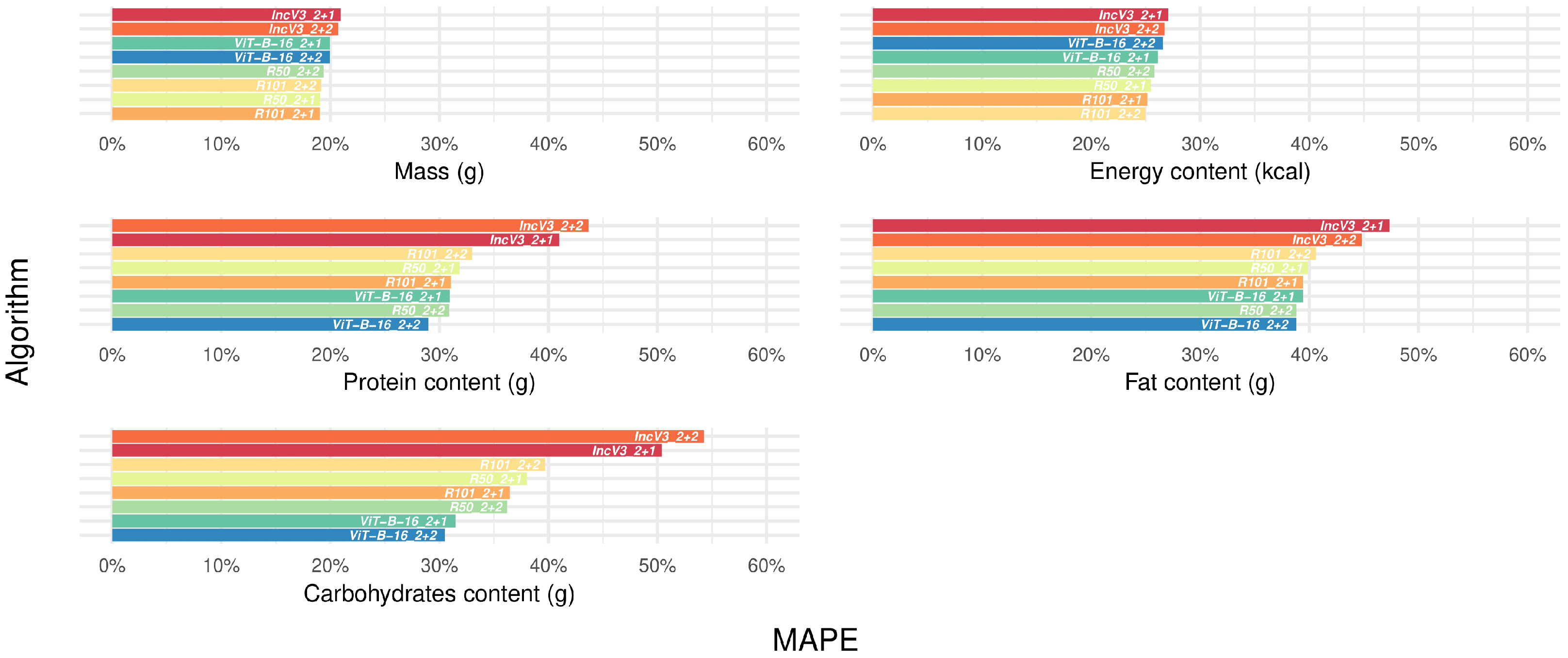
3.4. Selecting the Best-Performing Deep Learning Algorithms
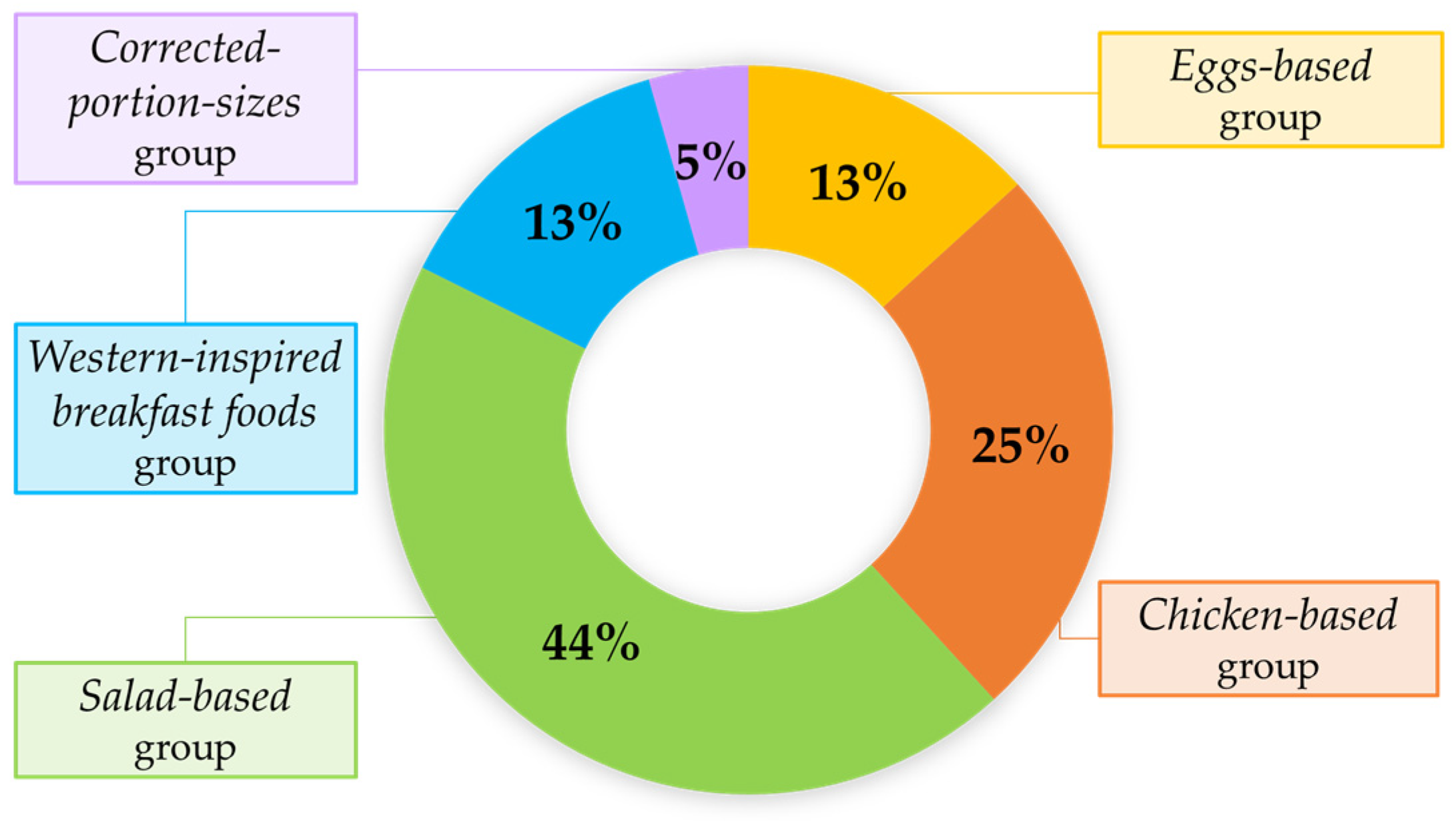
3.5. Incorrectly Predicted Dishes Across Target Variables and Datasets: A Focus on Nutritional Characteristics of the Ingredients After Manual Frame Filtering
- 7 dishes (dish_1563566909, dish_1566414291, dish_1566501575, dish_1566501594, dish_1566589933, dish_1563566939, and dish_1563566965) showed mismatches between visible ingredients and the (lower number of) ingredients listed in the metadata, mostly leading to overestimated predictions; the only reasonable exception was represented by carbohydrates, where dish ingredients were not entirely captured by most available images;
- 5 dishes (dish_1558630325, dish_1558720236, dish_1562703447, dish_1563389626, and dish_1566838351) suffered from image quality problems in all frames, including poor framing, blurriness, cropping, operator hands/feet in the frame, and background elements (e.g., chairs, tables, cables, and phones).
- Salad-based group (44%): Complex dishes, with at least two vegetables (e.g., raw and/or cooked), two protein sources (e.g., fish and/or meat), two cereal types, herbs, and dressings (median number of ingredients per dish: 16, IQR: 11.25–20.5);
- Chicken-based group (25%): Dishes primarily composed of chicken (grilled, breaded, or sliced in a cold salad), simpler in composition, but sometimes visually ambiguous (median number of ingredients per dish: 4, IQR: 3–5);
- Eggs-based group (13%): Dishes featuring mainly scrambled eggs or boiled egg whites, with few ingredients, but often showing partial occlusion (median number of ingredients per dish: 5, IQR: 3–6);
- Western-inspired breakfast foods group (13%): Hybrid category featuring dishes with a limited number of ingredients, blending elements of both sweet (e.g., fruit, almonds, or brownies) and savory (e.g., potatoes, sausages, or cheese) breakfasts without a single dominant component (median number of ingredients per dish: 2, IQR: 1–3).
