A Stress Reduction Intervention for Lactating Mothers Alters Maternal Gut, Breast Milk, and Infant Gut Microbiomes: Data from a Randomized Controlled Trial
Abstract
1. Introduction
2. Materials and Methods
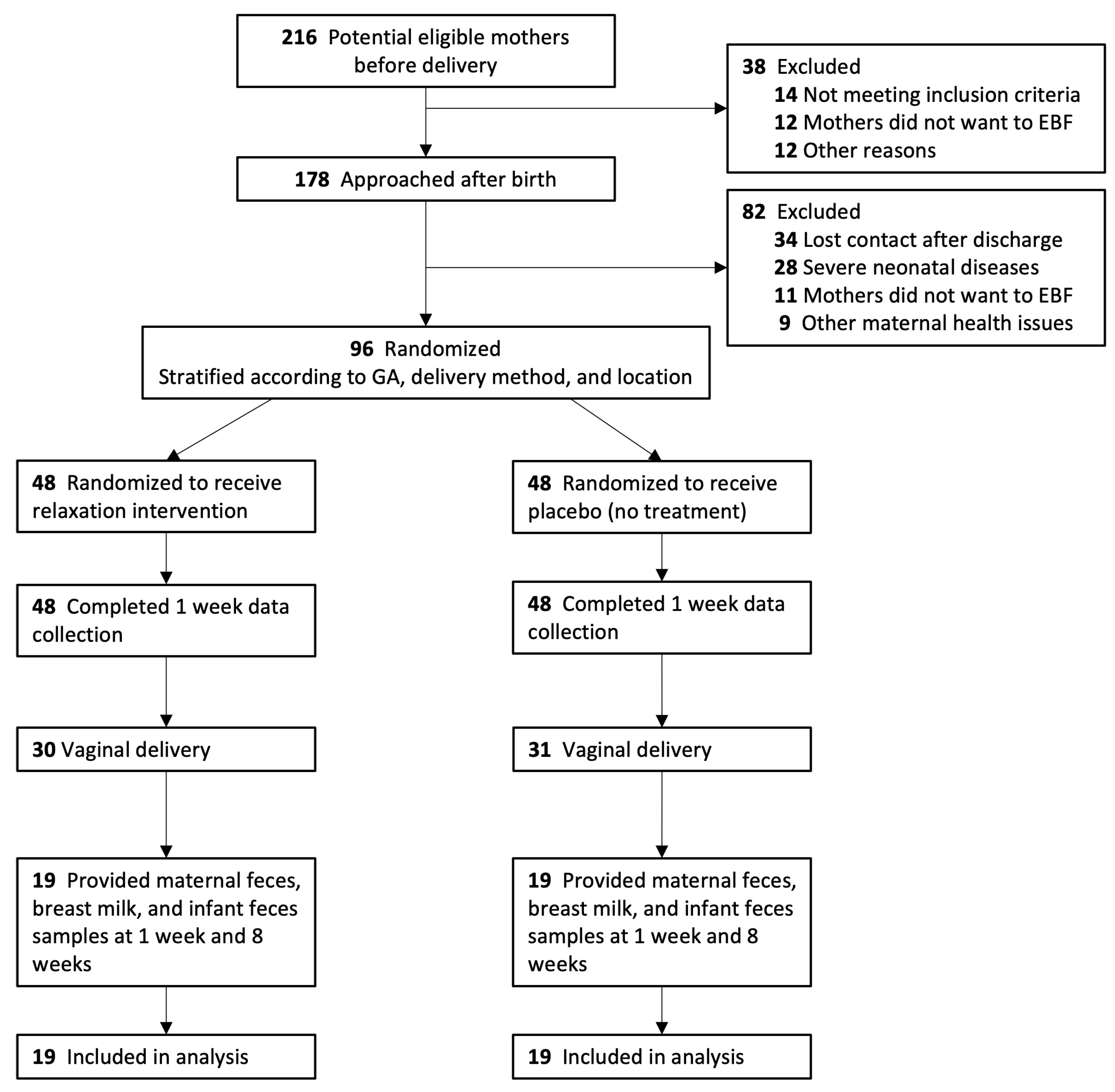
2.1. Study Design and Participants
2.2. Study Procedures and Randomization
2.3. Outcomes and Measures
2.4. DNA Extraction, Sequencing, and Data Processing
2.5. Bioinformatic Workflow of 16S rRNA Gene Amplicon
2.6. Statistical Analysis
3. Results
3.1. Study Population and Baseline Characteristics
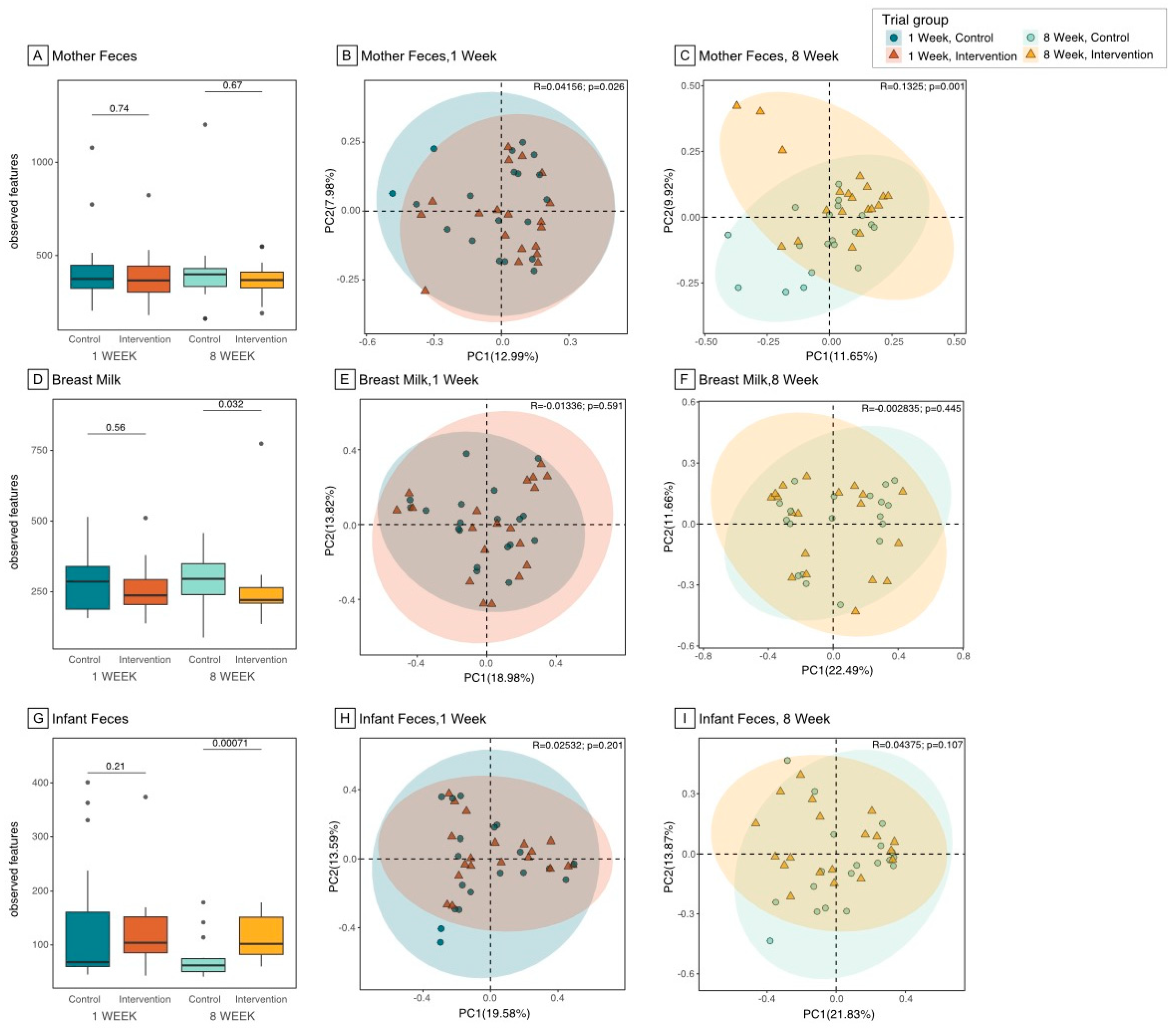
3.2. Microbiome Composition and Diversity in the Maternal Gut
3.3. Microbiome Composition and Diversity in Breast Milk
3.4. Microbiome Composition and Diversity in Infant Gut
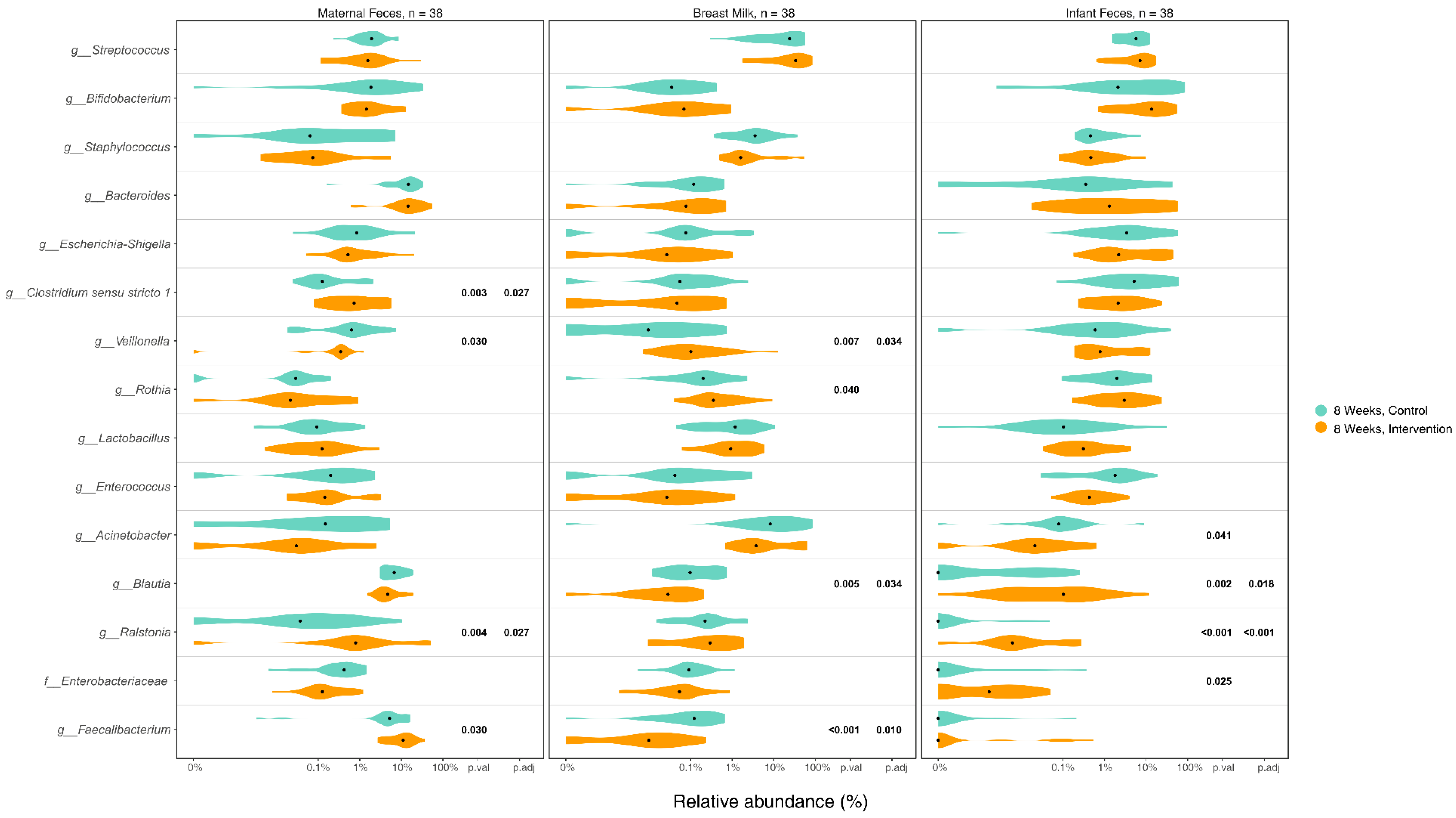
3.5. Differences in Microbiome Composition in Maternal Gut, Breast Milk, and Infant Gut Samples between Groups
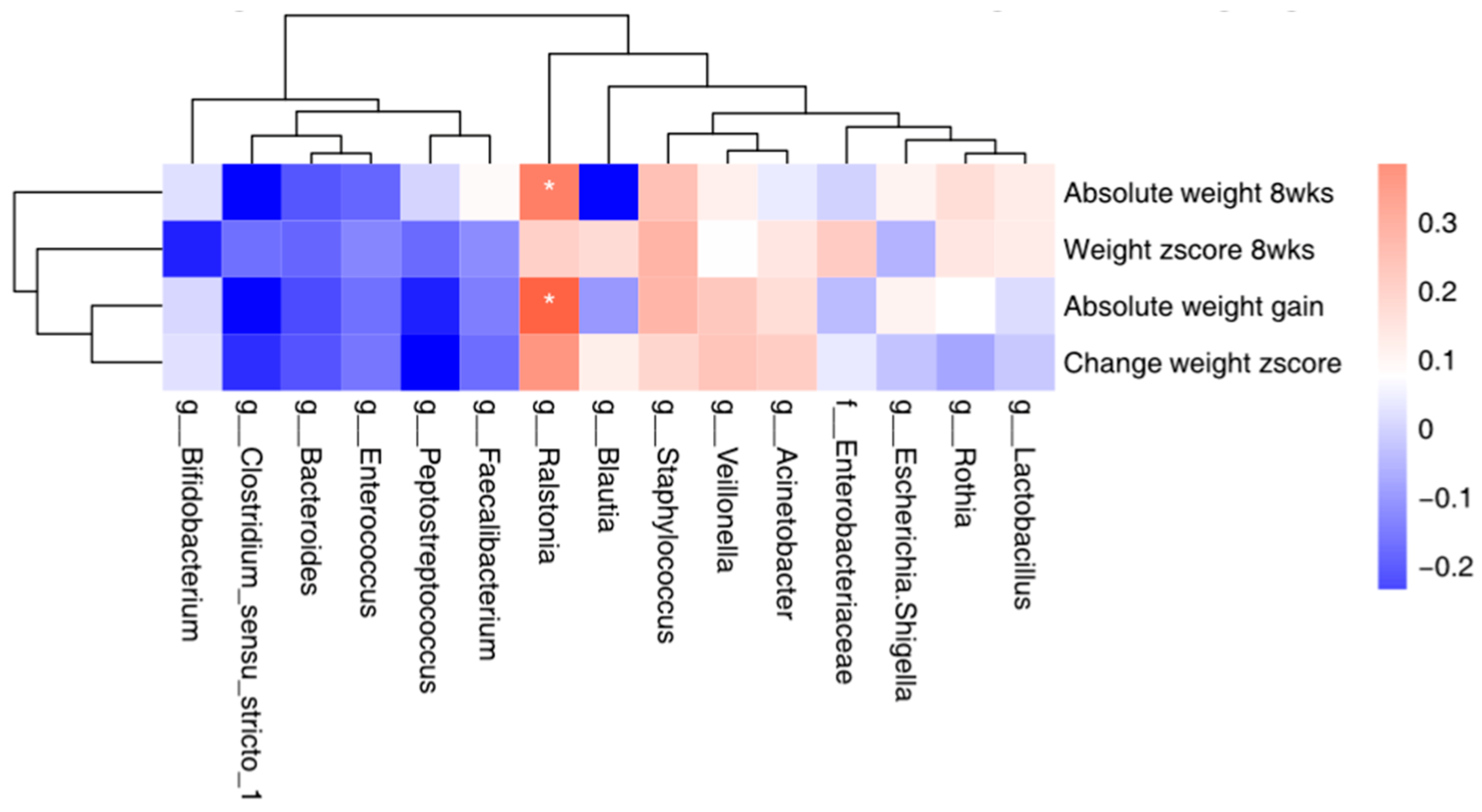
3.6. Correlation between the Top 15 Microbial and Maternal Stress/Infant Weight
3.7. Unintended Effects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Marchesi, J.R.; Scully, P.; Codling, C.; Ceolho, A.M.; Quigley, E.M.; Cryan, J.F.; Dinan, T.G. Early life stress alters behavior, immunity, and microbiota in rats: Implications for irritable bowel syndrome and psychiatric illnesses. Biol. Psychiatry 2009, 65, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 94. [Google Scholar] [CrossRef] [PubMed]
- Donnet-Hughes, A.; Perez, P.F.; Dore, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc. Nutr. Soc. 2010, 69, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, L.; Langa, S.; Martin, V.; Maldonado, A.; Jimenez, E.; Martin, R.; Rodriguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.F.; Dore, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. Bacterial imprinting of the neonatal immune system: Lessons from maternal cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef] [PubMed]
