Regular Exercise Training Induces More Changes on Intestinal Glucose Uptake from Blood and Microbiota Composition in Leaner Compared to Heavier Individuals in Monozygotic Twins Discordant for BMI
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Participants
2.3. Training Intervention
2.4. Euglycemic Hyperinsulinemic Clamp and FDG-PET Study
2.5. Magnetic Resonance Imaging
2.6. Body Composition and Peak Aerobic Exercise Capacity Test
2.7. Dietary Assessment
2.8. Analysis of Fecal Microbiota
2.9. Statistical Analysis and Modelling
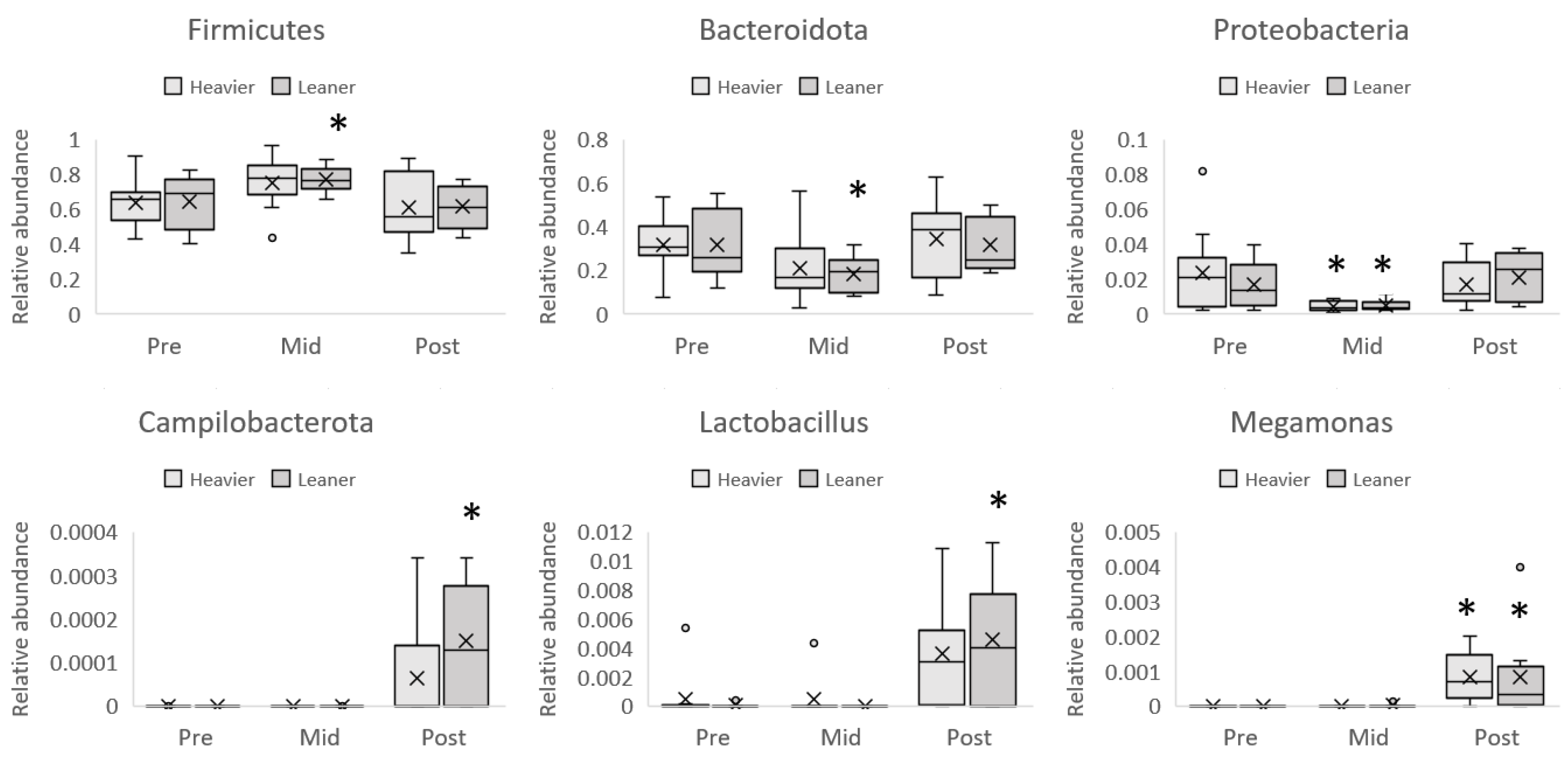
