Age-Dependent Gut Microbiome Dysbiosis in Autism Spectrum Disorder and the Role of Key Bacterial Ratios
Abstract
1. Introduction
2. Materials and Methods
Sample Collection and Processing
3. Results
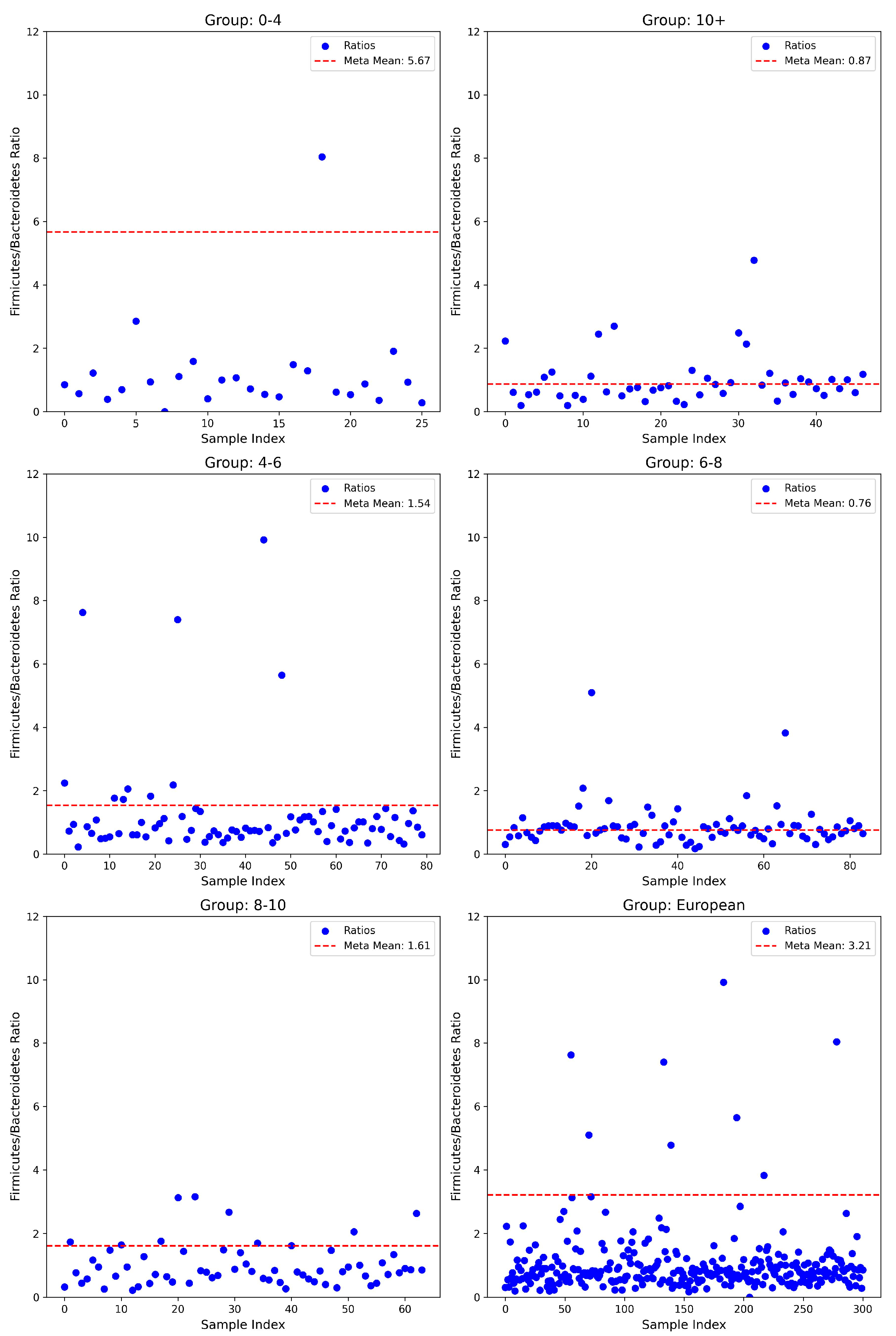
3.1. Firmicutes/Bacteroidetes Ratios