- Kapourchali, F.R.; Cresci, G.A. Early-Life gut microbiome—The importance of maternal and infant factors in its establishment. Nutr. Clin. Pract. 2020, 35, 386–405. [Google Scholar] [CrossRef] [PubMed]
- Mollova, D.; Vasileva, T.; Bivolarski, V.; Iliev, I. The Enzymatic Hydrolysis of Human Milk Oligosaccharides and Prebiotic Sugars from LAB Isolated from Breast Milk. Microorganisms 2023, 11, 1904. [Google Scholar] [CrossRef]
- Salli, K.; Hirvonen, J.; Anglenius, H.; Hibberd, A.A.; Ahonen, I.; Saarinen, M.T.; Maukonen, J.; Ouwehand, A.C. The Effect of Human Milk Oligosaccharides and Bifidobacterium longum subspecies infantis Bi-26 on Simulated Infant Gut Microbiome and Metabolites. Microorganisms 2023, 11, 1553. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Kurakawa, T.; Tsuji, H.; Takahashi, T.; Kawashima, K.; Nagata, S.; Nomoto, K.; Yamashiro, Y. Evolution of gut Bifidobacterium population in healthy Japanese infants over the first three years of life: A quantitative assessment. Sci. Rep. 2017, 7, 10097. [Google Scholar] [CrossRef] [PubMed]
- Galley, J.D.; Mashburn-Warren, L.; Blalock, L.C.; Lauber, C.L.; Carroll, J.E.; Ross, K.M.; Hobel, C.; Coussons-Read, M.; Schetter, C.D.; Gur, T.L. Maternal anxiety, depression and stress affects offspring gut microbiome diversity and bifidobacterial abundances. Brain Behav. Immun. 2023, 107, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, J.; Ren, E.; Su, Y.; Zhu, W. Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 2018, 49, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Hussain, N.; Hameed, Z.; Lin, L. Elucidating the role of diet in maintaining gut health to reduce the risk of obesity, cardiovascular and other age-related inflammatory diseases: Recent challenges and future recommendations. Gut Microbes 2024, 16, 2297864. [Google Scholar] [CrossRef] [PubMed]
- Aziz, T.; Khan, A.A.; Tzora, A.; Voidarou, C.; Skoufos, I. Dietary implications of the Bidirectional Relationship between the gut microflora and inflammatory diseases with special emphasis on irritable bowel disease: Current and future perspective. Nutrients 2023, 15, 2956. [Google Scholar] [CrossRef] [PubMed]
- Mady, E.A.; Doghish, A.S.; El-Dakroury, W.A.; Elkhawaga, S.Y.; Ismail, A.; El-Mahdy, H.A.; Elsakka, E.G.; El-Husseiny, H.M. Impact of the mother’s gut microbiota on infant microbiome and brain development. Neurosci. Biobehav. Rev. 2023, 150, 105195. [Google Scholar] [CrossRef]
- Roager, H.M.; Stanton, C.; Hall, L.J. Microbial metabolites as modulators of the infant gut microbiome and host-microbial interactions in early life. Gut Microbes 2023, 15, 2192151. [Google Scholar] [CrossRef]
- Marques, T.M.; Wall, R.; Ross, R.P.; Fitzgerald, G.F.; Ryan, C.A.; Stanton, C. Programming infant gut microbiota: Influence of dietary and environmental factors. Curr. Opin. Biotechnol. 2010, 21, 149–156. [Google Scholar] [CrossRef]
- Cong, X.; Xu, W.; Romisher, R.; Poveda, S.; Forte, S.; Starkweather, A.; Henderson, W.A. Focus: Microbiome: Gut microbiome and infant health: Brain-gut-microbiota axis and host genetic factors. Yale J. Biol. Med. 2016, 89, 299. [Google Scholar]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The human microbiome and child growth–first 1000 days and beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Dib, S.; Wells, J.C.; Eaton, S.; Fewtrell, M. A Breastfeeding Relaxation Intervention Promotes Growth in Late Preterm and Early Term Infants: Results from a Randomized Controlled Trial. Nutrients 2022, 14, 5041. [Google Scholar] [CrossRef] [PubMed]
- Mohd Shukri, N.H.; Wells, J.; Eaton, S.; Mukhtar, F.; Petelin, A.; Jenko-Pražnikar, Z.; Fewtrell, M. Randomized controlled trial investigating the effects of a breastfeeding relaxation intervention on maternal psychological state, breast milk outcomes, and infant behavior and growth. Am. J. Clin. Nutr. 2019, 110, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wei, Z.; Wells, J.C.; Fewtrell, M. Effects of relaxation therapy on maternal psychological status and infant growth following late preterm and early-term delivery: A randomized controlled trial. Am. J. Clin. Nutr. 2023, 117, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wells, J.; Wei, Z.; Fewtrell, M. Effects of relaxation therapy on maternal psychological state, infant growth and gut microbiome: Protocol for a randomised controlled trial investigating mother-infant signalling during lactation following late preterm and early term delivery. Int. Breastfeed. J. 2019, 14, 50. [Google Scholar] [CrossRef]
- Menelli, S. Breastfeeding Meditation; White Heart Publishing: Encinitas, CA, USA, 2004. [Google Scholar]
- Zhen, W.; Yuan, W.; Zhi-Guo, W.; Dan-dan, C.; Jue, C.; Ze-ping, X. Reliability and validity of the Chinese version of perceived stress scale. J. Shanghai Jiaotong Univ. Med. Sci. 2015, 35, 1448. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Robeson, M.S.; O’Rourke, D.R.; Kaehler, B.D.; Ziemski, M.; Dillon, M.R.; Foster, J.T.; Bokulich, N.A. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Comput. Biol. 2021, 17, e1009581. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Sun, X.; Cai, Y.; Dai, W.; Jiang, W.; Tang, W. The difference of gut microbiome in different biliary diseases in infant before operation and the changes after operation. BMC Pediatr. 2022, 22, 502. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Borewicz, K.; Suarez-Diez, M.; Hechler, C.; Beijers, R.; de Weerth, C.; Arts, I.; Penders, J.; Thijs, C.; Nauta, A.; Lindner, C. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci. Rep. 2019, 9, 2434. [Google Scholar] [CrossRef]
- Brink, L.R.; Mercer, K.E.; Piccolo, B.D.; Chintapalli, S.V.; Elolimy, A.; Bowlin, A.K.; Matazel, K.S.; Pack, L.; Adams, S.H.; Shankar, K. Neonatal diet alters fecal microbiota and metabolome profiles at different ages in infants fed breast milk or formula. Am. J. Clin. Nutr. 2020, 111, 1190–1202. [Google Scholar] [CrossRef]
- Ma, J.; Li, Z.; Zhang, W.; Zhang, C.; Zhang, Y.; Mei, H.; Zhuo, N.; Wang, H.; Wang, L.; Wu, D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: A study of 91 term infants. Sci. Rep. 2020, 10, 15792. [Google Scholar] [CrossRef]
- Odiase, E.; Frank, D.N.; Young, B.E.; Robertson, C.E.; Kofonow, J.M.; Davis, K.N.; Berman, L.M.; Krebs, N.F.; Tang, M. The Gut Microbiota Differ in Exclusively Breastfed and Formula-Fed United States Infants and are Associated with Growth Status. J. Nutr. 2023, 153, 2612–2621. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Zhou, Y.; Liu, X.; Wang, Q. Geographic patterns and environmental correlates of taxonomic, phylogenetic and functional β-diversity of wetland plants in the Qinghai-Tibet Plateau. Ecol. Indic. 2024, 160, 111889. [Google Scholar] [CrossRef]