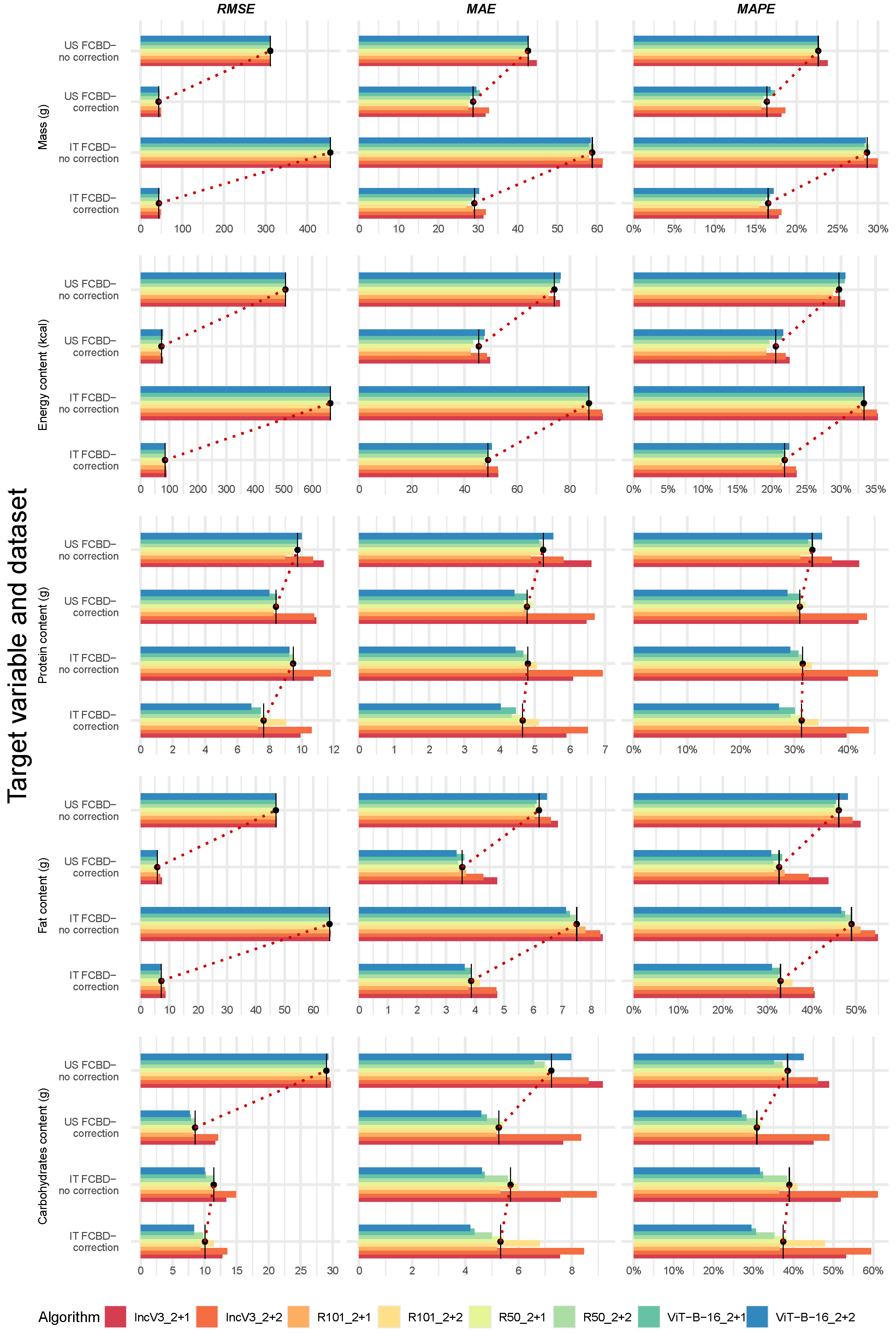
3.6. Confirming the Best-Performing Deep Learning Algorithms After Manual Frame Filtering
3.7. Sensitivity Analysis: Comparing Predicted Versus Calculated Energy Content Using the 4-Task and 5-Task Deep Learning Algorithms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| DL | Deep Learning |
| FCDB | Food Composition DataBase |
| Inc | Inception |
| MAE | Mean Absolute Error |
| MAPE | Mean Absolute Percentage Error |
| MLP | Multi-Layer Perceptron |
| ReLU | Rectified Linear Unit |
| RGB | Red, Green, and Blue |
| ResNet | Residual Network |
| RMSE | Root Mean Squared Error |
| ViT | Vision Transformer |
References
- Willett, W. Nutritional Epidemiology, 3rd ed.; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Pala, V.; Sieri, S.; Palli, D.; Salvini, S.; Berrino, F.; Bellegotti, M.; Frasca, G.; Tumino, R.; Sacerdote, C.; Fiorini, L.; et al. Diet in the Italian EPIC Cohorts: Presentation of Data and Methodological Issues. Tumori J. 2003, 89, 594–607. [Google Scholar] [CrossRef] [PubMed]
- Edefonti, V.; De Vito, R.; Parpinel, M.; Ferraroni, M. Dietary Patterns and Cancer Risk: An Overview with Focus on Methods. N. Engl. J. Stat. Data Sci. 2024, 2, 30–53. [Google Scholar] [CrossRef]
- Bianco, R.; Marinoni, M.; Coluccia, S.; Carioni, G.; Fiori, F.; Gnagnarella, P.; Edefonti, V.; Parpinel, M. Tailoring the Nutritional Composition of Italian Foods to the US Nutrition5k Dataset for Food Image Recognition: Challenges and a Comparative Analysis. Nutrients 2024, 16, 3339. [Google Scholar] [CrossRef] [PubMed]
- Kirk, D.; Catal, C.; Tekinerdogan, B. Precision Nutrition: A Systematic Literature Review. Comput. Biol. Med. 2021, 133, 104365. [Google Scholar] [CrossRef]
- Boushey, C.J.; Spoden, M.; Zhu, F.M.; Delp, E.J.; Kerr, D.A. New Mobile Methods for Dietary Assessment: Review of Image-Assisted and Image-Based Dietary Assessment Methods. Proc. Nutr. Soc. 2017, 76, 283–294. [Google Scholar] [CrossRef]
- Lu, Y.; Stathopoulou, T.; Vasiloglou, M.F.; Christodoulidis, S.; Blum, B.; Walser, T.; Meier, V.; Stanga, Z.; Mougiakakou, S.G. An Artificial Intelligence-Based System for Nutrient Intake Assessment of Hospitalised Patients. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Berlin, Germany, 23–27 July 2019; pp. 5696–5699. [Google Scholar]
