Low TAS1R2 Sweet Taste Receptor Expression in Skeletal Muscle of Genetically Diverse BXD Mice Mirrors Transcriptomic Signatures of Loss-of-Function Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model and Tissue Collection
2.2. RNA Isolation and Analysis
2.3. Transcriptomics
2.4. Comparative Transcriptomics in BL6 and BXD Models
2.5. Gene Set Enrichment and Overrepresentation Analysis
2.6. Statistics
3. Results
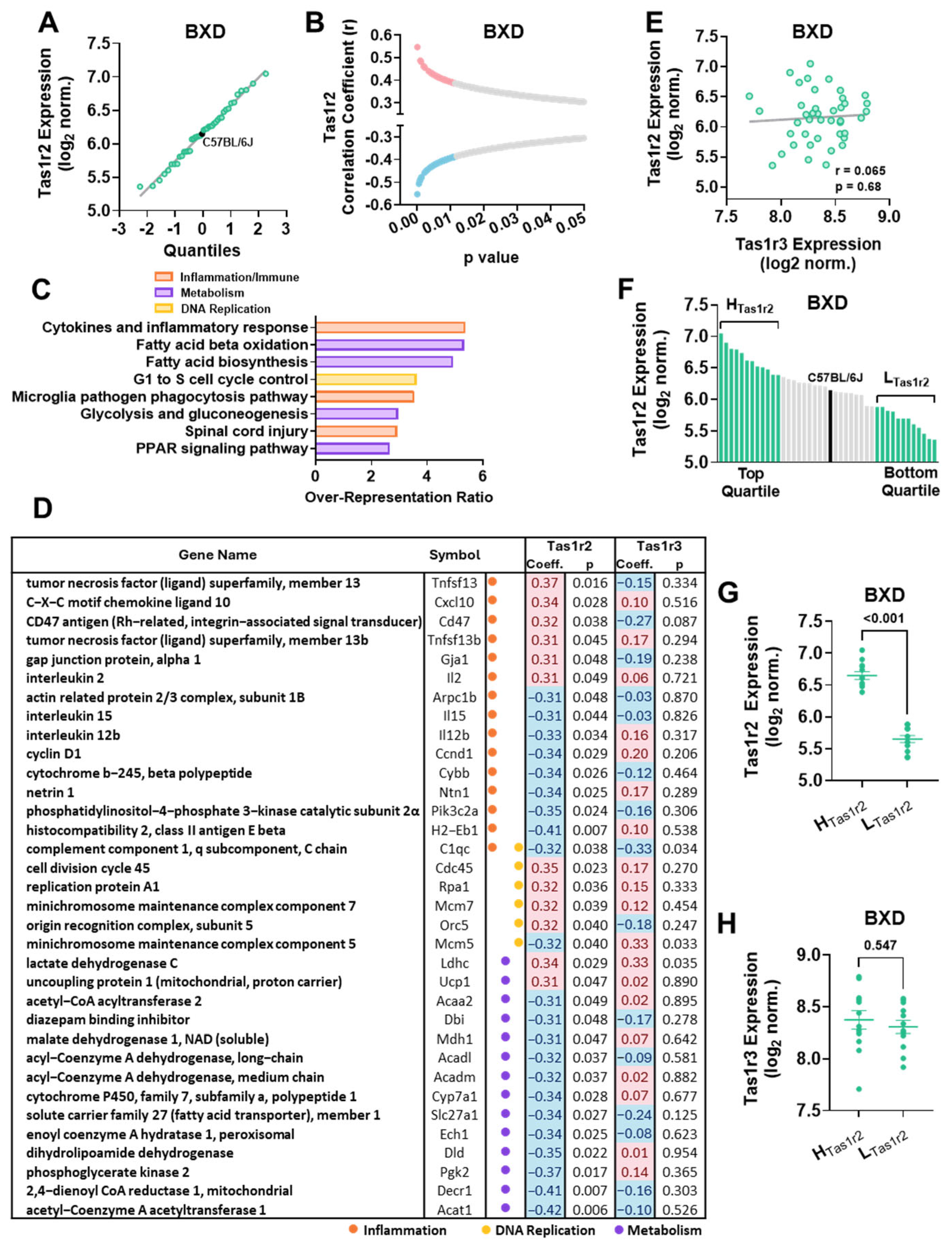
3.1. Tas1r2 Expression Ranking and Correlation Analysis in Skeletal Muscle of Genetically Diverse BXD Mice
3.2. Gene Set Enrichment Analysis (GSEA) in BXD Mice Stratified by Tas1r2 Expression
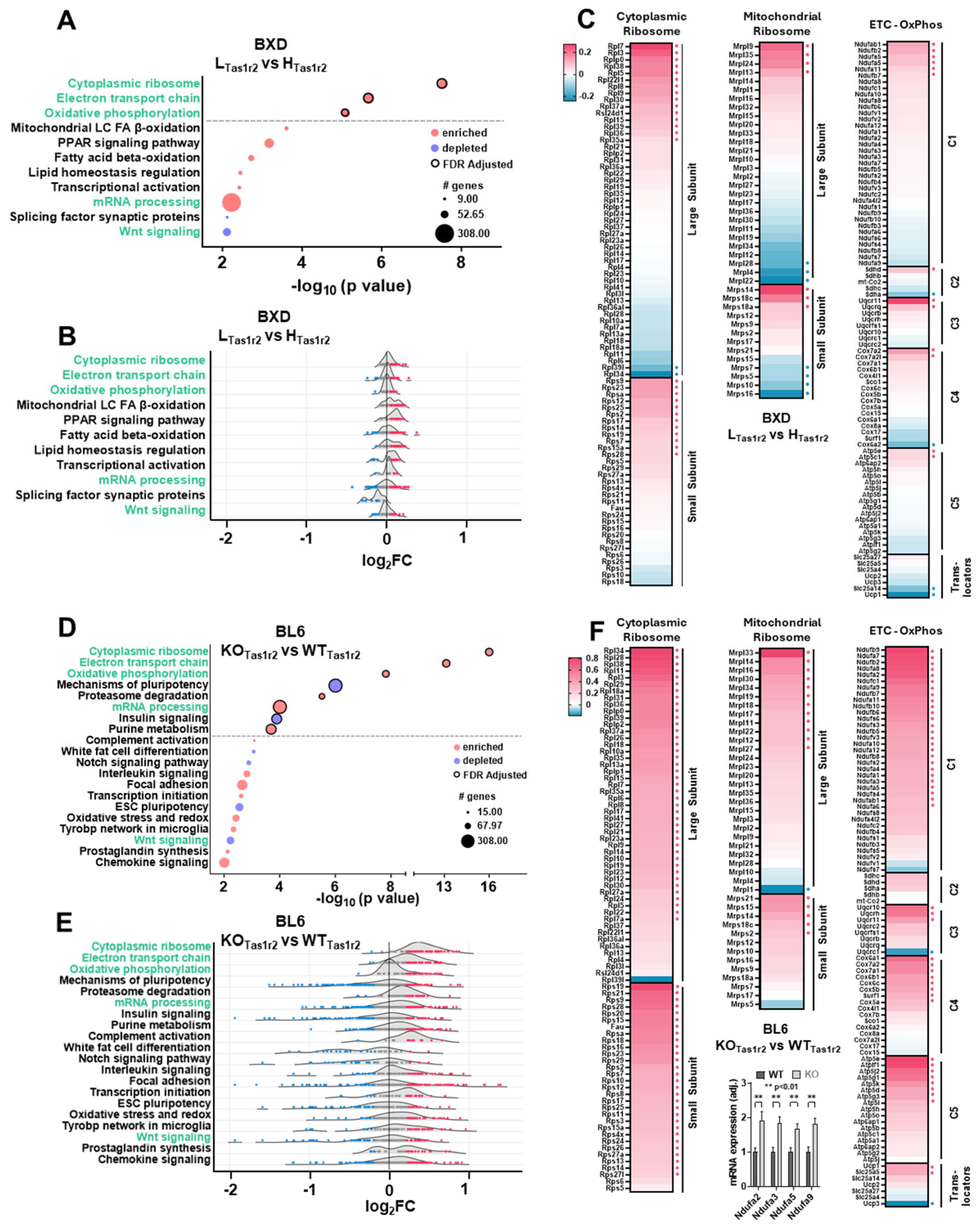
3.3. Comparison of Muscle Transcriptomics Between BXD LTas1r2 and BL6 KOTas1r2 Mice
3.4. Transcriptional Regulation of Shared Molecular Signatures in BL6 KOTas1r2 and BXD LTas1r2 Mice
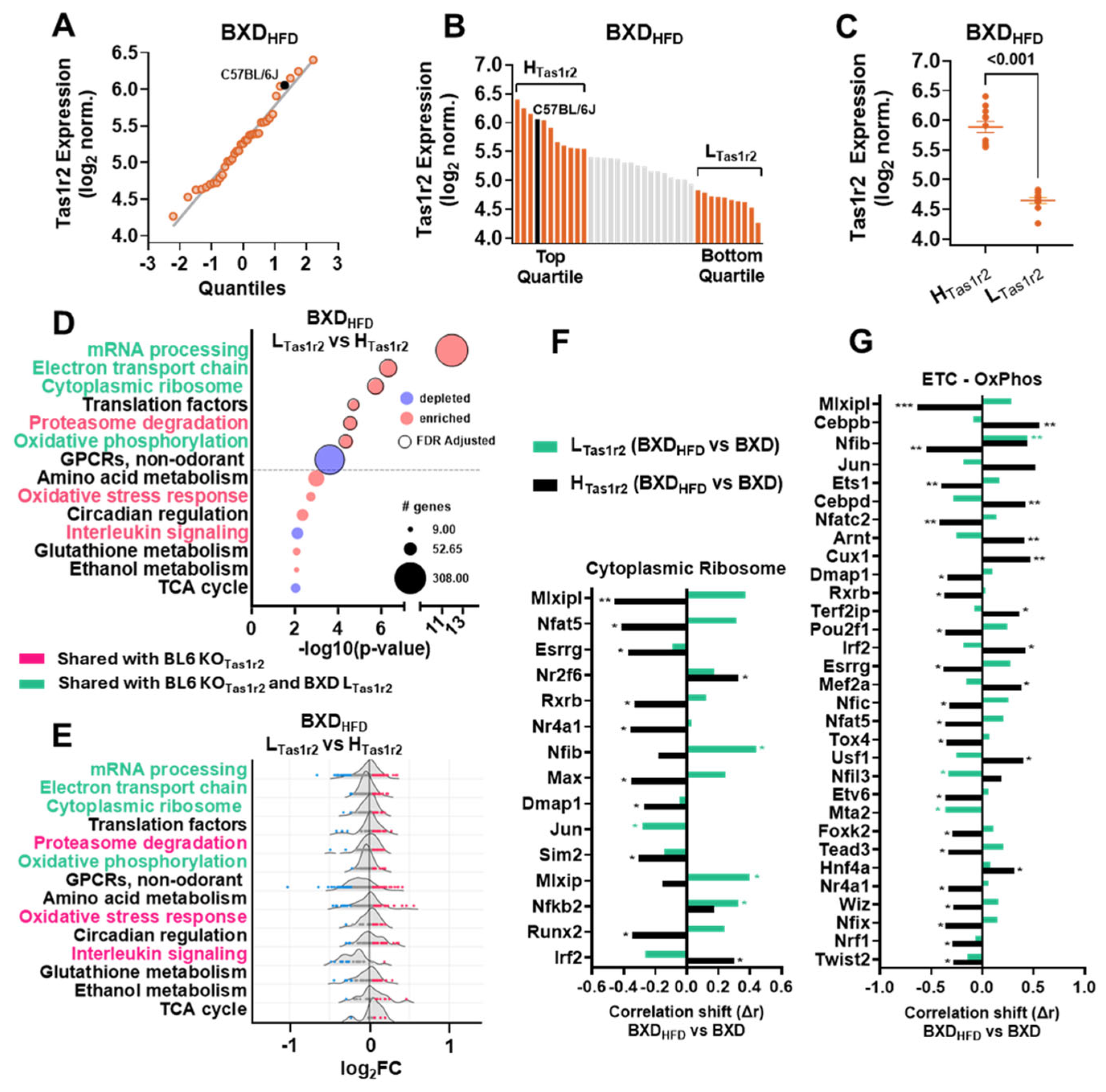
3.5. The Effects of a High-Fat Diet (HFD) on the Transcriptional Signatures in BXD Muscles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| LOF | Loss-of-function |
| SNP | Single nucleotide polymorphism |
| BXD | C57BL/6J × DBA/2J |
| RI | Recombinant Inbred |
| GRP | Genetic reference population |
| QTL | Quantitative trait locus |
| GWAS | Genome-wide association study |
| GPCR | G-protein coupled receptor |
| HFD | High-fat diet |
| KO | Knockout |
| WT | Wildtype |
| ORA | Over-representation analysis |
| OXPHOS | Oxidative Phosphorylation |
| GSEA | Gene Set Enrichment Analysis |
| ETC | Electron transport chain |
| TF | Transcription factor |
References
- MacArthur, D.G.; Balasubramanian, S.; Frankish, A.; Huang, N.; Morris, J.; Walter, K.; Jostins, L.; Habegger, L.; Pickrell, J.K.; Montgomery, S.B.; et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 2012, 335, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Monroe, J.G.; McKay, J.K.; Weigel, D.; Flood, P.J. The population genomics of adaptive loss of function. Heredity 2021, 126, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Doyle, A.; McGarry, M.P.; Lee, N.A.; Lee, J.J. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 2012, 21, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Sareila, O.; Hagert, C.; Rantakari, P.; Poutanen, M.; Holmdahl, R. Direct Comparison of a Natural Loss-Of-Function Single Nucleotide Polymorphism with a Targeted Deletion in the Ncf1 Gene Reveals Different Phenotypes. PLoS ONE 2015, 10, e0141974. [Google Scholar] [CrossRef]
- Liao, B.Y.; Zhang, J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 6987–6992. [Google Scholar] [CrossRef]
- Murthy, V.; Tebaldi, T.; Yoshida, T.; Erdin, S.; Calzonetti, T.; Vijayvargia, R.; Tripathi, T.; Kerschbamer, E.; Seong, I.S.; Quattrone, A.; et al. Hypomorphic mutation of the mouse Huntington’s disease gene orthologue. PLoS Genet. 2019, 15, e1007765. [Google Scholar] [CrossRef]
- Peirce, J.L.; Lu, L.; Gu, J.; Silver, L.M.; Williams, R.W. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004, 5, 7. [Google Scholar] [CrossRef]
- Ashbrook, D.G.; Arends, D.; Prins, P.; Mulligan, M.K.; Roy, S.; Williams, E.G.; Lutz, C.M.; Valenzuela, A.; Bohl, C.J.; Ingels, J.F.; et al. A platform for experimental precision medicine: The extended BXD mouse family. Cell Syst. 2021, 12, 235–247. [Google Scholar] [CrossRef]
- Andreux, P.A.; Williams, E.G.; Koutnikova, H.; Houtkooper, R.H.; Champy, M.-F.; Henry, H.; Schoonjans, K.; Williams, R.W.; Auwerx, J. Systems Genetics of Metabolism: The Use of the BXD Murine Reference Panel for Multiscalar Integration of Traits. Cell 2012, 150, 1287–1299. [Google Scholar] [CrossRef]
- Parker, C.C.; Dickson, P.E.; Philip, V.M.; Thomas, M.; Chesler, E.J. Systems genetic analysis in GeneNetwork.org. Curr. Protoc. Neurosci. 2017, 79, 8.39.1–8.39.20. [Google Scholar] [CrossRef]
- Molenhuis, R.T.; Bruining, H.; Brandt, M.J.V.; van Soldt, P.E.; Abu-Toamih Atamni, H.J.; Burbach, J.P.H.; Iraqi, F.A.; Mott, R.F.; Kas, M.J.H. Modeling the quantitative nature of neurodevelopmental disorders using Collaborative Cross mice. Mol. Autism 2018, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Voy, B.H. Systems Genetics: A Powerful Approach for Gene-Environment InteractionsSystems Genetics: A Powerful