- Tavalire, H.F.; Christie, D.M.; Leve, L.D.; Ting, N.; Cresko, W.A.; Bohannan, B.J. Shared environment and genetics shape the gut microbiome after infant adoption. MBio 2021, 12, e00548-21. [Google Scholar] [CrossRef] [PubMed]
- Rajilić-Stojanović, M.; De Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef] [PubMed]
- Latuga, M.S.; Stuebe, A.; Seed, P.C. A review of the source and function of microbiota in breast milk. Semin. Reprod. Med. 2014, 32, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Moloney, R.D.; Desbonnet, L.; Clarke, G.; Dinan, T.G.; Cryan, J.F. The microbiome: Stress, health and disease. Mamm. Genome 2014, 25, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, E.; Barranco, A.; Ramirez, M.; Gruart, A.; Delgado-Garcia, J.M.; Martinez-Lara, E.; Blanco, S.; Martin, M.J.; Castanys, E.; Buck, R.; et al. Effects of a human milk oligosaccharide, 2′-fucosyllactose, on hippocampal long-term potentiation and learning capabilities in rodents. J. Nutr. Biochem. 2015, 26, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Smilowitz, J.T.; O’sullivan, A.; Barile, D.; German, J.B.; Lönnerdal, B.; Slupsky, C.M. The human milk metabolome reveals diverse oligosaccharide profiles. J. Nutr. 2013, 143, 1709–1718. [Google Scholar] [CrossRef]
- Kortesniemi, M.; Slupsky, C.M.; Aatsinki, A.-K.; Sinkkonen, J.; Karlsson, L.; Linderborg, K.M.; Yang, B.; Karlsson, H.; Kailanto, H.-M. Human milk metabolome is associated with symptoms of maternal psychological distress and milk cortisol. Food Chem. 2021, 356, 129628. [Google Scholar] [CrossRef]