- Oka, R.; Nomura, A.; Yasugi, A.; Kometani, M.; Gondoh, Y.; Yoshimura, K.; Yoneda, T. Study Protocol for the Effects of Artificial Intelligence (AI)-Supported Automated Nutritional Intervention on Glycemic Control in Patients with Type 2 Diabetes Mellitus. Diabetes Ther. 2019, 10, 1151–1161. [Google Scholar] [CrossRef]
- Chen, G.; Jia, W.; Zhao, Y.; Mao, Z.H.; Lo, B.; Anderson, A.K.; Frost, G.; Jobarteh, M.L.; McCrory, M.A.; Sazonov, E.; et al. Food/Non-Food Classification of Real-Life Egocentric Images in Low- and Middle-Income Countries Based on Image Tagging Features. Front. Artif. Intell. 2021, 4, 644712. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, Q.; Siddiquie, B.; Divakaran, A.; Sawhney, H. “snap-n-Eat”: Food Recognition and Nutrition Estimation on a Smartphone. J. Diabetes Sci. Technol. 2015, 9, 525–533. [Google Scholar] [CrossRef]
- Mezgec, S.; Seljak, B.K. Deep Neural Networks for Image-Based Dietary Assessment. J. Vis. Exp. 2021, 2021, e61906. [Google Scholar] [CrossRef]
- Lo, F.P.W.; Sun, Y.; Qiu, J.; Lo, B. Image-Based Food Classification and Volume Estimation for Dietary Assessment: A Review. IEEE J. Biomed. Health Inform. 2020, 24, 1926–1939. [Google Scholar] [CrossRef]
- Tahir, G.A.; Loo, C.K. Explainable Deep Learning Ensemble for Food Image Analysis on Edge Devices. Comput. Biol. Med. 2021, 139, 104972. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.; Wook, J.; Chris, K.; Aditya, H.; Gabriel, R.; Sandhini, G.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning Transferable Visual Models From Natural Language Supervision. In Proceedings of the 38th International Conference on Machine Learning, PMLR, Virtual, 18–24 July 2021. [Google Scholar]
- Sahoo, D.; Hao, W.; Ke, S.; Xiongwei, W.; Le, H.; Achananuparp, P.; Lim, E.P.; Hoi, S.C.H. FoodAI: Food Image Recognition via Deep Learning for Smart Food Logging. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 2260–2268. [Google Scholar]
- Dillet, R. Foodvisor Automatically Tracks What You Eat Using Deep Learning. Available online: https://techcrunch.com/2019/10/14/foodvisor-automatically-tracks-what-you-eat-using-deep-learning/ (accessed on 20 June 2025).