Approach for Gene-Environment Interactions. J. Nutr. 2011, 141, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.; Zuker, C.S. Mammalian sweet taste receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Mueller, K.L.; Cook, B.; Wu, D.; Zuker, C.S.; Ryba, N.J. Coding of sweet, bitter, and umami tastes: Different receptor cells sharing similar signaling pathways. Cell 2003, 112, 293–301. [Google Scholar] [CrossRef]
- Calvo, S.S.; Egan, J.M. The endocrinology of taste receptors. Nat. Rev. Endocrinol. 2015, 11, 213–227. [Google Scholar] [CrossRef]
- Kyriazis, G.A.; Smith, K.R.; Tyrberg, B.; Hussain, T.; Pratley, R.E. Sweet taste receptors regulate basal insulin secretion and contribute to compensatory insulin hypersecretion during the development of diabetes in male mice. Endocrinology 2014, 155, 2112–2121. [Google Scholar] [CrossRef]
- Serrano, J.; Meshram, N.N.; Soundarapandian, M.M.; Smith, K.R.; Mason, C.; Brown, I.S.; Tyrberg, B.; Kyriazis, G.A. Saccharin Stimulates Insulin Secretion Dependent on Sweet Taste Receptor-Induced Activation of PLC Signaling Axis. Biomedicines 2022, 10, 120. [Google Scholar] [CrossRef]
- Smith, K.; Karimian Azari, E.; LaMoia, T.E.; Hussain, T.; Vargova, V.; Karolyi, K.; Veldhuis, P.P.; Arnoletti, J.P.; de la Fuente, S.G.; Pratley, R.E.; et al. T1R2 receptor-mediated glucose sensing in the upper intestine potentiates glucose absorption through activation of local regulatory pathways. Mol. Metab. 2018, 17, 98–111. [Google Scholar] [CrossRef]
- Smith, K.R.; Hussain, T.; Karimian Azari, E.; Steiner, J.L.; Ayala, J.E.; Pratley, R.E.; Kyriazis, G.A. Disruption of the sugar-sensing receptor T1R2 attenuates metabolic derangements associated with diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E688–E698. [Google Scholar] [CrossRef]
- Eny, K.M.; Wolever, T.M.; Corey, P.N.; El-Sohemy, A. Genetic variation in TAS1R2 (Ile191Val) is associated with consumption of sugars in overweight and obese individuals in 2 distinct populations. Am. J. Clin. Nutr. 2010, 92, 1501–1510. [Google Scholar] [CrossRef]