- Mikami, K.; Kimura, M.; Takahashi, H. Influence of maternal bifidobacteria on the development of gut bifidobacteria in infants. Pharmaceuticals 2012, 5, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Kubota, H.; Gawad, A.; Sakai, T.; Oishi, K.; Martin, R.; Ben-Amor, K.; Knol, J. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS ONE 2013, 8, e78331. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Ruiz, L.; Jara, J.; Orgaz, B.; Rodríguez, J.M. Strategies for the preservation, restoration and modulation of the human milk microbiota. Implications for human milk banks and neonatal intensive care units. Front. Microbiol. 2018, 9, 2676. [Google Scholar] [CrossRef] [PubMed]
- LeBouder, E.; Rey-Nores, J.E.; Raby, A.-C.; Affolter, M.; Vidal, K.; Thornton, C.A.; Labéta, M.O. Modulation of neonatal microbial recognition: TLR-mediated innate immune responses are specifically and differentially modulated by human milk. J. Immunol. 2006, 176, 3742–3752. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Páez, A.; Gómez del Pugar, E.M.; López-Almela, I.; Moya-Pérez, Á.; Codoñer-Franch, P.; Sanz, Y. Depletion of Blautia species in the microbiota of obese children relates to intestinal inflammation and metabolic phenotype worsening. Msystems 2020, 5, e00857-19. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]
| Characteristics | Total N = 38 | Control N = 19 | Intervention N = 19 |
|---|---|---|---|
| Mean (SD) | |||
| Maternal age (yr) | 31 (3) | 30 (1.6) | 31 (3.8) |
| Maternal education (yr) | 16.0 (1) | 15.5 (0.9) | 16.4 (1.6) |
| Maternal BMI (kg/m2) a | 23.3 (3) | 22.6 (1.9) | 24.0 (3.7) |
| Maternal stress b | 20.3 (8) | 20.2 (7.1) | 20.5 (8.9) |
| Infant weight (kg) c | 2.70 (0.3) | 2.69 (0.3) | 2.71 (0.4) |
| Infant length (cm) c | 47.7(2) | 47.5(2) | 48.0(2) |
| N (%) | |||
| Gestational week | |||
| 34 | 1 (3) | 0 (0) | 1 (5.3) |
| 35 | 4 (11) | 2 (10.5) | 2 (10.5) |
| 36 | 17 (45) | 8 (42.1) | 9 (47.4) |