- Min, W.; Jiang, S.; Liu, L.; Rui, Y.; Jain, R. A Survey on Food Computing. ACM Comput. Surv. 2019, 52, 92. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, N.H.; Jo, D.; Won, C.S. Multimodal Food Image Classification with Large Language Models. Electronics 2024, 13, 4552. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, P.; Jiang, Y.; Xia, K. Visual Nutrition Analysis: Leveraging Segmentation and Regression for Food Nutrient Estimation. Front. Nutr. 2024, 11, 1469878. [Google Scholar] [CrossRef]
- Xu, M.; Wang, J.; Tao, M.; Bao, B.K.; Xu, C. CookGALIP: Recipe Controllable Generative Adversarial CLIPs with Sequential Ingredient Prompts for Food Image Generation. IEEE Trans. Multimed. 2025, 27, 2772–2782. [Google Scholar] [CrossRef]
- Salvador, A.; Drozdzal, M.; Giro-I-Nieto, X.; Romero, A. Inverse Cooking: Recipe Generation from Food Images. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 10445–10454. [Google Scholar]
- Su, Z.; Shen, J.; Zhou, Q.; Yong, B. FoodFlavorNet: A Multimodal Deep Learning Model for Food Flavor Recognition. IEEE Trans. Consum. Electron. 2024, 1. [Google Scholar] [CrossRef]
- Cao, Y.; Kementchedjhieva, Y.; Cui, R.; Karamolegkou, A.; Zhou, L.; Dare, M.; Donatelli, L.; Hershcovich, D. Cultural Adaptation of Recipes. Trans. Assoc. Comput. Linguist. 2024, 12, 80–99. [Google Scholar] [CrossRef]
- Wu, X.; Fu, X.; Liu, Y.; Lim, E.P.; Hoi, S.C.H.; Sun, Q. A Large-Scale Benchmark for Food Image Segmentation. In Proceedings of the 29th ACM International Conference on Multimedia, Virtual, 20–24 October 2021; pp. 506–515. [Google Scholar]
- Liu, Y.; Min, W.; Jiang, S.; Rui, Y. Convolution-enhanced bi-branch adaptive transformer with cross-task interaction for food category and ingredient recognition. IEEE Trans. Image Process. 2024, 33, 2572–2586. [Google Scholar] [CrossRef]
- Konstantakopoulos, F.S.; Georga, E.I.; Fotiadis, D.I. A Novel Approach to Estimate the Weight of Food Items Based on Features Extracted from an Image Using Boosting Algorithms. Sci. Rep. 2023, 13, 21040. [Google Scholar] [CrossRef]
- Kagaya, H.; Aizawa, K. Highly Accurate Food/Non-Food Image Classification Based on a Deep Convolutional Neural Network. In Proceedings of the ICIAP 2015 International Workshops, BioFor, CTMR, RHEUMA, ISCA, MADiMa, SBMI, and QoEM, Genoa, Italy, 7–8 September 2015; pp. 350–357. [Google Scholar]
- Martinel, N.; Foresti, G.L.; Micheloni, C. Wide-Slice Residual Networks for Food Recognition. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision, Lake Tahoe, NV, USA, 12–15 March 2018; pp. 567–576. [Google Scholar]
- Wang, H.; Min, W.; Xiangyang, L.; Jiang, S. Where and What to Eat: Simultaneous Restaurant and Dish Recognition from Food Image. In Proceedings of the Advances in Multimedia Information Processing, Xi’an, China, 15–16 September 2016; Springer International Publishing: Beijing, China, 2016; pp. 520–528. [Google Scholar]
- Yanai, K.; Kawano, Y. Food Image Recognition Using Deep Convolutional Network with Pre-Training and Fine-Tuning. In Proceedings of the IEEE International Conference on Multimedia and Expo Workshops, ICMEW 2015, Turin, Italy, 29 June–3 July 2015; IEEE: Turin, Italy, 2015; pp. 1–6. [Google Scholar]
- Bossard, L.; Guillaumin, M.; Van Gool, L. Food-101—Mining Discriminative Components with Random Forests. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Volume 8694, pp. 446–461. [Google Scholar]