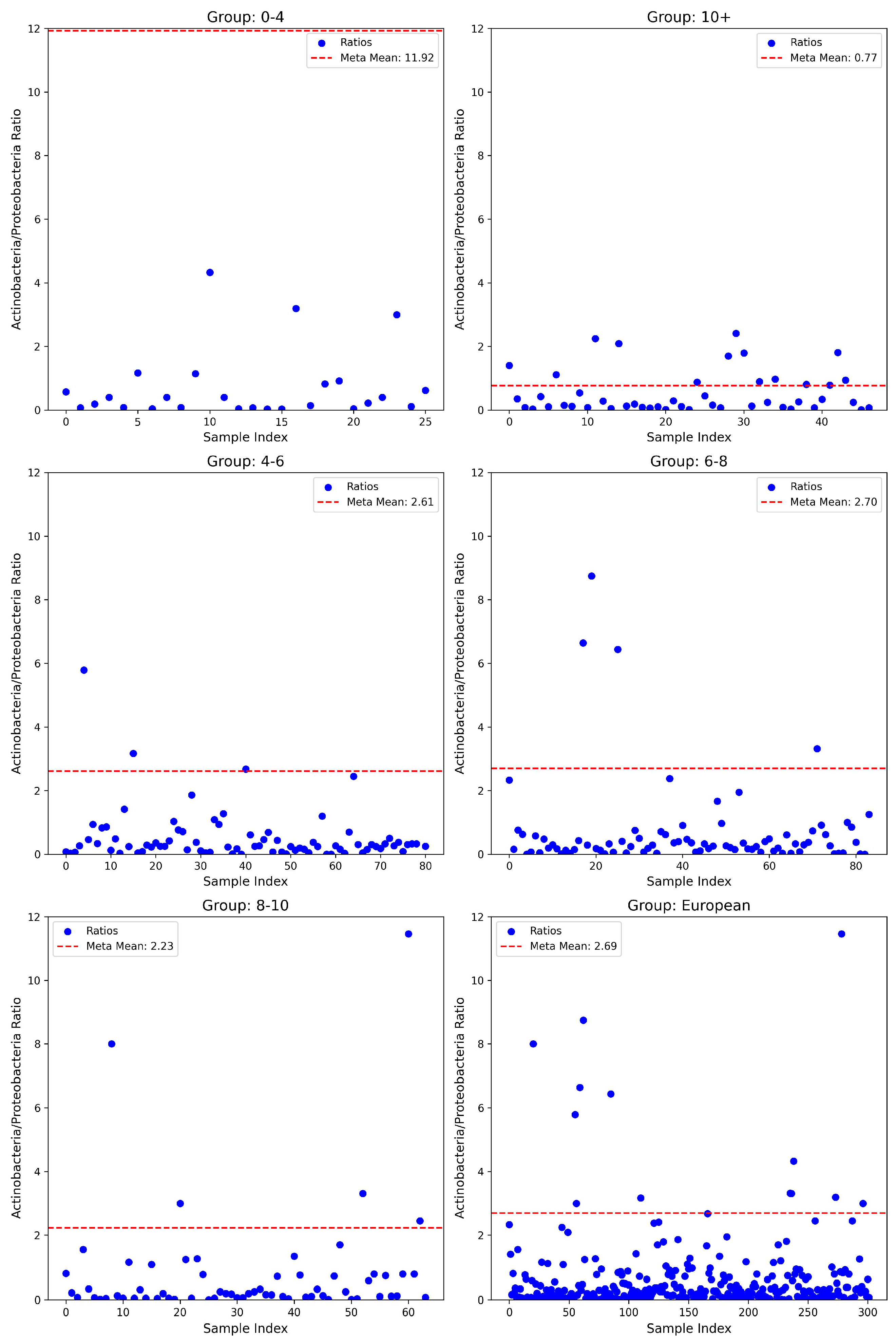
3.2. Actinobacteria/Proteobacteria Ratios
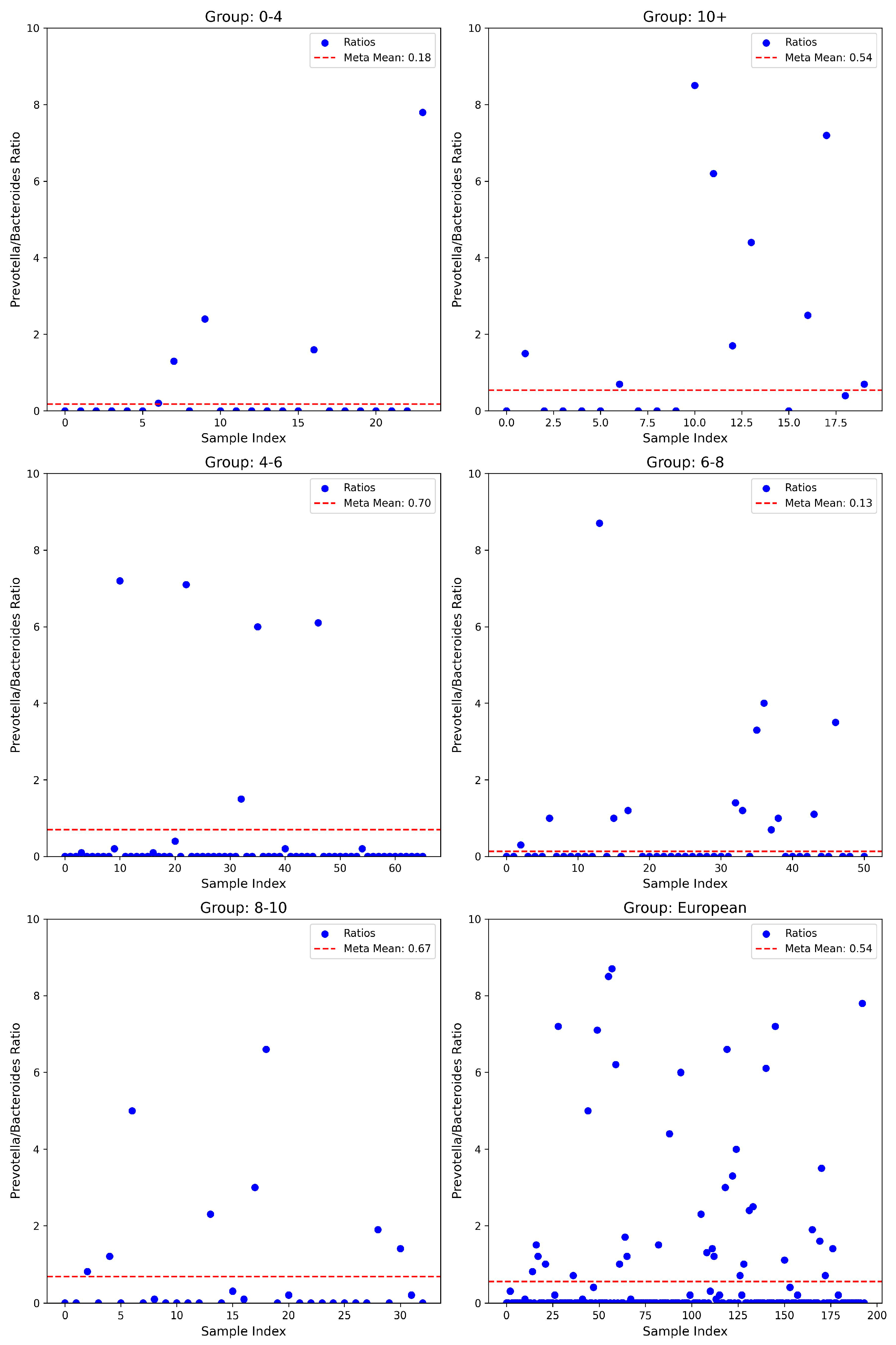
3.3. Prevotella/Bacteroides Ratios
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, D.; Letchumanan, V.; Thurairajasingam, S.; Lee, L.-H. A Revolutionizing Approach to Autism Spectrum Disorder Using the Microbiome. Nutrients 2020, 12, 1983. [Google Scholar] [CrossRef] [PubMed]
- Autism Rates by Country. 2025. Available online: https://worldpopulationreview.com/country-rankings/autism-rates-by-country (accessed on 3 April 2025).