3. Results
Dietary Assessment
4. Discussion
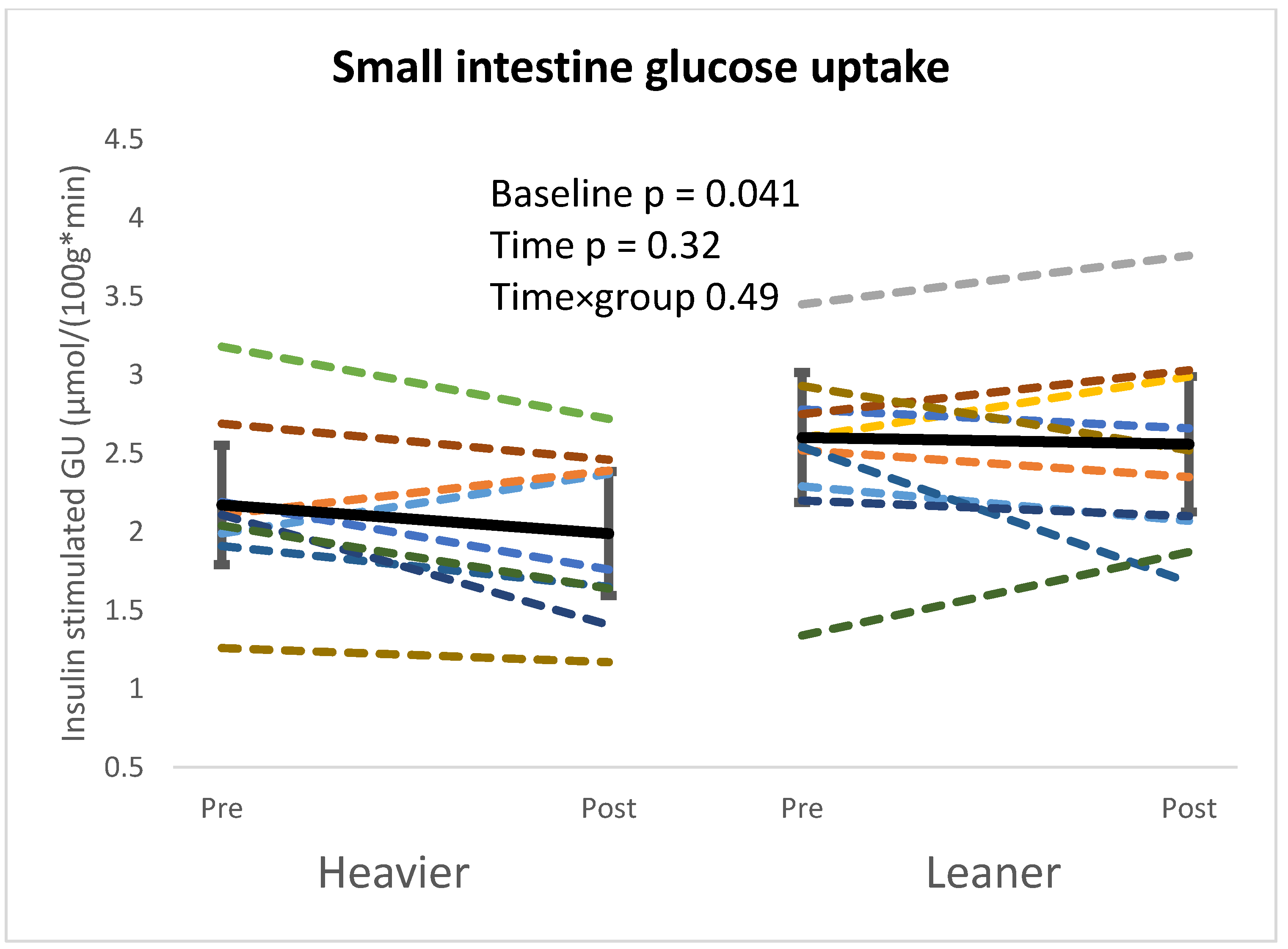
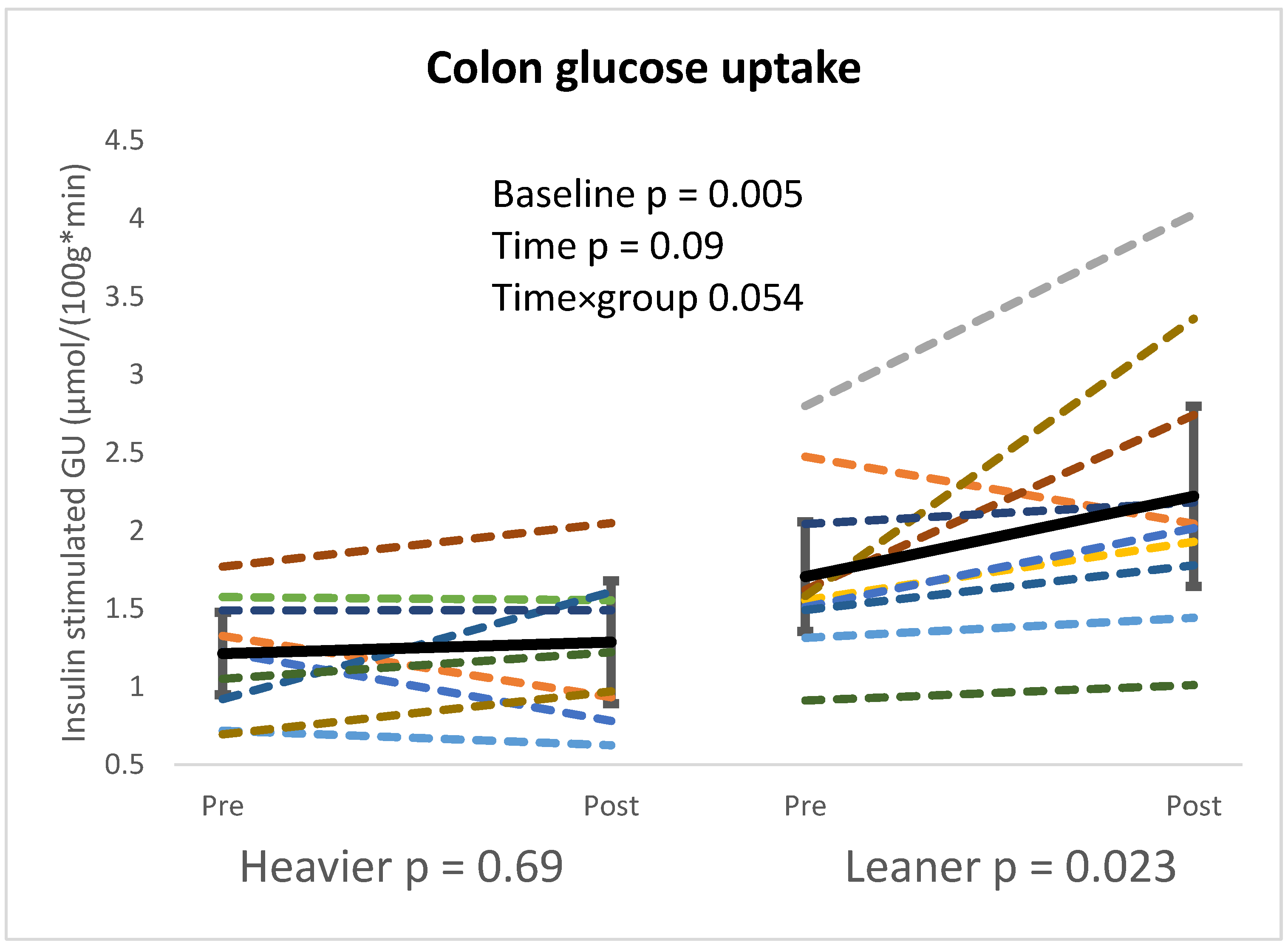
4.1. Intestinal Glucose Metabolism
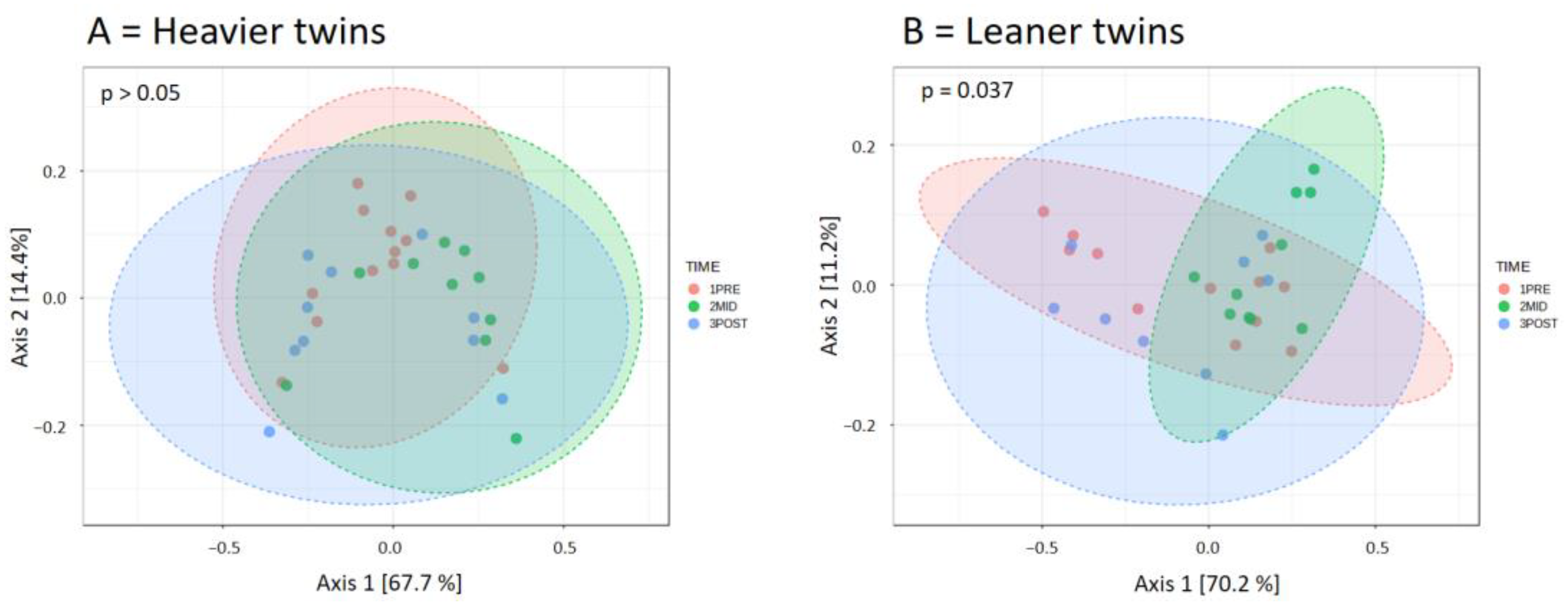
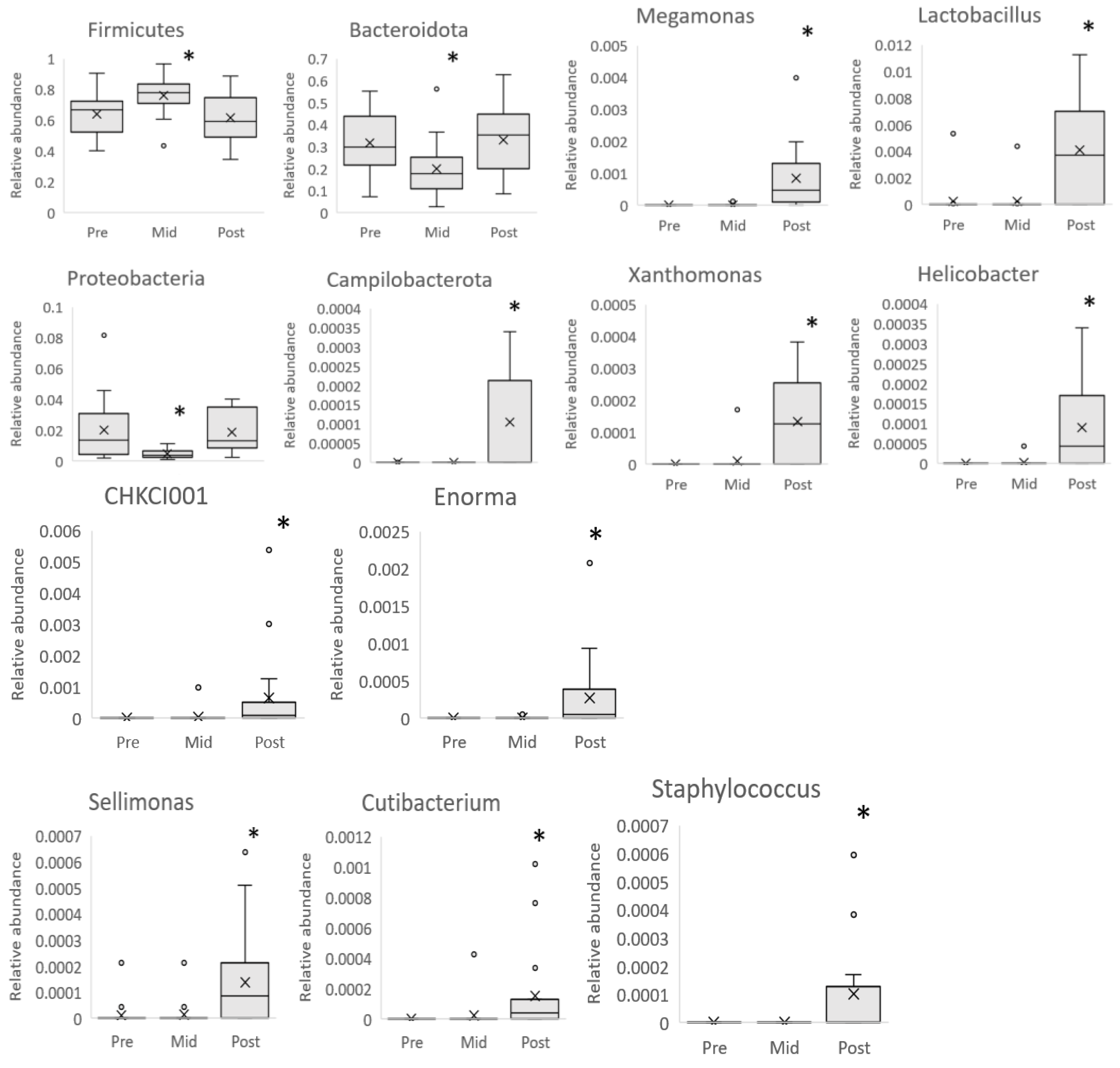
4.2. Microbiota Composition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GU | Glucose uptake |
| MZ | Monozygotic |
| BMI | Body mass index |
| PET | Positron emission tomography |
| ASV | Amplicon sequencing variant |
| IFG | Impaired fasting glucose |
| IGT | Impaired glucose tolerance |
| ROI | Region of interest |