- Melo, S.V.; Agnes, G.; Vitolo, M.R.; Mattevi, V.S.; Campagnolo, P.D.B.; Almeida, S. Evaluation of the association between the TAS1R2 and TAS1R3 variants and food intake and nutritional status in children. Genet. Mol. Biol. 2017, 40, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Lopez, O.; Panduro, A.; Martinez-Lopez, E.; Roman, S. Sweet Taste Receptor TAS1R2 Polymorphism (Val191Val) Is Associated with a Higher Carbohydrate Intake and Hypertriglyceridemia among the Population of West Mexico. Nutrients 2016, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Hwang, L.D.; Lin, C.; Gharahkhani, P.; Cuellar-Partida, G.; Ong, J.S.; An, J.; Gordon, S.D.; Zhu, G.; MacGregor, S.; Lawlor, D.A.; et al. New insight into human sweet taste: A genome-wide association study of the perception and intake of sweet substances. Am. J. Clin. Nutr. 2019, 109, 1724–1737. [Google Scholar] [CrossRef] [PubMed]
- Stevens, H.; Graham, C.A.; Concas, M.P.; Piluso, F.; Mavrommatis, Y.; King, A.; Pilic, L.; Gasparini, P. TAS1R2 rs35874116 Associations with Taste, Diet, and Health in an Italian Population. Nutrients 2025, 17, 329. [Google Scholar] [CrossRef]
- Ponnusamy, V.; Subramanian, G.; Vasanthakumar, K.; Muthuswamy, K.; Panneerselvan, P.; Krishnan, V.; Subramaniam, S. T1R2/T1R3 polymorphism affects sweet and fat perception: Correlation between SNP and BMI in the context of obesity development. Hum. Genet. 2025, 144, 15–30. [Google Scholar] [CrossRef]
- Belloir, C.; Jeannin, M.; Karolkowski, A.; Briand, L. TAS1R2/TAS1R3 Single-Nucleotide Polymorphisms Affect Sweet Taste Receptor Activation by Sweeteners: The SWEET Project. Nutrients 2025, 17, 949. [Google Scholar] [CrossRef]
- Koc, G.; Soyocak, A.; Andac-Ozturk, S. TAS1R2 rs35874116 and TRPM5 rs886277 polymorphisms are not related with risk of obesity. Int. J. Clin. Pract. 2021, 75, e14562. [Google Scholar] [CrossRef]
- Serrano, J.; Seflova, J.; Park, J.; Pribadi, M.; Sanematsu, K.; Shigemura, N.; Serna, V.; Yi, F.; Mari, A.; Procko, E.; et al. The Ile191Val is a partial loss-of-function variant of the TAS1R2 sweet-taste receptor and is associated with reduced glucose excursions in humans. Mol. Metab. 2021, 54, 101339. [Google Scholar] [CrossRef]
