Iron Supplementation Increases Tumor Burden and Alters Protein Expression in a Mouse Model of Human Intestinal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Study Design
2.2. Diet
2.3. Tissue Collection
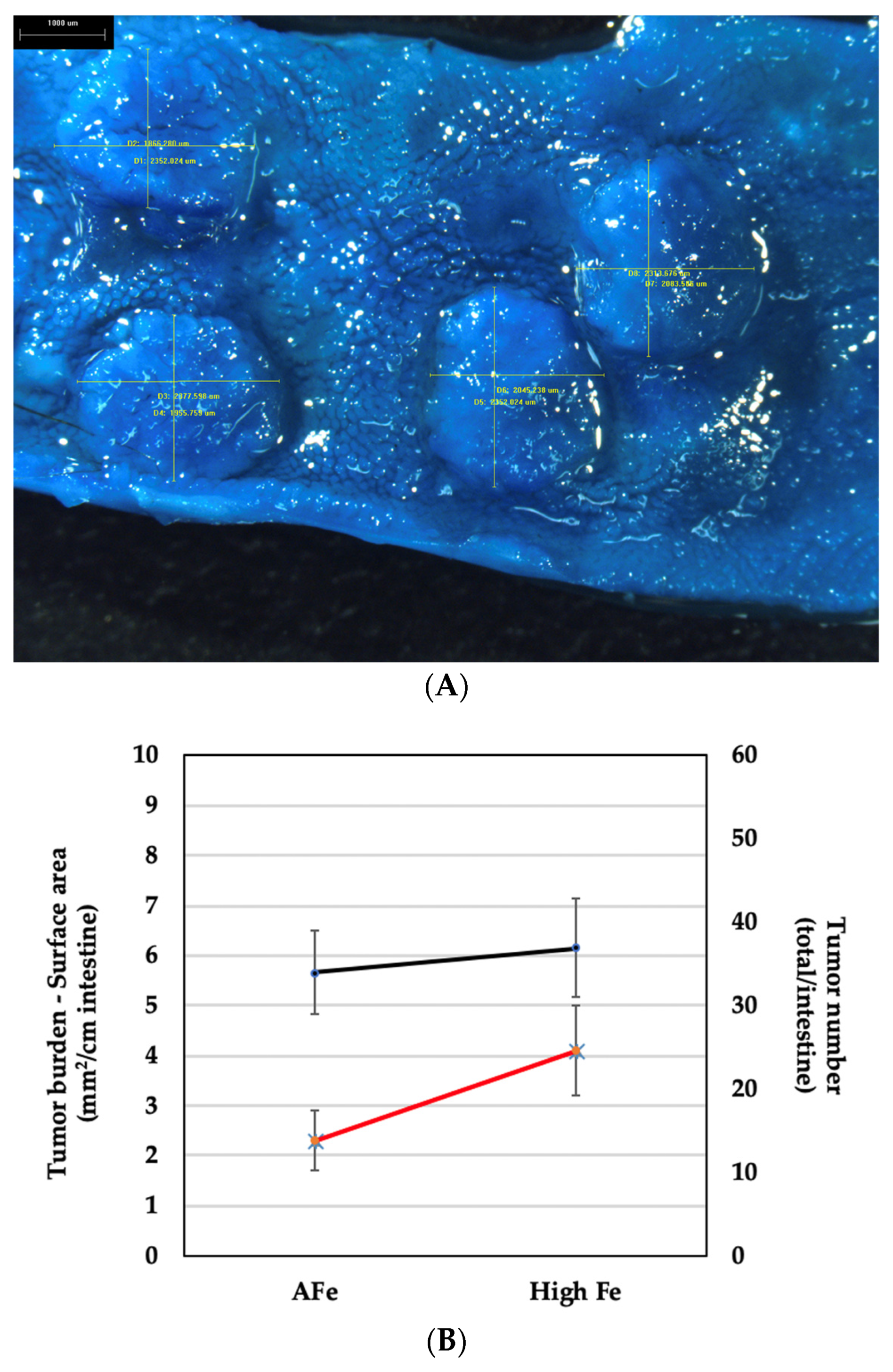
2.4. Tumor Burden
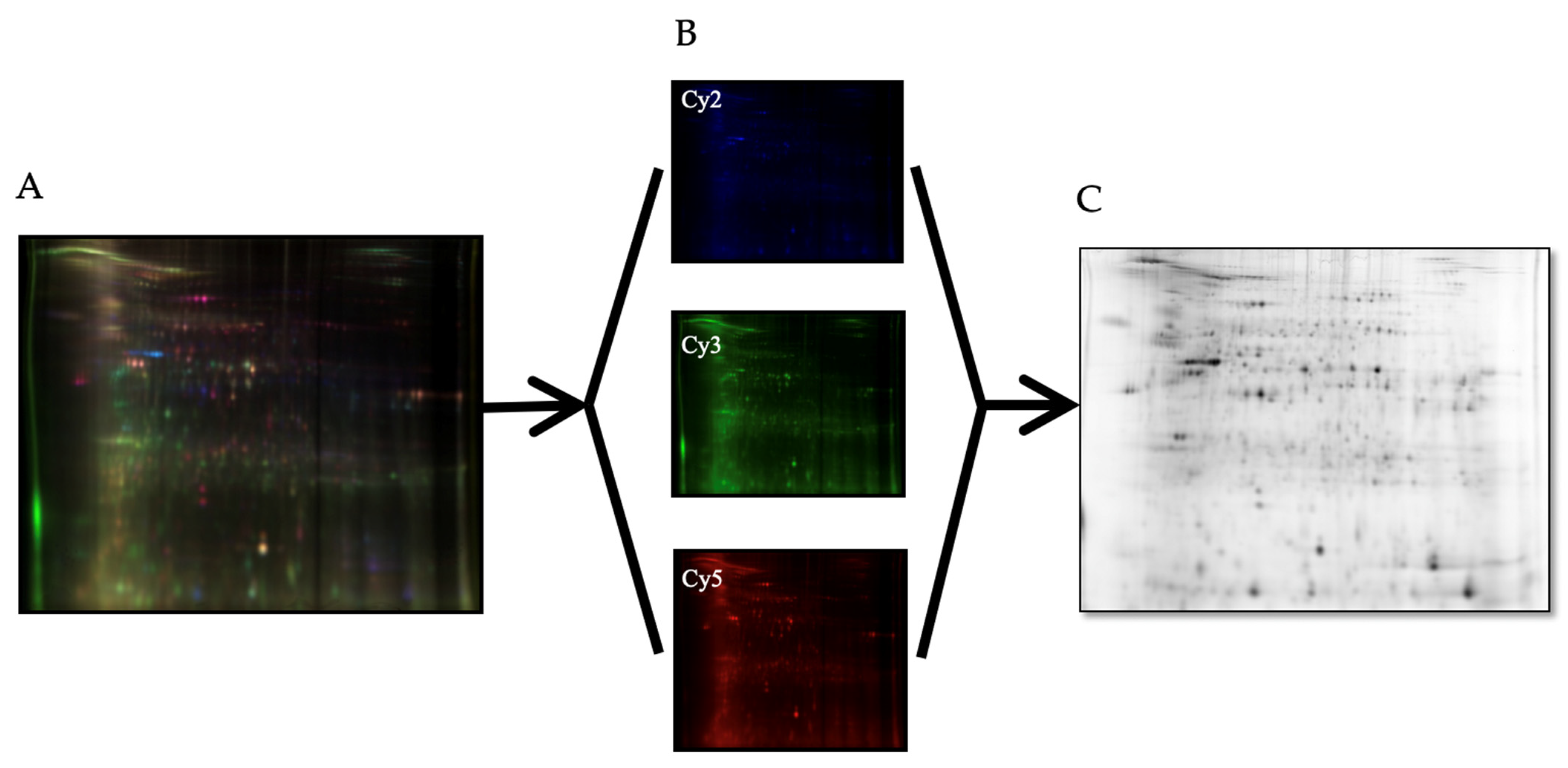
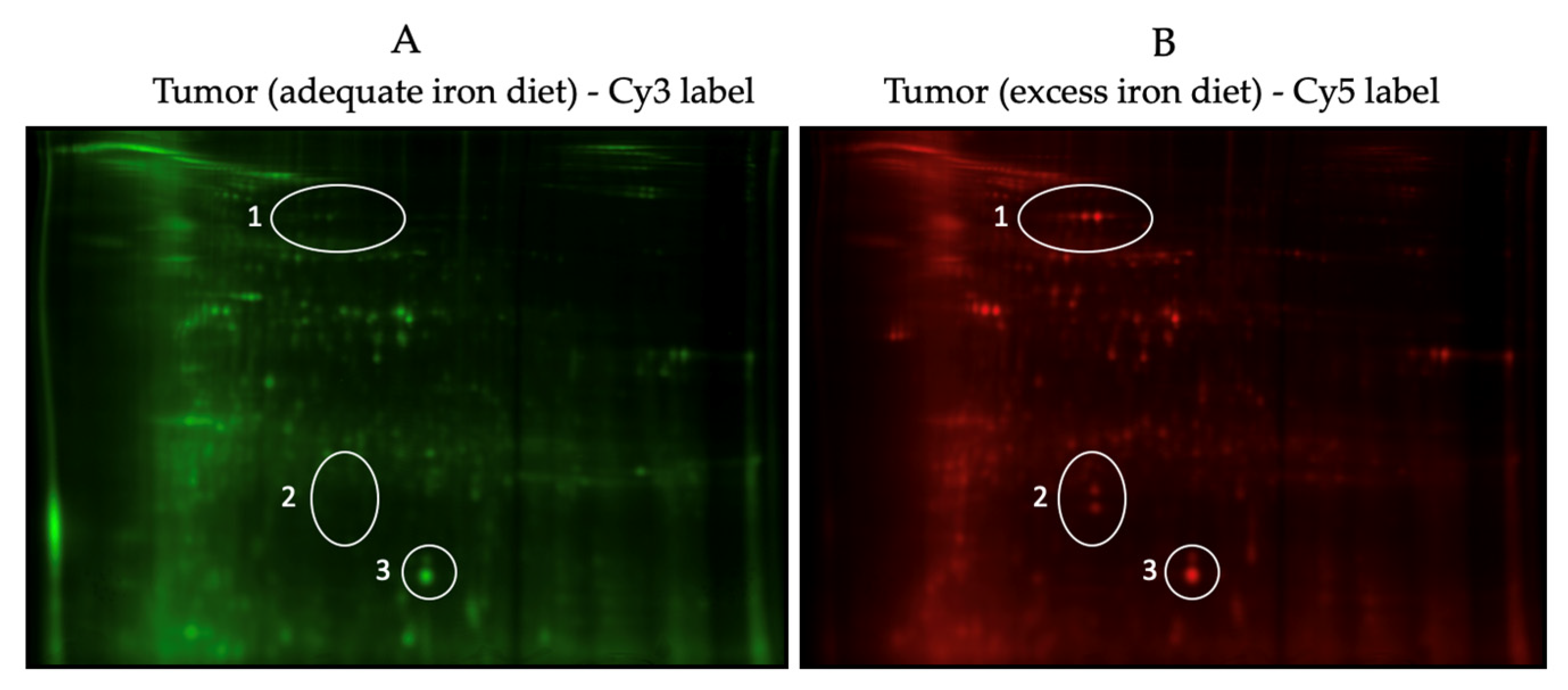
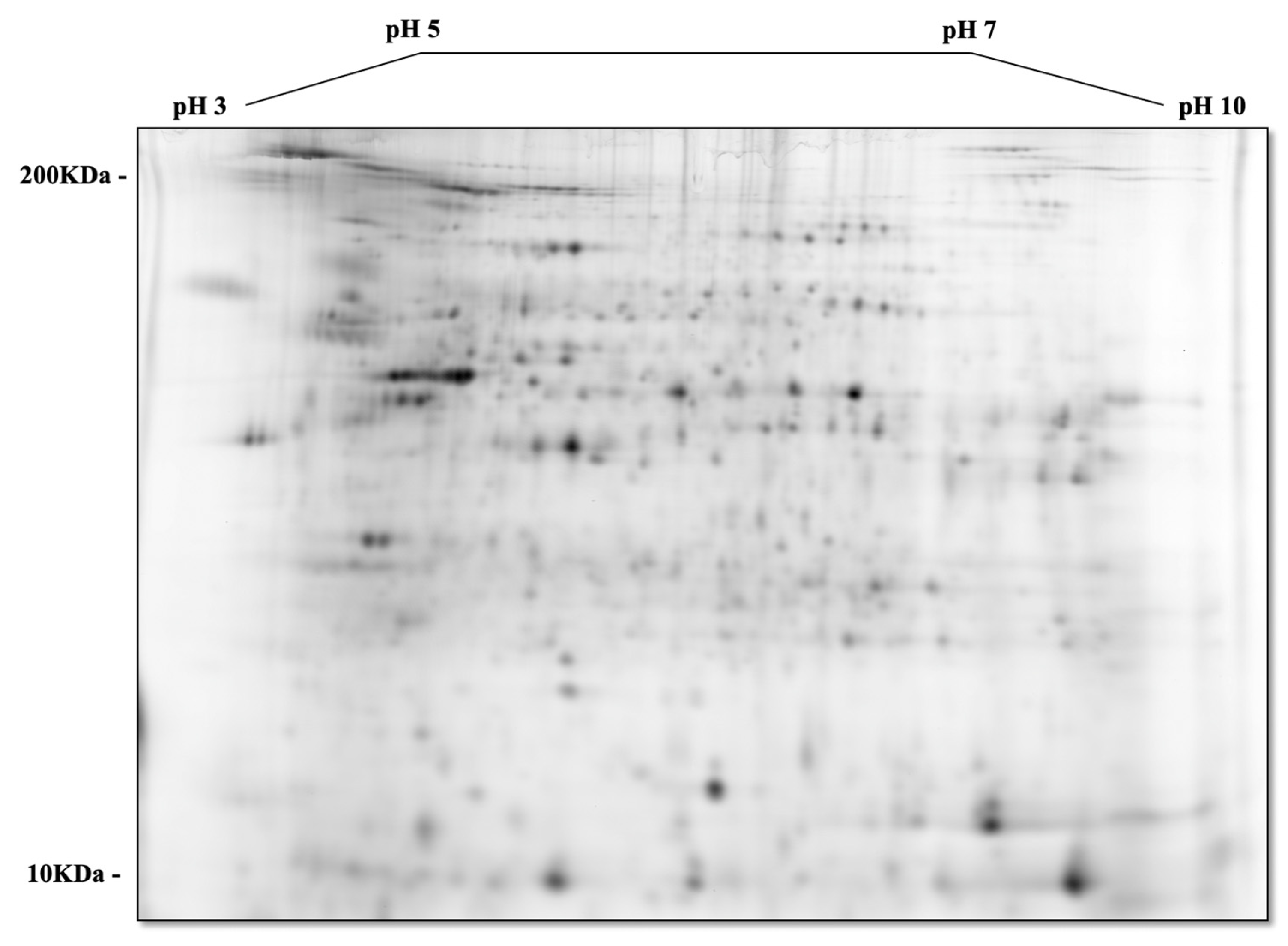
2.5. Proteomic Analysis—2D-DIGE (2-Dimensional Fluorescence Difference Gel Electrophoresis)
2.6. Pathway Mapping
2.7. Statistical Analyses
3. Results
3.1. Animal Weight and Food Intake
3.2. Tumor Identification, Measurements, and Burden Calculation
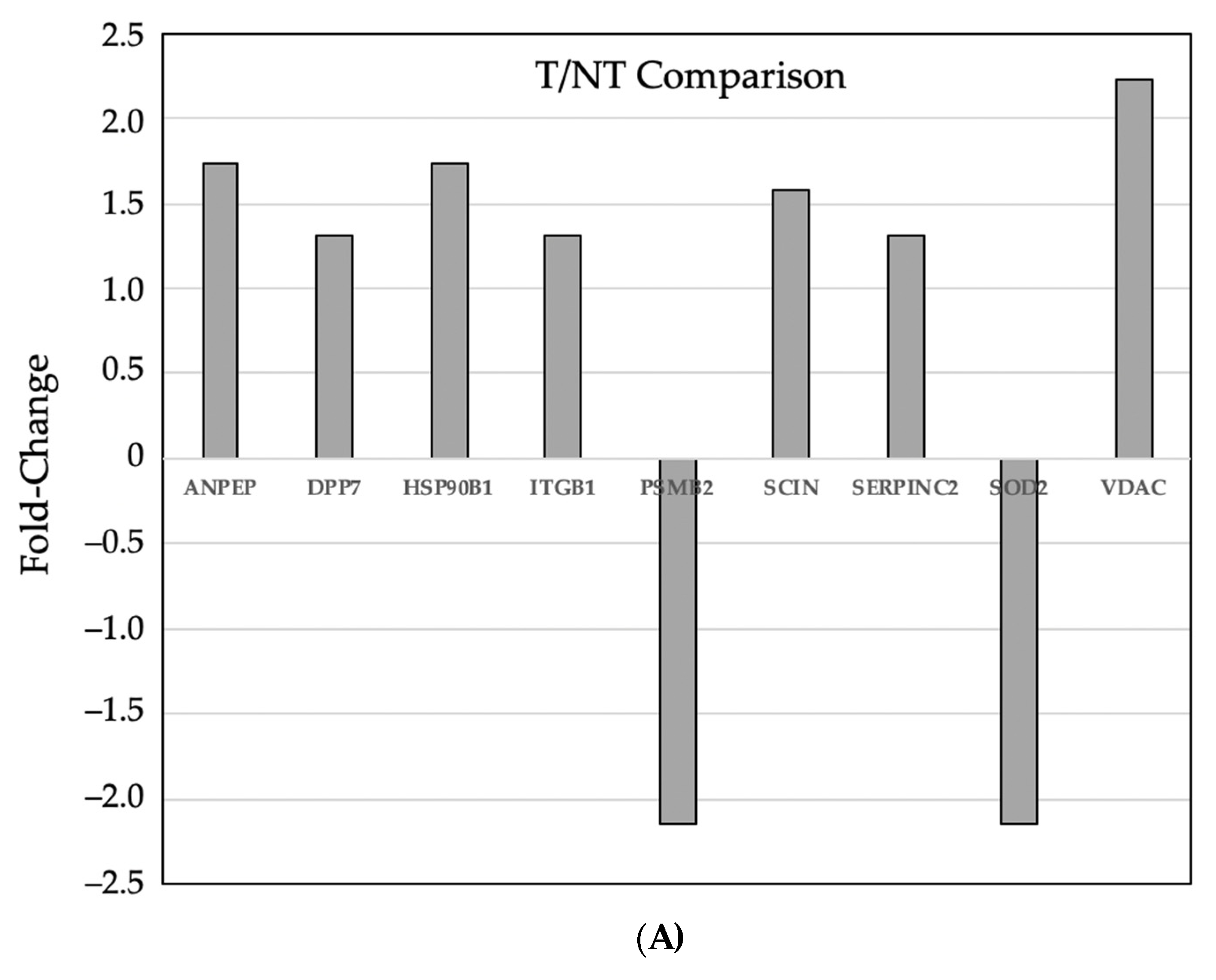
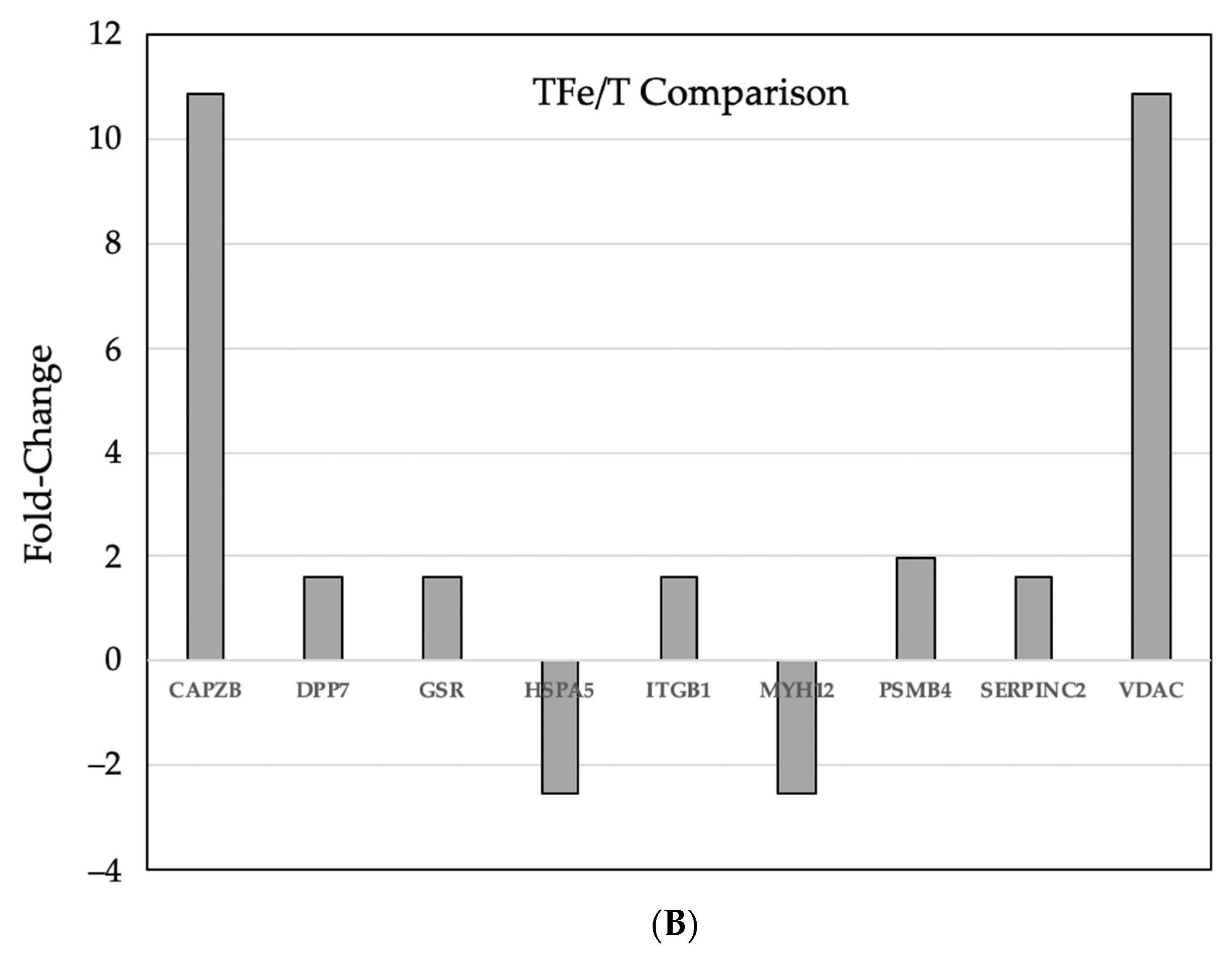
3.3. Comparative Proteomic Analysis of Tumor and Non-Tumor Intestinal Tissue
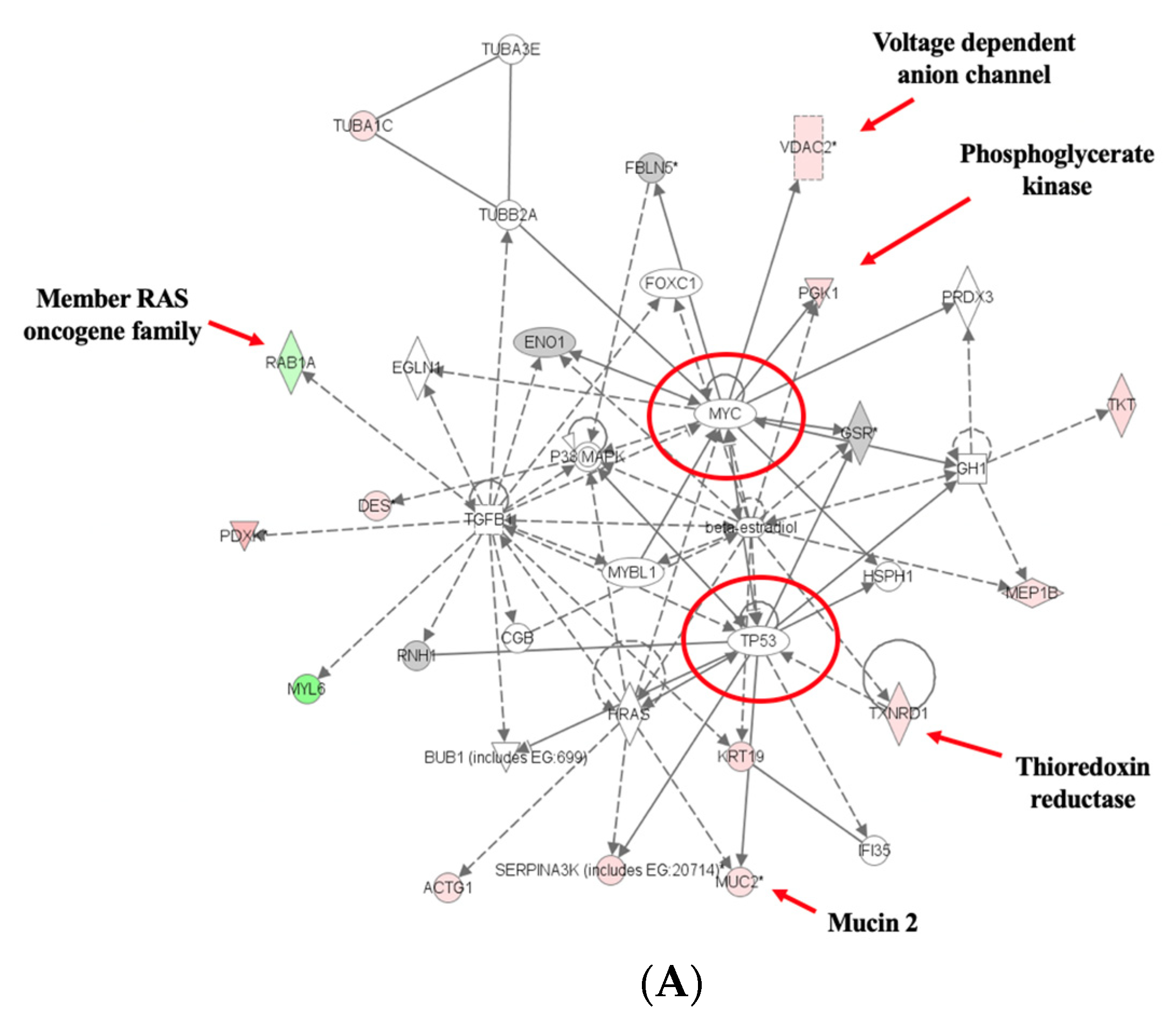
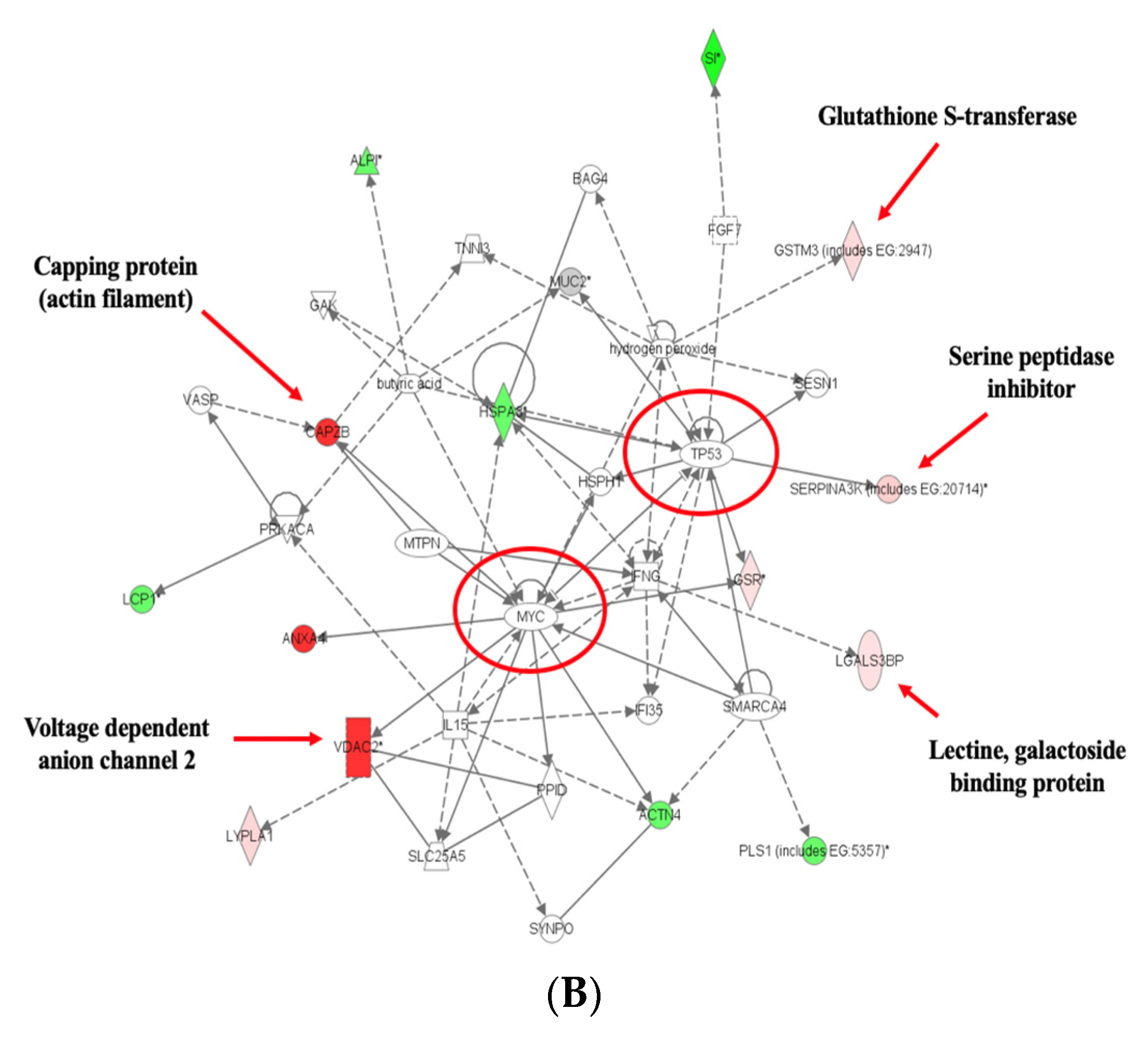
3.4. Pathway Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.; Miller, K.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.L.; Gahche, J.J.; Lentino, C.V.; Dwyer, J.T.; Engel, J.S.; Thomas, P.R.; Betz, J.M.; Sempos, C.T.; Picciano, M.F. Dietary supplement use in the United States, 2003–2006. J. Nutr. 2011, 141, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Ems, T.; St Lucia, K.; Huecker, M.R. Biochemistry, Iron Absorption. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar] [PubMed]
- Kontoghiorghes, G.J.; Weinberg, E.D. Iron: Mammalian defense systems, mechanisms of disease, and chelation therapy approaches. Blood Rev. 1995, 9, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Cherayil, B.J. The role of iron in the immune response to bacterial infection. Immunol. Res. 2011, 50, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.D. The role of iron in cancer. Eur. J. Cancer Prev. 1996, 5, 19–36. [Google Scholar] [PubMed]
- Connor, J.R.; Beard, J.L. Dietary iron supplements in the elderly: To use or not to use? Nutr. Today 1997, 32, 102–109. [Google Scholar] [CrossRef]
- Tuomainen, T.-P.; Punnonen, K.; Nyyssonen, K.; Salonen, J.T. Association between body iron stores and the risk of acute myocardial infarction in men. Circulation 1998, 97, 1461–1466. [Google Scholar] [CrossRef]
- Blakeborough, M.H.; Owen, R.W.; Bilton, R.F. Free radical generating mechanisms in the colon: Their role in the induction and promotion of colorectal cancer? Free Radic. Res. Commun. 1989, 6, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Babbs, C.F. Free radicals and the etiology of colon cancer. Free Radic. Biol. Med. 1990, 8, 191–200. [Google Scholar] [CrossRef]
- Toyokuni, S. Iron-induced carcinogenesis: The role of redox regulation. Free Radic. Biol. Med. 1996, 20, 553–566. [Google Scholar] [CrossRef]
- Glade, M.J. Food, nutrition, and the prevention of cancer: A global perspective. American Institute for Cancer Research/World Cancer Research Fund, American Institute for Cancer Research, 1997. Nutrition 1999, 15, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.B. Nutritional chemoprevention of colon cancer. Semin. Gastrointest. Dis. 2002, 13, 143–153. [Google Scholar] [PubMed]
- Howell, M.A. Deit as an etiological factor in the development of cancers of the colon and rectum. J. Chronic Dis. 1975, 28, 67–80. [Google Scholar] [CrossRef]