| 37 | 16 (42) | 9 (47.4) | 7 (36.8) |
| N (%) | |||
| Infant sex | |||
| Male | 16 (42) | 7 (36.8) | 9 (47.4) |
| Female | 22 (58) | 12 (63.2) | 10 (52.6) |
| EBF | |||
| 1 week d | 38 (100) | 19 (100) | 19(100) |
| 8 weeks e | 20 (52.6) | 9 (47.4) | 11 (57.9) |
| Use of antibiotics f | |||
| During hospital stay | 3 (7.9) | 2 (10.5) | 1 (5.3) |
| During 1–8 weeks | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Zhang, Y.; Wells, J.C.K.; Wei, Z.; Bajaj-Elliott, M.; Nielsen, D.S.; Fewtrell, M.S. A Stress Reduction Intervention for Lactating Mothers Alters Maternal Gut, Breast Milk, and Infant Gut Microbiomes: Data from a Randomized Controlled Trial. Nutrients 2024, 16, 1074. https://doi.org/10.3390/nu16071074
Yu J, Zhang Y, Wells JCK, Wei Z, Bajaj-Elliott M, Nielsen DS, Fewtrell MS. A Stress Reduction Intervention for Lactating Mothers Alters Maternal Gut, Breast Milk, and Infant Gut Microbiomes: Data from a Randomized Controlled Trial. Nutrients. 2024; 16(7):1074. https://doi.org/10.3390/nu16071074
Chicago/Turabian StyleYu, Jinyue, Yan Zhang, Jonathan C. K. Wells, Zhuang Wei, Mona Bajaj-Elliott, Dennis Sandris Nielsen, and Mary S. Fewtrell. 2024. "A Stress Reduction Intervention for Lactating Mothers Alters Maternal Gut, Breast Milk, and Infant Gut Microbiomes: Data from a Randomized Controlled Trial" Nutrients 16, no. 7: 1074. https://doi.org/10.3390/nu16071074
APA StyleYu, J., Zhang, Y., Wells, J. C. K., Wei, Z., Bajaj-Elliott, M., Nielsen, D. S., & Fewtrell, M. S. (2024). A Stress Reduction Intervention for Lactating Mothers Alters Maternal Gut, Breast Milk, and Infant Gut Microbiomes: Data from a Randomized Controlled Trial. Nutrients, 16(7), 1074. https://doi.org/10.3390/nu16071074