- Park, J.; Selvam, B.; Sanematsu, K.; Shigemura, N.; Shukla, D.; Procko, E. Structural architecture of a dimeric class C GPCR based on co-trafficking of sweet taste receptor subunits. J. Biol. Chem. 2019, 294, 4759–4774. [Google Scholar] [CrossRef]
- Shimizu, M.; Goto, M.; Kawai, T.; Yamashita, A.; Kusakabe, Y. Distinct human and mouse membrane trafficking systems for sweet taste receptors T1r2 and T1r3. PLoS ONE 2014, 9, e100425. [Google Scholar] [CrossRef]
- Serrano, J.; Kondo, S.; Link, G.M.; Brown, I.S.; Pratley, R.E.; Baskin, K.K.; Goodpaster, B.H.; Coen, P.M.; Kyriazis, G.A. A partial loss-of-function variant (Ile191Val) of the TAS1R2 glucose receptor is associated with enhanced responses to exercise training in older adults with obesity: A translational study. Metabolism 2024, 162, 156045. [Google Scholar] [CrossRef] [PubMed]
- Serrano, J.; Yi, F.; Smith, J.; Pratley, R.E.; Kyriazis, G.A. The Ile191Val Variant of the TAS1R2 Subunit of Sweet Taste Receptors Is Associated With Reduced HbA1c in a Human Cohort With Variable Levels of Glucose Homeostasis. Front. Nutr. 2022, 9, 896205. [Google Scholar] [CrossRef] [PubMed]
- Serrano, J.; Boyd, J.; Brown, I.S.; Mason, C.; Smith, K.R.; Karolyi, K.; Maurya, S.K.; Meshram, N.N.; Serna, V.; Link, G.M.; et al. The TAS1R2 G-protein-coupled receptor is an ambient glucose sensor in skeletal muscle that regulates NAD homeostasis and mitochondrial capacity. Nat. Commun. 2024, 15, 4915. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Erlenbach, I.; Ryba, N.J.; Zuker, C.S. The receptors for mammalian sweet and umami taste. Cell 2003, 115, 255–266. [Google Scholar] [CrossRef]
- Pirinen, E.; Canto, C.; Jo, Y.S.; Morato, L.; Zhang, H.; Menzies, K.J.; Williams, E.G.; Mouchiroud, L.; Moullan, N.; Hagberg, C.; et al. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab. 2014, 19, 1034–1041. [Google Scholar] [CrossRef]
- Quackenbush, J. Microarray data normalization and transformation. Nat. Genet. 2002, 32, 496–501. [Google Scholar] [CrossRef]