- Armstrong, B.; Doll, R. Environmental factors and cancer incidence and mortality in different countries, with special reference to dietary practices. Int. J. Cancer 1975, 15, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.G.; Jones, D.Y.; Micozzi, M.S.; Taylor, P.R. Body iron stores and the risk of cancer. N. Engl. J. Med. 1988, 319, 1047–1052. [Google Scholar] [CrossRef]
- Selby, J.V.; Friedman, G.D. Epidemiologic evidence of an association between body iron stores and risk of cancer. Int. J. Cancer 1988, 41, 677–682. [Google Scholar] [CrossRef]
- Stevens, R.G. Iron and the risk of cancer. Med. Oncol. Tumor Pharmacother. 1990, 7, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Sussman, H.H. Iron in cancer. Pathobiology 1992, 60, 2–9. [Google Scholar] [CrossRef]
- Knekt, P.; Reunanen, A.; Takkunen, H.; Aromaa, A.; Heliövaara, M.; Hakulinen, T. Body iron stores and risk of cancer. Int. J. Cancer 1994, 56, 379–382. [Google Scholar] [CrossRef]
- Mainous, A.G.; Gill, J.M.; Everett, C.J. Transferrin saturation, dietary iron intake, and risk of cancer. Ann. Fam. Med. 2005, 3, 131–137. [Google Scholar] [CrossRef]
- Freudenheim, J.L.; Graham, S.; Marshall, J.R.; Haughey, B.P.; Wilkinson, G. A case-control study of diet and rectal cancer in western New York. Am. J. Epidemiol. 1990, 131, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Wurzelmann, J.I.; Silver, A.; Schreinemachers, D.M.; Sandler, R.S.; Everson, R.B. Iron intake and the risk of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 1996, 5, 503–507. [Google Scholar] [PubMed]
- Erhardt, J.G.; Lim, S.S.; Bode, J.C.; Bode, C. A diet rich in fat and poor in dietary fiber increases the in vitro formation of reactive oxygen species in human feces. J. Nutr. 1997, 127, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, M. The second World Cancer Research Fund/American Institute for Cancer Research expert report. Food, nutrition, physical activity, and the prevention of cancer: A global perspective. Proc. Nutr. Soc. 2008, 67, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Luongo, C.; Gould, K.A.; Su, L.K.; Kinzler, K.W.; Vogelstein, B.; Dietrich, W.; Lander, E.S.; Moser, A.R. Mapping of multiple intestinal neoplasia (Min) to proximal chromosome 18 of the mouse. Genomics 1993, 15, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Nishisho, I.; Kinzler, K.W.; Vogelstein, B.; Miyoshi, Y.; Miki, Y.; Ando, H.; Horii, A. Mutations of the APC (adenomatous polyposis coli) gene in FAP (familial polyposis coli) patients and in sporadic colorectal tumors. Tohoku J. Exp. Med. 1992, 168, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Nishisho, I.; Kinzler, K.W.; Vogelstein, B.; Miyoshi, Y.; Miki, Y.; Ando, H.; Horii, A.; Nagase, H. Mutations of the adenomatous polyposis coli gene in familial polyposis coli patients and sporadic colorectal tumors. Princess Takamatsu. Symp. 1991, 22, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Cotrell, S.; Bicknell, D.; Kaklamanis, L.; Bodmer, W.F. Molecular analysis of APC mutations in familial adenomatous polyposis and sporadic colon carcinomas. Lancet 1992, 340, 626–630. [Google Scholar] [CrossRef]