- Warreman, E.; Nooteboom, L.; Terry, M.; Hoek, H.; Leenen, P.; van Rossum, E.; Ramlal, D.; Vermeiren, R.; Ester, W. Psychological, behavioural and biological factors associated with gastrointestinal symptoms in autistic adults and adults with autistic traits. Autism 2023, 27, 2173–2186. [Google Scholar] [CrossRef]
- Lefter, R.; Ciobica, A.; Timofte, D.; Stanciu, C.; Trifan, A. A Descriptive Review on the Prevalence of Gastrointestinal Disturbances and Their Multiple Associations in Autism Spectrum Disorder. Medicina 2019, 56, 11. [Google Scholar] [CrossRef]
- Madra, M.; Ringel, R.; Margolis, K.G. Gastrointestinal Issues and Autism Spectrum Disorder. Psychiatr. Clin. N. Am. 2021, 44, 69–81. [Google Scholar] [CrossRef]
- Loh, J.S.; Mak, W.Q.; Tan, L.K.S.; Ng, C.X.; Chan, H.H.; Yeow, S.H.; Foo, J.B.; Ong, Y.S.; How, C.W.; Khaw, K.Y. Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases. Signal Transduct. Target. Ther. 2024, 9, 37. [Google Scholar]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Dash, S.; Syed, Y.A.; Khan, M.R. Understanding the Role of the Gut Microbiome in Brain Development and Its Association with Neurodevelopmental Psychiatric Disorders. Front. Cell Dev. Biol. 2022, 10, 880544. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.-G.; Li, J.; Cheng, J.; Zhou, D.-D.; Wu, S.-X.; Huang, S.-Y.; Saimaiti, A.; Yang, Z.-J.; Gan, R.-Y.; Li, H.-B. The Role of Gut Microbiota in Anxiety, Depression, and Other Mental Disorders as Well as the Protective Effects of Dietary Components. Nutrients 2023, 15, 3258. [Google Scholar] [CrossRef]
- Wen, L.; Duffy, A. Factors Influencing the Gut Microbiota, Inflammation, and Type 2 Diabetes. J. Nutr. 2017, 147, 1468S–1475S. [Google Scholar] [CrossRef]
- Peralta-Marzal, L.N.; Rojas-Velazquez, D.; Rigters, D.; Prince, N.; Garssen, J.; Kraneveld, A.D.; Perez-Pardo, P.; Lopez-Rincon, A. A robust microbiome signature for autism spectrum disorder across different studies using machine learning. Sci. Rep. 2024, 14, 814. [Google Scholar] [CrossRef]
- Dargenio, V.N.; Dargenio, C.; Castellaneta, S.; De Giacomo, A.; Laguardia, M.; Schettini, F.; Francavilla, R.; Cristofori, F. Intestinal Barrier Dysfunction and Microbiota–Gut–Brain Axis: Possible Implications in the Pathogenesis and Treatment of Autism Spectrum Disorder. Nutrients 2023, 15, 1620. [Google Scholar] [CrossRef] [PubMed]
- Młynarska, E.; Barszcz, E.; Budny, E.; Gajewska, A.; Kopeć, K.; Wasiak, J.; Rysz, J.; Franczyk, B. The Gut–Brain–Microbiota Connection and Its Role in Autism Spectrum Disorders. Nutrients 2025, 17, 1135. [Google Scholar] [CrossRef] [PubMed]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar] [PubMed]
- Fillier, T.A.; Shah, S.; Doody, K.M.; Pham, T.H.; Aubry, I.; Tremblay, M.L.; Cheema, S.K.; Blundell, J.; Thomas, R.H. Brief exposure of neuronal cells to levels of SCFAs observed in human systemic circulation impair lipid metabolism resulting in apoptosis. Sci. Rep. 2022, 12, 14355. [Google Scholar] [CrossRef]