- Durán, I.; Leandro, R.; Guevara-Coto, J. Analysis of Different Pre-Processing Techniques to the Development of Machine Learning Predictors with Gene Expression Profiles. In Proceedings of the 2019 IV Jornadas Costarricenses de Investigación en Computación e Informática (JoCICI), San Pedro, Costa Rica, 19–20 August 2019; pp. 1–6. [Google Scholar]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2021, 060012. [Google Scholar] [CrossRef]
- Berger, V.W.; Zhou, Y. Kolmogorov–Smirnov Test: Overview. In Wiley Statsref: Statistics Reference Online; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 1165–1188. [Google Scholar] [CrossRef]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. WebGestalt 2024: Faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef]
- Karp, P.D.; Midford, P.E.; Caspi, R.; Khodursky, A. Pathway size matters: The influence of pathway granularity on over-representation (enrichment analysis) statistics. BMC Genom. 2021, 22, 191. [Google Scholar] [CrossRef]
- Zhang, Y.; Szustakowski, J.; Schinke, M. Bioinformatics analysis of microarray data. Cardiovasc. Genom. Methods Protoc. 2009, 573, 259–284. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, M.; Xia, X.; Gong, T.; Feng, J.; Liu, W.; Liu, Y.; Zhen, B.; Wang, Y.; Ding, C.; et al. A mouse tissue transcription factor atlas. Nat. Commun. 2017, 8, 15089. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.C.; Lopez-Granero, C.; Ferrer, B.; Tinkov, A.A.; Skalny, A.V.; Paoliello, M.M.B.; Aschner, M. BXD Recombinant Inbred Mice as a Model to Study Neurotoxicity. Biomolecules 2021, 11, 1762. [Google Scholar] [CrossRef] [PubMed]
- Loos, M.; Verhage, M.; Spijker, S.; Smit, A.B. Complex Genetics of Behavior: BXDs in the Automated Home-Cage. Methods Mol. Biol. 2017, 1488, 519–530. [Google Scholar] [CrossRef]
- Boutilier, J.K.; Taylor, R.L.; Ram, R.; McNamara, E.; Nguyen, Q.; Goullee, H.; Chandler, D.; Mehta, M.; Balmer, L.; Laing, N.G.; et al. Variable cardiac alpha-actin (Actc1) expression in early adult skeletal muscle correlates with promoter methylation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1025–1036. [Google Scholar] [CrossRef]