- Augenlicht, L.H.; Velcich, A.; Klampfer, L.; Jie, H.; Georgia, C.; Maria, A.; Laboisse, C.; Basil, R.; Martin, L.; Kan, Y.; et al. Application of gene expression profiling to colon cell maturation, transformation and chemoprevention. J. Nutr. 2003, 133 (Suppl. S7), 2410S–2416S. [Google Scholar] [CrossRef]
- Milner, J.A.; Allison, R.G.; Elliott, J.G.; Go, V.J.; Miller, G.A.; Rock, C.; Rabindra, R.; Wargovich, M.J. Opportunities and challenges for future nutrition research in cancer prevention: A panel discussion. J. Nutr. 2003, 133 (Suppl. S7), 2502S–2504S. [Google Scholar] [CrossRef]
- Deng, S.S.; Xing, T.Y.; Zhou, H.Y.; Xiong, R.H.; Lu, Y.G.; Web, B.; Liu, S.Q.; Yang, H.J. Comparative proteome analysis of breast cancer and adjacent normal breast tissues in human. Genom. Proteom. Bioinform. 2006, 4, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Varambally, S.; Chinnaiyan, A.M. Differential proteomic alterations between localised and metastatic prostate cancer. Br. J. Cancer 2006, 95, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Alexe, G.; Alexe, S.; Liotta, L.A.; Petricoin, E.; Reiss, M.; Hammer, P.L. Ovarian cancer detection by logical analysis of proteomic data. Proteomics 2004, 4, 766–783. [Google Scholar] [CrossRef] [PubMed]
- Mazzanti, R.; Solazzo, M.; Fantappié, O.; Elfering, S.; Pantaleo, P.; Cianchi, F.; Ettl, A.; Giulivi, C. Differential expression proteomics of human colon cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G1329–G1338. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C., Jr. AIN-93 purified diets for laboratory rodents: Final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef] [PubMed]
- Zielińska-Dawidziak, M.; Hertig, I.; Staniek, H.; Piasecka-Kwiatkowska, D.; Nowak, K.W. Effect of iron status in rats on the absorption of metal ions from plant ferritin. Plant Foods Hum. Nutr. 2014, 69, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.R.; Bothwell, T.; SUSTAIN Task Force on Iron Powders. A comparison of physical properties, screening procedures and a human efficacy trial for predicting the bioavailability of commercial elemental iron powders used for food fortification. Int. J. Vitam. Nutr. Res. 2007, 77, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US). Panel on Micronutrients. In Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc; National Academies Press: Washington, DC, USA, 2001; Bookshelf ID: NBK222310. [Google Scholar] [CrossRef]
- Hurrell, R.; Egli, I. Iron bioavailability and dietary reference values. Am. J. Clin. Nutr. 2010, 91, 1461S–1467S. [Google Scholar] [CrossRef] [PubMed]
- Pickhardt, P.J.; Halberg, R.B.; Taylor, A.J.; Weichert, J.P. Microcomputed tomography colonography for polyp detection in an in vivo mouse tumor model. Proc. Natl. Acad. Sci. USA 2005, 102, 3419–3422. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar]
- Fricker, L.D.; Tashima, A.K.; Fakira, A.K.; Hochgeschwender, U.; Wetsel, W.C.; Devi, L.A. Neuropeptidomic Analysis of a Genetically Defined Cell Type in Mouse Brain and Pituitary. Cell Chem. Biol. 2021, 28, 105–112.e4. [Google Scholar] [CrossRef]
- Liu, C.; Gong, J.; Zhang, Q.; Chen, G.; Yin, S.; Luo, Z.; Zeng, W.; Yu, W.; Lan, P.; He, Z. Dietary iron modulates gut microbiota and induces SLPI secretion to promote colorectal tumorigenesis. Gut Microbes. 2023, 15, 2221978. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, N.; Adam, J.; Steinberg, P.; Wirtz, S.; Schwerdtle, T.; Adams-Quack, P.; Hovelmeyer, N.; Kaina, B.; Foersch, S.; Fahrer, J. Chronic intestinal inflammation drives colorectal tumor formation triggered by dietary heme iron in vivo. Arch. Toxicol. 2021, 95, 2507–2522. [Google Scholar] [CrossRef]