- Killingsworth, J.; Sawmiller, D.; Shytle, R.D. Propionate and Alzheimer’s Disease. Front. Aging Neurosci. 2020, 12, 580001. [Google Scholar] [CrossRef] [PubMed]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; De Caro, C.; Comegna, M.; et al. Gut Microbiota Features in Young Children with Autism Spectrum Disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef]
- Yunes, R.; Poluektova, E.; Dyachkova, M.; Klimina, K.; Kovtun, A.; Averina, O.; Orlova, V.; Danilenko, V. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 2016, 42, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Duranti, S.; Ruiz, L.; Lugli, G.A.; Tames, H.; Milani, C.; Mancabelli, L.; Mancino, W.; Longhi, G.; Carnevali, L.; Sgoifo, A.; et al. Bifidobacterium adolescentis as a key member of the human gut microbiota in the production of GABA. Sci. Rep. 2020, 10, 14112. [Google Scholar] [CrossRef]
- Li, Q.; Han, Y.; Dy, A.B.C.; Hagerman, R.J. The Gut Microbiota and Autism Spectrum Disorders. Front. Cell. Neurosci. 2017, 11, 120. [Google Scholar] [CrossRef]
- Lin, Q.; Lin, S.; Fan, Z.; Liu, J.; Ye, D.; Guo, P. A Review of the Mechanisms of Bacterial Colonization of the Mammal Gut. Microorganisms 2024, 12, 1026. [Google Scholar] [CrossRef]
- Pantazi, A.C.; Balasa, A.L.; Mihai, C.M.; Chisnoiu, T.; Lupu, V.V.; Kassim, M.A.K.; Mihai, L.; Frecus, C.E.; Chirila, S.I.; Lupu, A.; et al. Development of Gut Microbiota in the First 1000 Days after Birth and Potential Interventions. Nutrients 2023, 15, 3647. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What Is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Fattorusso, A.; Di Genova, L.; Dell’Isola, G.B.; Mencaroni, E.; Esposito, S. Autism Spectrum Disorders and the Gut Microbiota. Nutrients 2019, 11, 521. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Nie, Q.; He, H.; Tan, H.; Geng, F.; Ji, H.; Hu, J.; Nie, S. Gut firmicutes: Relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 2023, 63, 12073–12088. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Liu, F.; Li, J.; Wu, F.; Zheng, H.; Peng, Q.; Zhou, H. Altered composition and function of intestinal microbiota in autism spectrum disorders: A systematic review. Transl. Psychiatry 2019, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Elhefnawy, A.M.; Azouz, H.G.; Roshdy, Y.S.; Ashry, M.H.; Ibrahim, A.E.; Meheissen, M.A. Study of the gut Microbiome Profile in Children with Autism Spectrum Disorder: A Single Tertiary Hospital Experience. J. Mol. Neurosci. 2020, 70, 887–896. [Google Scholar] [CrossRef]
- Tomova, A.; Husarova, V.; Lakatosova, S.; Bakos, J.; Vlkova, B.; Babinska, K.; Ostatnikova, D. Gastrointestinal microbiota in children with autism in Slovakia. Physiol. Behav. 2015, 138, 179–187. [Google Scholar] [CrossRef]
- Zhou, M.; Niu, B.; Ma, J.; Ge, Y.; Han, Y.; Wu, W.; Yue, C. Intervention and research progress of gut microbiota-immune-nervous system in autism spectrum disorders among students. Front. Microbiol. 2025, 16, 1535455. [Google Scholar] [CrossRef] [PubMed]
- Pulikkan, J.; Mazumder, A.; Grace, T. Role of the Gut Microbiome in Autism Spectrum Disorders. Adv. Exp. Med. Biol. 2019, 1118, 253–269. [Google Scholar] [PubMed]
- Iglesias-Vázquez, L.; Van Ginke Riba, G.; Arija, V.; Canals, J. Composition of Gut Microbiota in Children with Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 792. [Google Scholar] [CrossRef]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef]