- Chen, J.; Ngo, C.; Chen, J. Deep-Based Ingredient Recognition for Cooking Recipe Retrieval. In Proceedings of the 24th ACM International conference on Multimedia, Amsterdam, The Netherlands, 15–19 October 2016; pp. 32–41. [Google Scholar]
- Chen, J.; Zhu, B.; Ngo, C.W.; Chua, T.S.; Jiang, Y.G. A Study of Multi-Task and Region-Wise Deep Learning for Food Ingredient Recognition. IEEE Trans. Image Process. 2021, 30, 1514–1526. [Google Scholar] [CrossRef] [PubMed]
- Tai, C.A.; Nair, S.; Markham, O.; Keller, M.; Wu, Y.; Chen, Y.; Wong, A. NutritionVerse-Real: An Open Access Manually Collected 2D Food Scene Dataset for Dietary Intake Estimation. In Proceedings of the 37th Conference on Neural Information Processing Systems, New Orleans, LA, USA, 10–16 December 2023; Volume 2, pp. 1–4. [Google Scholar]
- Nair, S.; Tai, C.A.; Chen, Y.; Wong, A. NutritionVerse-Synth: An Open Access Synthetically Generated 2D Food Scene Dataset for Dietary Intake Estimation. In Proceedings of the 37th Conference on Neural Information Processing Systems, New Orleans, LA, USA, 10–16 December 2023. [Google Scholar]
- Chen, Y.; He, J.; Vinod, G.; Raghavan, S.; Czarnecki, C.; Ma, J.; Mahmud, T.I.; Coburn, B.; Mao, D.; Nair, S.; et al. MetaFood3D: 3D Food Dataset with Nutrition Values. arXiv 2024, arXiv:2409.01966. [Google Scholar]
- Thames, Q.; Karpur, A.; Norris, W.; Xia, F.; Panait, L.; Weyand, T.; Sim, J. Nutrition5k: Towards Automatic Nutritional Understanding of Generic Food. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 8899–8907. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture. Agricultural Research Service. Available online: https://fdc.nal.usda.gov/ (accessed on 20 June 2025).
- Aldoseri, A.; Al-Khalifa, K.N.; Hamouda, A.M. Re-Thinking Data Strategy and Integration for Artificial Intelligence: Concepts, Opportunities, and Challenges. Appl. Sci. 2023, 13, 7082. [Google Scholar] [CrossRef]
- Budach, L.; Feuerpfeil, M.; Ihde, N.; Nathansen, A.; Noack, N.; Patzlaff, H.; Naumann, F.; Harmouch, H. The Effects of Data Quality on Machine Learning Performance. arXiv 2022, arXiv:2207.14529. [Google Scholar]
- Greenfield, H.; Southgate, D.A. Food Composition Data: Production, Management, and Use; Food & Agriculture Organization: Rome, Italy, 2003; Volume II, ISBN 9251049491. [Google Scholar]
- Finglas, P.M.; Berry, R.; Astley, S. Assessing and Improving the Quality of Food Composition Databases for Nutrition and Health Applications in Europe: The Contribution of EuroFIR. Adv. Nutr. 2014, 5, 608S–614S. [Google Scholar] [CrossRef]
- Drewnowski, A.; Heber, D. Nutrient Composition of Foods: The First Step in Precision Nutrition. In Precision Nutrition; Academic Press: Cambridge, MA, USA, 2024; pp. 3–22. [Google Scholar]
- Nutrition 5k Dataset. Available online: https://paperswithcode.com/dataset/nutrition5k (accessed on 20 June 2025).
- Gnagnarella, P.; Parpinel, M.; Salvini, S. Banca Dati Di Composizione Degli Alimenti per Studi Epidemiologici in Italia. Edizione Compatta; Libreriauniversitaria.it Edizioni: Padova, Italy, 2022. [Google Scholar]
- Gnagnarella, P.; Salvini, S.; Parpinel, M. Food Composition Database for Epidemiological Studies in Italy. Available online: https://bda.ieo.it/ (accessed on 20 June 2025).
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Pytorch. Available online: https://pytorch.org/vision/stable/models.html (accessed on 20 June 2025).