- Xue, X.; Ramakrishnan, S.K.; Weisz, K.; Triner, D.; Xie, L.; Attili, D.; Pant, A.; Gyorffy, B.; Zhan, M.; Carter-Su, C.; et al. Iron Uptake via DMT1 Integrates Cell Cycle with JAK-STAT3 Signaling to Promote Colorectal Tumorigenesis. Cell Metab. 2016, 24, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.J.; Goyert, J.W.; Solanki, S.; Kerk, S.A.; Chen, B.; Castillo, C.; Hsu, P.P.; Do, B.T.; Singhal, R.; Dame, M.K.; et al. Hepcidin sequesters iron to sustain nucleotide metabolism and mitochondrial function in colorectal cancer epithelial cells. Nat. Metab. 2021, 3, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Brookes, M.J.; Hughes, S.; Turner, F.E.; Reynolds, G.; Sharma, N.; Ismail, T.; Berx, G.; McKie, A.T.; Hotchin, N.; Anderson, G.J.; et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut 2006, 55, 1449–1460. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Wang, X.; Zhang, G.; Xia, T.; Zhu, R.; Tou, J. Dihydromyricetin functions as a tumor suppressor in hepatoblastoma by regulating SOD1/ROS pathway. Front. Oncol. 2023, 13, 1160548. [Google Scholar] [CrossRef]
- Wang, Y.C.; Leng, X.X.; Zhou, C.B.; Lu, S.Y.; Tsang, C.K.; Xu, J.; Zhang, M.M.; Chen, H.M.; Fang, J.Y. Non-enzymatic role of SOD1 in intestinal stem cell growth. Cell Death Dis. 2022, 13, 882. [Google Scholar] [CrossRef]
- Zhang, Z.; Lang, J.; Cao, Z.; Li, R.; Wang, X.; Wang, W. Radiation-induced SOD2 overexpression sensitizes colorectal cancer to radiation while protecting normal tissue. Oncotarget 2017, 8, 7791–7800. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, H.; Ding, C.; Jiang, D.; Zhao, Z.; Li, Y.; Ding, X.; Gao, J.; Zhou, H.; Luo, C.; et al. Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1. Signal Transduct. Target. Ther. 2023, 8, 51. [Google Scholar] [CrossRef]
- Wu, N.; Du, X.; Peng, Z.; Zhang, Z.; Cui, L.; Li, D.; Wang, R. Silencing of peroxiredoxin 1 expression ameliorates ulcerative colitis in a rat model. J. Int. Med. Res. 2021, 49, 300060520986313. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, S.; Yu, W.; Zheng, Y.; Wu, Y. Inhibition of ITGB1 enhance the anti-tumor effect of cetuximab in colorectal cancer cell. Medicine 2020, 99, e20944. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Sakai, N.; Furukawa, K.; Takayishiki, T.; Kuboki, S.; Takano, S.; Ohira, G.; Matsubara, H.; Ohtsuka, M. Yin Yang 1 regulates ITGAV and ITGB1, contributing to improved prognosis of colorectal cancer. Oncol. Rep. 2022, 47, 87. [Google Scholar] [CrossRef]
- Kung, Y.J.; Lam, B.; Tseng, S.H.; MacDonald, A.; Tu, H.F.; Wang, S.; Lin, J.; Tsai, Y.C.; Wu, T.C.; Hung, C.F. Localization of Salmonella and albumin-IL-2 to the tumor microenvironment augments anticancer T cell immunity. J. Biomed. Sci. 2022, 29, 57. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Luo, M.; Zhang, K.; Zhang, J.; Gao, T.; O’Connell, D.; Yao, F.; Mu, C.; Cai, B.; Shang, Y.; et al. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020, 11, 433. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, A.M.; Sacco, A.; Vecchio, E.; Scicchitano, S.; Petriaggi, L.; Giorgio, E.; Bulotta, S.; Levi, S.; Faniello, C.M.; Biamonte, F.; et al. Iron affects the sphere-forming ability of ovarian cancer cells in non-adherent culture conditions. Front. Cell Dev. Biol. 2023, 11, 1272667. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Schubert, D.; Maher, P. Oxytosis: A novel form of programmed cell death. Curr. Top. Med. Chem. 2001, 1, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef]