- Lionikas, A.; Blizard, D.A.; Gerhard, G.S.; Vandenbergh, D.J.; Stout, J.T.; Vogler, G.P.; McClearn, G.E.; Larsson, L. Genetic determinants of weight of fast- and slow-twitch skeletal muscle in 500-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol. Genom. 2005, 21, 184–192. [Google Scholar] [CrossRef]
- Lionikas, A.; Meharg, C.; Derry, J.M.; Ratkevicius, A.; Carroll, A.M.; Vandenbergh, D.J.; Blizard, D.A. Resolving candidate genes of mouse skeletal muscle QTL via RNA-Seq and expression network analyses. BMC Genom. 2012, 13, 592. [Google Scholar] [CrossRef]
- Ahn, B. The Function of MondoA and ChREBP Nutrient-Sensing Factors in Metabolic Disease. Int. J. Mol. Sci. 2023, 24, 8811. [Google Scholar] [CrossRef]
- Pistocchi, A.; Gaudenzi, G.; Foglia, E.; Monteverde, S.; Moreno-Fortuny, A.; Pianca, A.; Cossu, G.; Cotelli, F.; Messina, G. Conserved and divergent functions of Nfix in skeletal muscle development during vertebrate evolution. Development 2013, 140, 1528–1536. [Google Scholar] [CrossRef]
- Lefebvre, P.; Benomar, Y.; Staels, B. Retinoid X receptors: Common heterodimerization partners with distinct functions. Trends Endocrinol. Metab. 2010, 21, 676–683. [Google Scholar] [CrossRef]
- Berkes, C.A.; Bergstrom, D.A.; Penn, B.H.; Seaver, K.J.; Knoepfler, P.S.; Tapscott, S.J. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 2004, 14, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Hoy, A.J. Lipid metabolism in skeletal muscle: Generation of adaptive and maladaptive intracellular signals for cellular function. Am. J. Physiol. Endocrinol. Metab. 2012, 302, E1315–E1328. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.J.; Grant, J.N.; Moon, J.I.; So, S.S.; Finch, A.M. Critically evaluating sweet taste receptor expression and signaling through a molecular pharmacology lens. FEBS J. 2021, 288, 2660–2672. [Google Scholar] [CrossRef] [PubMed]
- Kyriazis, G.A.; Soundarapandian, M.M.; Tyrberg, B. Sweet taste receptor signaling in beta cells mediates fructose-induced potentiation of glucose-stimulated insulin secretion. Proc. Natl. Acad. Sci. USA 2012, 109, E524–E532. [Google Scholar] [CrossRef]