- Jia, H.; Zhang, J.; Ma, K.; Qiao, X.; Ren, L.; Shi, X. Application of Convolutional Neural Networks in Medical Images: A Bibliometric Analysis. Quant. Imaging Med. Surg. 2024, 14, 3501–3518. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Virtual, 3–7 May 2021; pp. 1–21. [Google Scholar]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting Unreasonable Effectiveness of Data in Deep Learning Era. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 843–852. [Google Scholar]
- ResNet. Available online: https://pytorch.org/vision/stable/models/generated/torchvision.models.resnet50.html#torchvision.models.ResNet50_Weights (accessed on 20 June 2025).
- Chai, T.; Draxler, R.R. Root Mean Square Error (RMSE) or Mean Absolute Error (MAE)?—Arguments against Avoiding RMSE in the Literature. Geosci. Model Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef]
- Caruana, R. Multitask Learning. Mach. Learn. 1997, 28, 41–75. [Google Scholar] [CrossRef]
- Van Puyvelde, H.; Perez-cornago, A.; Casagrande, C.; Versele, V.; Skeie, G.; Schulze, M.B.; Johansson, I. Comparing Calculated Nutrient Intakes Using Different Food Composition Databases: Results from the European Prospective Investigation into Cancer and Nutrition (EPIC) Cohort. Nutrients 2020, 12, 2906. [Google Scholar] [CrossRef]
- Westenbrink, S.; Roe, M.; Oseredczuk, M.; Castanheira, I.; Finglas, P. EuroFIR Quality Approach for Managing Food Composition Data; Where Are We in 2014? Food Chem. 2016, 193, 69–74. [Google Scholar] [CrossRef]
- ECUSTFD-Resized. Available online: https://github.com/Liang-yc/ECUSTFD-resized- (accessed on 20 June 2025).
- Food-101-CNN. Available online: https://github.com/prajwalsable99/Food-101-CNN/blob/main/food-nutrients.csv (accessed on 20 June 2025).
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A Simple Framework for Contrastive Learning of Visual Representations. In Proceedings of the 37th International Conference on Machine Learning, Online, 13–18 July 2020; pp. 1575–1585. [Google Scholar]
- Caron, M.; Misra, I.; Mairal, J.; Goyal, P.; Bojanowski, P.; Joulin, A. Unsupervised Learning of Visual Features by Contrasting Cluster Assignments. Adv. Neural Inf. Process. Syst. 2020, 33, 9912–9924. [Google Scholar]
- Tai, C.A.; Keller, M.; Kerrigan, M.; Chen, Y.; Xi, P. NutritionVerse-3D: A 3D Food Model Dataset for Nutritional Intake Estimation. arXiv 2021, arXiv:2304.05619. [Google Scholar]
- Pre-NeRF 360. Available online: https://amughrabi.github.io/prenerf/ (accessed on 20 June 2025).
- Tosi, M.; Radice, D.; Carioni, G.; Vecchiati, T.; Fiori, F.; Parpinel, M.; Gnagnarella, P. Accuracy of Applications to Monitor Food Intake: Evaluation by Comparison with 3-d Food Diary. Nutrition 2021, 84, 111018. [Google Scholar] [CrossRef] [PubMed]
| Original Model Identifier 1 | Label Used in Tables and Figures 2 |
|---|---|
| Inception_V3_IMAGENET1K_V1_2+1 | IncV3_2+1 |
| Inception_V3_IMAGENET1K_V1_2+2 | IncV3_2+2 |
| ResNet101_IMAGENET1K_V2_2+1 | R101_2+1 |
| ResNet101_IMAGENET1K_V2_2+2 | R101_2+2 |
| ResNet50_IMAGENET1K_V2_2+1 | R50_2+1 |
| ResNet50_IMAGENET1K_V2_2+2 | R50_2+2 |
| ViT_B_16_IMAGENET1K_SWAG_E2E_V1_2+1 | ViT-B-16_2+1 |
| ViT_B_16_IMAGENET1K_SWAG_E2E_V1_2+2 | ViT-B-16_2+2 |