- Sorensen, K.D.; Abildgaard, M.O.; Haldrup, C.; Ulhoi, B.P.; Kristensen, H.; Strand, S.; Parker, C.; Hoyer, S.; Borre, M.; Orntoft, T.F. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br. J. Cancer 2013, 108, 420–428. [Google Scholar] [CrossRef]
- Fischer, S.; Tahoun, M.; Klaan, B.; Thierfelder, K.M.; Weber, M.A.; Krause, B.J.; Hakenberg, O.; Fuellen, G.; Hamed, M. A Radiogenomic Approach for Decoding Molecular Mechanisms Underlying Tumor Progression in Prostate Cancer. Cancers 2019, 11, 1293. [Google Scholar] [CrossRef]
- Ahluwalia, P.; Mondal, A.K.; Bloomer, C.; Fulzele, S.; Jones, K.; Ananth, S.; Gahlay, G.K.; Heneidi, S.; Rojiani, A.M.; Kota, V. Identification and Clinical Validation of a Novel 4 Gene-Signature with Prognostic Utility in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 3818. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Kroemer, G.; Tang, D. The tumor suppressor protein p53 and the ferroptosis network. Free Radic. Biol. Med. 2019, 133, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Lu, Y.; Shangguan, J.; Shu, X. PSMA1 mediates tumor progression and poor prognosis of gastric carcinoma by deubiquitinating and stabilizing TAZ. Cell Death Dis. 2022, 13, 989. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, W.; Zhou, Y.; Li, L.; Zhou, W. PSMA1, a Poor Prognostic Factor, Promotes Tumor Growth in Lung Squamous Cell Carcinoma. Dis. Markers. 2023, 2023, 5386635. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Roehrl, M.H.; Wang, J.Y. Proteomic profiling of antibody-inducing immunogens in tumor tissue identifies PSMA1, LAP3, ANXA3, and maspin as colon cancer markers. Oncotarget 2017, 9, 3996–4019. [Google Scholar] [CrossRef] [PubMed]
- He, Y.J.; Li, W.L.; Liu, B.H.; Dong, H.; Mou, Z.R.; Wu, Y.Z. Identification of differential proteins in colorectal cancer cells treated with caffeic acid phenethyl ester. World J. Gastroenterol. 2014, 20, 11840–11849. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Peng, X.; Nian, Z.; Yuan, S.; Wang, Z.; Song, Y.; Shamsnur, R.; Wang, H. UCHL-3 as a potential biomarker of ovarian cancer. Gynecol. Oncol. 2024, 182, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Ma, J.; Tian, Y.; Liu, Q.; Zhang, J. An immune-related exosome signature predicts the prognosis and immunotherapy response in ovarian cancer. BMC Womens Health 2024, 24, 49. [Google Scholar] [CrossRef]
- Qi, J.; Hu, Z.; Liu, S.; Li, F.; Wang, S.; Wang, W.; Sheng, X.; Feng, L. Comprehensively Analyzed Macrophage-Regulated Genes Indicate That PSMA2 Promotes Colorectal Cancer Progression. Front. Oncol. 2021, 10, 618902. [Google Scholar] [CrossRef]
- Peng, P.; Wang, Y.; Wang, B.L.; Song, Y.H.; Fang, Y.; Ji, H.; Huangfu, C.N.; Wang, K.M.; Zheng, Q. LncRNA PSMA3-AS1 promotes colorectal cancer cell migration and invasion via regulating miR-4429. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11594–11601. [Google Scholar] [CrossRef]