- Young, R.L.; Sutherland, K.; Pezos, N.; Brierley, S.M.; Horowitz, M.; Rayner, C.K.; Blackshaw, L.A. Expression of taste molecules in the upper gastrointestinal tract in humans with and without type 2 diabetes. Gut 2009, 58, 337–346. [Google Scholar] [CrossRef]
- Malki, A.; Fiedler, J.; Fricke, K.; Ballweg, I.; Pfaffl, M.W.; Krautwurst, D. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J. Leukoc. Biol. 2015, 97, 533–545. [Google Scholar] [CrossRef]
- Sishi, B.; Loos, B.; Ellis, B.; Smith, W.; du Toit, E.F.; Engelbrecht, A.M. Diet-induced obesity alters signalling pathways and induces atrophy and apoptosis in skeletal muscle in a prediabetic rat model. Exp. Physiol. 2011, 96, 179–193. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
King, K.; Serrano, J.; Meshram, N.N.; Saadi, M.; Moreira, L.; Papachristou, E.G.; Kyriazis, G.A. Low TAS1R2 Sweet Taste Receptor Expression in Skeletal Muscle of Genetically Diverse BXD Mice Mirrors Transcriptomic Signatures of Loss-of-Function Mice. Nutrients 2025, 17, 1918. https://doi.org/10.3390/nu17111918
King K, Serrano J, Meshram NN, Saadi M, Moreira L, Papachristou EG, Kyriazis GA. Low TAS1R2 Sweet Taste Receptor Expression in Skeletal Muscle of Genetically Diverse BXD Mice Mirrors Transcriptomic Signatures of Loss-of-Function Mice. Nutrients. 2025; 17(11):1918. https://doi.org/10.3390/nu17111918
Chicago/Turabian StyleKing, Kendall, Joan Serrano, Nishita N. Meshram, Mahdiye Saadi, Lynn Moreira, Evaggelia G. Papachristou, and George A. Kyriazis. 2025. "Low TAS1R2 Sweet Taste Receptor Expression in Skeletal Muscle of Genetically Diverse BXD Mice Mirrors Transcriptomic Signatures of Loss-of-Function Mice" Nutrients 17, no. 11: 1918. https://doi.org/10.3390/nu17111918
APA StyleKing, K., Serrano, J., Meshram, N. N., Saadi, M., Moreira, L., Papachristou, E. G., & Kyriazis, G. A. (2025). Low TAS1R2 Sweet Taste Receptor Expression in Skeletal Muscle of Genetically Diverse BXD Mice Mirrors Transcriptomic Signatures of Loss-of-Function Mice. Nutrients, 17(11), 1918. https://doi.org/10.3390/nu17111918