| Single Available Datasets 1 | Merged Dataset 1 | ||||
|---|---|---|---|---|---|
| Target Variable | US FCDB—No Correction | US FCDB—Correction | IT FCDB—No Correction | IT FCDB—Correction | |
| Mass (g) | |||||
| Median (Q1, Q3) | 142.0 (70.5, 250.0) | 142.0 (70.0, 249.0) | 143.0 (70.5, 251.5) | 142.0 (70.0, 249.0) | 142.0 (70.0, 250.0) |
| Mean (SD) | 189.1 (332.1) | 177.3 (142.9) | 205.9 (467.1) | 177.4 (142.9) | 187.4 (304) |
| Min, Max | 1.0, 7974.0 | 1.0, 871.0 | 1.0, 8094.0 | 1.0, 871.0 | 1.0, 8094.0 |
| Energy content (kcal) | |||||
| Median (Q1, Q3) | 161.9 (61.5, 343.4) | 160.0 (61.2, 339.7) | 167.2 (60.6, 333.3) | 165.6 (60.3, 329.8) | 164.5 (60.8, 335.3) |
| Mean (SD) | 252.1 (536.2) | 224.0 (205.6) | 264.7 (687.8) | 227.5 (218.1) | 242.1 (461.0) |
| Min, Max | 0.0, 9485.8 | 0.0, 1050.5 | 0.0, 12,376.1 | 0.0, 1332.6 | 0.0, 12,376.1 |
| Protein content (g) | |||||
| Median (Q1, Q3) | 8.7 (1.6, 23.0) | 8.5 (1.6, 22.7) | 7.9 (1.6, 22.0) | 7.9 (1.6, 21.8) | 8.3 (1.6, 22.2) |
| Mean (SD) | 15.7 (19.3) | 15.4 (18.9) | 15.2 (18.7) | 14.9 (17.9) | 15.3 (18.7) |
| Min, Max | 0.0, 105.6 | 0.0, 105.6 | 0.0, 123.6 | 0.0, 89.8 | 0.0, 123.6 |
| Fat content (g) | |||||
| Median (Q1, Q3) | 6.6 (0.6, 17.6) | 6.6 (0.6, 17.4) | 7.5 (0.4, 17.4) | 7.4 (0.4, 17.2) | 6.9 (0.5, 17.4) |
| Mean (SD) | 13.6 (48.3) | 11.1 (13.5) | 15.4 (66.8) | 11.9 (15.1) | 13.0 (42.4) |
| Min, Max | 0.0, 875.5 | 0.0, 84.2 | 0.0, 1221.8 | 0.0, 115.8 | 0.0, 1221.8 |
| Carbohydrates content (g) | |||||
| Median (Q1, Q3) | 13.1 (4.4, 25.7) | 13.0 (4.4, 25.4) | 9.1 (2.3, 20.5) | 9.0 (2.2, 20.5) | 11.3 (3.3, 23.5) |
| Mean (SD) | 19.2 (31.5) | 17.5 (16.4) | 15.0 (17.9) | 14.6 (16.9) | 16.6 (21.7) |
| Min, Max | 0.0, 506.1 | 0.0, 85.8 | 0.0, 138.6 | 0.0, 116.5 | 0.0, 506.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianco, R.; Coluccia, S.; Marinoni, M.; Falcon, A.; Fiori, F.; Serra, G.; Ferraroni, M.; Edefonti, V.; Parpinel, M. 2D Prediction of the Nutritional Composition of Dishes from Food Images: Deep Learning Algorithm Selection and Data Curation Beyond the Nutrition5k Project. Nutrients 2025, 17, 2196. https://doi.org/10.3390/nu17132196
Bianco R, Coluccia S, Marinoni M, Falcon A, Fiori F, Serra G, Ferraroni M, Edefonti V, Parpinel M. 2D Prediction of the Nutritional Composition of Dishes from Food Images: Deep Learning Algorithm Selection and Data Curation Beyond the Nutrition5k Project. Nutrients. 2025; 17(13):2196. https://doi.org/10.3390/nu17132196
Chicago/Turabian StyleBianco, Rachele, Sergio Coluccia, Michela Marinoni, Alex Falcon, Federica Fiori, Giuseppe Serra, Monica Ferraroni, Valeria Edefonti, and Maria Parpinel. 2025. "2D Prediction of the Nutritional Composition of Dishes from Food Images: Deep Learning Algorithm Selection and Data Curation Beyond the Nutrition5k Project" Nutrients 17, no. 13: 2196. https://doi.org/10.3390/nu17132196
APA StyleBianco, R., Coluccia, S., Marinoni, M., Falcon, A., Fiori, F., Serra, G., Ferraroni, M., Edefonti, V., & Parpinel, M. (2025). 2D Prediction of the Nutritional Composition of Dishes from Food Images: Deep Learning Algorithm Selection and Data Curation Beyond the Nutrition5k Project. Nutrients, 17(13), 2196. https://doi.org/10.3390/nu17132196