- Biao, T.; Cai-Feng, H.; Xiao-Hong, L.; Xiao-Li, C.; Wen-Bei, L.; Jun, W.; Chai, C.; Tao, Y. From Bowen disease to cutaneous squamous cell carcinoma: Eight markers were verified from transcriptomic and proteomic analyses. J. Transl. Med. 2022, 20, 416. [Google Scholar] [CrossRef] [PubMed]
- Lerman, I.; Ma, X.; Seger, C.; Maolake, A.; Garcia-Hernandez, M.; Rangel-Moreno, J.; Ackerman, J.; Nastiuk, K.L.; Susiarjo, M.; Hammes, S.R. Epigenetic Suppression of SERPINB1 Promotes Inflammation-Mediated Prostate Cancer Progression. Mol. Cancer Res. 2019, 17, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.C.; Yang, B.L.; Chen, W.C.; Ho, A.S.; Sie, Z.L.; Lin, H.C.; Chang, C.C. STAT3 Mediated miR-30a-5p Inhibition Enhances Proliferation and Inhibits Apoptosis in Colorectal Cancer Cells. Int. J. Mol. Sci. 2020, 21, 7315. [Google Scholar] [CrossRef] [PubMed]
- Giampieri, R.; Scartozzi, M.; Loretelli, C.; Piva, F.; Mandolesi, A.; Lezoche, G.; Prete, M.D.; Bitonni, A.; Faloppi, L.; Bianconi, M.; et al. Cancer stem cell gene profile as predictor of relapse in high risk stage II and stage III, radically resected colon cancer patients. PLoS ONE. 2013, 8, e72843. [Google Scholar] [CrossRef]
- Yang, L.; Bhattacharya, A.; Li, Y.; Sexton, S.; Ling, X.; Li, F.; Zhang, Y. Depleting receptor tyrosine kinases EGFR and HER2 overcomes resistance to EGFR inhibitors in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 184. [Google Scholar] [CrossRef] [PubMed]
- Mukaihara, K.; Suehara, Y.; Kohsaka, S.; Kubota, D.; Toda-Ishii, M.; Akaike, K.; Fujimara, T.; Kobayashi, E.; Yao, T.; Ladanyi, M.; et al. Expression of F-actin-capping protein subunit beta, CAPZB, is associated with cell growth and motility in epithelioid sarcoma. BMC Cancer 2016, 16, 206. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Wang, D.; Lu, Y.; He, Y.; Yu, J.; Wei, W.; Chen, C.; Wang, R.; Zhang, L.; Zhang, L.; et al. Vacuolin-1 inhibits endosomal trafficking and metastasis via CapZβ. Oncogene 2021, 40, 1775–1791. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Niu, X.; Li, Y.; Zhang, J.R.; Zhu, S.J.; Yang, Q.Y.; Zhang, W.; Gong, L. Role of the mucin-like glycoprotein FCGBP in mucosal immunity and cancer. Front. Immunol. 2022, 13, 863317. [Google Scholar] [CrossRef]
- Lang, T.; Klasson, S.; Larsson, E.; Johansson, M.E.; Hansson, G.C.; Samuelsson, T. Searching the Evolutionary Origin of Epithelial Mucus Protein Components-Mucins and FCGBP. Mol. Biol. Evol. 2016, 33, 1921–1936. [Google Scholar] [CrossRef]
- Yan, T.; Tian, D.; Chen, J.; Tan, Y.; Cheng, Y.; Ye, L.; Deng, G.; Liu, B.; Yuan, F.; Zhang, S.; et al. FCGBP Is a Prognostic Biomarker and Associated With Immune Infiltration in Glioma. Front. Oncol. 2022, 11, 769033. [Google Scholar] [CrossRef]
- Lane, D.J.R.; Bae, D.H.; Siafakas, A.R.; Rahmanto, Y.S.; Al-Akra, L.; Jansson, P.J.; Casero, R.A.; Richardson, D.R. Coupling of the polyamine and iron metabolism pathways in the regulation of proliferation: Mechanistic links to alterations in key polyamine biosynthetic and catabolic enzymes. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 2793–2813. [Google Scholar] [CrossRef] [PubMed]