- Marras, L.; Caputo, M.; Bisicchia, S.; Soato, M.; Bertolino, G.; Vaccaro, S.; Inturri, R. The Role of Bifidobacteria in Predictive and Preventive Medicine: A Focus on Eczema and Hypercholesterolemia. Microorganisms 2021, 9, 836. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Cantabrana, C.; Delgado, S.; Ruiz, L.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Health-Promoting Effects. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Mehra, A.; Arora, G.; Sahni, G.; Kaur, M.; Singh, H.; Singh, B.; Kaur, S. Gut microbiota and Autism Spectrum Disorder: From pathogenesis to potential therapeutic perspectives. J. Tradit. Complement. Med. 2023, 13, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Mu, C.-L.; Farzi, A.; Zhu, W.-Y. Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, A. Microbial pathogenesis in inflammatory bowel diseases. Microb. Pathog. 2022, 163, 105383. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Plaza-Díaz, J.; Gómez-Fernández, A.; Chueca, N.; De La Torre-Aguilar, M.J.; Gil, Á.; Perez-Navero, J.L.; Flores-Rojas, K.; Martín-Borreguero, P.; Solis-Urra, P.; Ruiz-Ojeda, F.J.; et al. Autism Spectrum Disorder (ASD) with and without Mental Regression is Associated with Changes in the Fecal Microbiota. Nutrients 2019, 11, 337. [Google Scholar] [CrossRef] [PubMed]
- Demarquoy, J.; Othman, H.; Demarquoy, C. Modify gut microbiome in autism: A promising strategy? Explor. Neurosci. 2023, 2, 140–152. [Google Scholar] [CrossRef]
- Alharthi, A.; Alhazmi, S.; Alburae, N.; Bahieldin, A. The Human Gut Microbiome as a Potential Factor in Autism Spectrum Disorder. Int. J. Mol. Sci. 2022, 23, 1363. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Wong, O.W.; Tun, H.M.; Su, Q.; Xu, Z.; Tang, W.; Ma, S.L.; Chan, S.; Chan, F.K.L.; Ng, S.C. Fecal microbial marker panel for aiding diagnosis of autism spectrum disorders. Gut Microbes 2024, 16, 2418984. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Xu, F.; Wang, Y.; Duan, M.; Guo, M.; Zhang, Q.; Zhao, H.; Zheng, H. Changes in the Gut Microbiota of Children with Autism Spectrum Disorder. Autism Res. Off. J. Int. Soc. Autism Res. 2020, 13, 1614–1625. [Google Scholar] [CrossRef]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Renzi, D.; Calabrò, A.; et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 2017, 5, 24. [Google Scholar] [CrossRef]
- Ronan, V.; Yeasin, R.; Claud, E.C. Childhood Development and the Microbiome—The Intestinal Microbiota in Maintenance of Health and Development of Disease During Childhood Development. Gastroenterology 2021, 160, 495–506. [Google Scholar] [CrossRef]
- Wong, O.W.; Lam, A.M.; Or, B.P.; Mo, F.Y.; Shea, C.K.; Lai, K.Y.; Ma, S.L.; Hung, S.F.; Chan, S.; Kwong, T.N.; et al. Disentangling the relationship of gut microbiota, functional gastrointestinal disorders and autism: A case–control study on prepubertal Chinese boys. Sci. Rep. 2022, 12, 10659. [Google Scholar] [CrossRef]
- Finegold, S.M.; Dowd, S.E.; Gontcharova, V.; Liu, C.; Henley, K.E.; Wolcott, R.D.; Youn, E.; Summanen, P.H.; Granpeesheh, D.; Dixon, D.; et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe 2010, 16, 444–453. [Google Scholar] [CrossRef]