- Kontoghiorghes, G.J. New Iron Metabolic Pathways and Chelation Targeting Strategies Affecting the Treatment of All Types and Stages of Cancer. Int. J. Mol. Sci. 2022, 23, 13990. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Yu, D.; Zeller, K.I.; Kim, J.W.; Racke, F.; Thomas-Tikhonenko, A.; Dang, C.V. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol. Cell Biol. 2006, 26, 2373–2386. [Google Scholar] [CrossRef] [PubMed]
- Saletta, F.; Rahmanto, Y.S.; Siafakas, A.R.; Richardson, D.R. Cellular iron depletion and the mechanisms involved in the iron-dependent regulation of the growth arrest and DNA damage family of genes. J. Biol. Chem. 2011, 286, 35396–35406. [Google Scholar] [CrossRef]
- Nurtjahja-Tjendraputra, E.; Fu, D.; Phang, J.M.; Richardson, D.R. Iron chelation regulates cyclin D1 expression via the proteasome: A link to iron deficiency-mediated growth suppression. Blood 2007, 109, 4045–4054. [Google Scholar] [CrossRef]
| Formula | g/Kg | |
|---|---|---|
| Casein, low Cu and Fe | 200.0 | |
| DL-Methionine | 3.0 | |
| Sucrose | 545.19 | |
| Corn Starch | 150.0 | |
| Corn Oil | 50.0 | |
| Alphacel (low mineral fiber) | 50.0 | |
| Mineral Mix, Fe Deficient (81062) *** | 35.0 | |
| Vitamin Mix, AIN-76A (40077) *** | 14.0 | |
| Choline Bitartrate | 2.8 | |
| Ethoxyquin, antioxidant | 0.01 | |
| Macronutrient | % dry weight | % kcal |
| Protein | 17.7 | 17.8 |
| Carbohydrate | 69.8 | 70.4 |
| Fat | 5.2 | 11.8 |
| Gene | GI ID | Spot | Fold Change | Description | |
|---|---|---|---|---|---|
| T/NT | TFe/T | ||||
| ACTG1 | 809561 | 1246 | 1.74 | actin, gamma 1 | |
| ACTN4 | 11230802 | 741 | −2.55 | actinin, alpha 4 | |
| ALB | 26986064 | 1728 | 2.87 | albumin | |
| 33859506 | 1455 | 3.47 | |||
| ALDOB | 1619606 | 1856 | 2.16 | aldolase B | |
| 15723268 | 1682 | 3.07 | |||
| ANPEP | 6678664 | 543 | 1.73 | alanyl (membrane) aminopeptidase | |
| AOC3 | 4185817 | 543 | 1.73 | amine oxidase, copper containing 3 | |
| ARG2 | 6753110 | 1698 | 2.75 | arginase, type Il | |
| CAPZB | 83649737 | 1484 | 10.86 | capping protein (actin filament) muscle -line, beta | |
| CPS1 | 8393186 | 909 | 1.77 | carbamoyl-phosphate synthetase 1, mitochondrial | |
| DES | 33563250 | 1046 | 1.76 | desmin | |
| DPP7 | 13626390 | 933 | 1.31 | 1.59 | dipeptidyl-peptidase 7 |
| FBN1 | 118197277 | 543 | 1.73 | fibrillin 1 | |
| FCGBP | 21410127 | 485 | −3.83 | Fc fragment of IgG binding protein | |
| 94381948 | 1372 | 15.21 | |||
| FLNA | 38257560 | 1728 | 2.87 | filamin A, alpha (actin binding protein 280) | |
| 47847514 | 909 | 1.77 | |||
| FLNB | 38257404 | 579 | 1.58 | filamin B, beta (actin binding protein 278) | |
| FLNC | 94377129 | 578 | 1.79 | 1.72 | filamin C, gamma (actin binding protein 280) |
| 94377129 | 579 | 1.58 | |||
| GPD1 | 387177 | 1682 | 3.07 | glycerol-3-phosphate dehydrogenase 1 (soluble) | |
| GSR | 13624751 | 933 | 1.31 | 1.59 | glutathione reductase |
| GSTM3 | 6754086 | 1809 | 1.96 | glutathione S-transferase M3 | |
| HPX | 23956086 | 1032 | 2.3 | hemopexin | |
| HSP90A81 | 40556608 | 543 | 1.73 | heat shock protein 90 kDa alpha, class B member 1 | |
| HSP90B1 | 6755863 | 543 | 1.73 | heat shock protein 90 kDa beta (Grp94), member 1 | |
| HSPA5 | 1304157 | 741 | −2.55 | heat shock 70 kDa protein 5 | |
| HSPA9 | 42542422 | 741 | −2.55 | heat shock 70 kDa protein 8 | |
| IGH | 62027409 | 909 | 1.77 | immunoglobulin heavy chain complex | |
| ITGB1 | 762977 | 933 | 1.31 | 1.59 | integrin, beta 1 |
| KHK | 31982229 | 1468 | −2.8 | ketohexokinase | |
| KRT19 | 6680606 | 1698 | 2.75 | keratin 19 | |
| LAMC1 | 31791057 | 933 | 1.31 | 1.59 | laminin, gamma 1 |
| LCP2 | 31543113 | 741 | −2.55 | lymphocyte cytosolic protein 1 | |
| LCT | 74192292 | 263 | −1.95 | lactase | |
| 74192292 | 652 | −2.86 | |||
| LGALS3BP | 397800 | 933 | 1.31 | 1.59 | lectin, galactoside-binding, soluble, 3 binding protein |
| MUC2 | 28865873 | 1052 | 1.84 | mucin 2, oligomeric mucus/gel-forming | |
| MYH12 | 50510675 | 741 | −2.55 | myosin, heavy chain 11, smooth muscle | |
| PEPD | 6679279 | 933 | 1.31 | 1.59 | peptidase D |
| PGK1 | 129903 | 1698 | 2.75 | phosphoglycerate kinase 1 | |
| PRDX1 | 6754976 | 1953 | −2.15 | peroxiredoxin 1 | |
| PRDX4 | 7948999 | 1698 | 2.75 | peroxiredoxin 4 | |
| PSMA1 | 33563282 | 1574 | 3.49 | proteasome subunit, alpha type, 1 | |
| 33563282 | 1640 | 2.23 | |||
| PSMA3 | 31981534 | 2155 | −3.53 | proteasome subunit, alpha type, 3 | |
| PSMB2 | 31981327 | 1953 | −2.15 | proteasome subunit, beta type, 2 | |
| PSMB4 | 3914439 | 1809 | 1.96 | proteasome subunit, beta type, 4 | |
| RAB1A | 206553 | 1909 | −1.68 | RAB1A, member RAS oncogene family | |
| SCIN | 2851563 | 579 | 1.58 | scinderin | |
| SERPINA3K | 54173 | 861 | 2.39 | 2.21 | serine peptidase inhibitor, clade A, member 3K |
| 54173 | 862 | 2.54 | 2.6 | ||
| 54173 | 863 | 2.33 | |||
| 54173 | 868 | 2.19 | |||
| SERPINB1 | 114158675 | 1296 | 1.58 | serpin peptidase inhibitor, clade B, member 1 | |
| 114158675 | 1698 | 2.75 | |||
| SERPINB6 | 6678097 | 1246 | 1.74 | serpin peptidase inhibitor, clade B, member 6 | |
| 6678097 | 1247 | 1.73 | |||
| SERPINC2 | 18252782 | 933 | 1.31 | 1.59 | serpin peptidase inhibitor, clade C. member 1 |
| SOD1 | 45597447 | 2137 | −6.52 | superoxide dismutase 1 | |
| SOD2 | 53450 | 1953 | −2.15 | superoxide dismutase 2 | |
| TNC | 220610 | 543 | 1.75 | tenascin C | |
| 29290613 | 868 | 2.19 | |||
| TPI1 | 54855 | 1682 | 3.07 | triosephosphate isomerase 1 | |
| TUBA1C | 6678469 | 1032 | 2.3 | tubulin, alpha 1c | |
| TXNRD1 | 13569841 | 909 | 1.77 | thioredoxin reductase 1 | |
| VDAC2 | 6755965 | 1484 | 10.86 | voltage-dependent anion channel 2 | |
| 6755965 | 1640 | 2.23 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Swain, I.X.; Kresak, A.M. Iron Supplementation Increases Tumor Burden and Alters Protein Expression in a Mouse Model of Human Intestinal Cancer. Nutrients 2024, 16, 1316. https://doi.org/10.3390/nu16091316
Swain IX, Kresak AM. Iron Supplementation Increases Tumor Burden and Alters Protein Expression in a Mouse Model of Human Intestinal Cancer. Nutrients. 2024; 16(9):1316. https://doi.org/10.3390/nu16091316
Chicago/Turabian StyleSwain, Ian X., and Adam M. Kresak. 2024. "Iron Supplementation Increases Tumor Burden and Alters Protein Expression in a Mouse Model of Human Intestinal Cancer" Nutrients 16, no. 9: 1316. https://doi.org/10.3390/nu16091316
APA StyleSwain, I. X., & Kresak, A. M. (2024). Iron Supplementation Increases Tumor Burden and Alters Protein Expression in a Mouse Model of Human Intestinal Cancer. Nutrients, 16(9), 1316. https://doi.org/10.3390/nu16091316