- Argou-Cardozo, I.; Zeidán-Chuliá, F. Clostridium Bacteria and Autism Spectrum Conditions: A Systematic Review and Hypothetical Contribution of Environmental Glyphosate Levels. Med. Sci. 2018, 6, 29. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Brain-Gut-Microbiota Axis and Mental Health. Psychosom. Med. 2017, 79, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Bruijning, M.; Ayroles, J.F.; Henry, L.P.; Koskella, B.; Meyer, K.M.; Metcalf, C.J.E. Relative abundance data can misrepresent heritability of the microbiome. Microbiome 2023, 11, 222. [Google Scholar] [CrossRef] [PubMed]
- Carmel, J.; Ghanayem, N.; Mayouf, R.; Saleev, N.; Chaterjee, I.; Getselter, D.; Tikhonov, E.; Turjeman, S.; Shaalan, M.; Khateeb, S.; et al. Bacteroides is increased in an autism cohort and induces autism-relevant behavioral changes in mice in a sex-dependent manner. NPJ Biofilms Microbiomes 2023, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Laue, H.E.; Coker, M.O.; Madan, J.C. The Developing Microbiome from Birth to 3 Years: The Gut-Brain Axis and Neurodevelopmental Outcomes. Front. Pediatr. 2022, 10, 815885. [Google Scholar] [CrossRef]
- Savage, J.S.; Fisher, J.O.; Birch, L.L. Parental Influence on Eating Behavior: Conception to Adolescence. J. Law Med. Ethics 2007, 35, 22–34. [Google Scholar] [CrossRef]
- Inchingolo, F.; Inchingolo, A.M.; Latini, G.; Ferrante, L.; de Ruvo, E.; Campanelli, M.; Longo, M.; Palermo, A.; Inchingolo, A.D.; Dipalma, G. Difference in the Intestinal Microbiota between Breastfeed Infants and Infants Fed with Artificial Milk: A Systematic Review. Pathogens 2024, 13, 533. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadiyska, T.; Vassilev, D.; Tourtourikov, I.; Ciurinskiene, S.; Madzharova, D.; Savcheva, M.; Stoynev, N.; Mileva-Popova, R.; Tafradjiiska-Hadjiolova, R.; Mitev, V. Age-Dependent Gut Microbiome Dysbiosis in Autism Spectrum Disorder and the Role of Key Bacterial Ratios. Nutrients 2025, 17, 1775. https://doi.org/10.3390/nu17111775
Kadiyska T, Vassilev D, Tourtourikov I, Ciurinskiene S, Madzharova D, Savcheva M, Stoynev N, Mileva-Popova R, Tafradjiiska-Hadjiolova R, Mitev V. Age-Dependent Gut Microbiome Dysbiosis in Autism Spectrum Disorder and the Role of Key Bacterial Ratios. Nutrients. 2025; 17(11):1775. https://doi.org/10.3390/nu17111775
Chicago/Turabian StyleKadiyska, Tanya, Dimitar Vassilev, Ivan Tourtourikov, Stanislava Ciurinskiene, Dilyana Madzharova, Maria Savcheva, Nikolay Stoynev, Rene Mileva-Popova, Radka Tafradjiiska-Hadjiolova, and Vanyo Mitev. 2025. "Age-Dependent Gut Microbiome Dysbiosis in Autism Spectrum Disorder and the Role of Key Bacterial Ratios" Nutrients 17, no. 11: 1775. https://doi.org/10.3390/nu17111775
APA StyleKadiyska, T., Vassilev, D., Tourtourikov, I., Ciurinskiene, S., Madzharova, D., Savcheva, M., Stoynev, N., Mileva-Popova, R., Tafradjiiska-Hadjiolova, R., & Mitev, V. (2025). Age-Dependent Gut Microbiome Dysbiosis in Autism Spectrum Disorder and the Role of Key Bacterial Ratios. Nutrients, 17(11), 1775. https://doi.org/10.3390/nu17111775