| VOI | Volume of interest |
| MRI | Magnetic resonance imaging |
| VO2peak | Peak exercise capacity |
| M-value | Whole-body insulin-stimulated glucose uptake |
| PCoA | Principal Coordinate Analysis |
| FDR | False discovery rate |
References
- Gribble, F.M.; Reimann, F. Function and Mechanisms of Enteroendocrine Cells and Gut Hormones in Metabolism. Nat. Rev. Endocrinol. 2019, 15, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Holst, J.J.; Vilsbøll, T.; Deacon, C.F. The Incretin System and Its Role in Type 2 Diabetes Mellitus. Mol. Cell Endocrinol. 2009, 297, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, J.; Hannukainen, J.C.; Karmi, A.; Immonen, H.M.; Soinio, M.; Nelimarkka, L.; Savisto, N.; Helmiö, M.; Ovaska, J.; Salminen, P.; et al. Obesity-Associated Intestinal Insulin Resistance Is Ameliorated after Bariatric Surgery. Diabetologia 2015, 58, 1055–1062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Chow, J.; Lee, S.M.; Shen, Y.; Khosravi, A.; Mazmanian, S.K. Host-Bacterial Symbiosis in Health and Disease. Adv. Immunol. 2010, 107, 243–274. [Google Scholar] [CrossRef]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Cronin, O.; O’Sullivan, O.; Barton, W.; Cotter, P.D.; Molloy, M.G.; Shanahan, F. Gut Microbiota: Implications for Sports and Exercise Medicine. Br. J. Sports Med. 2017, 51, 700–701. [Google Scholar] [CrossRef]
- Lee, C.J.; Sears, C.L.; Maruthur, N. Gut Microbiome and Its Role in Obesity and Insulin Resistance. Ann. N. Y. Acad. Sci. 2020, 1461, 37–52. [Google Scholar] [CrossRef]
- Motiani, K.K.; Savolainen, A.M.; Eskelinen, J.-J.; Toivanen, J.; Ishizu, T.; Yli-Karjanmaa, M.; Virtanen, K.A.; Parkkola, R.; Kapanen, J.; Grönroos, T.J.; et al. Two Weeks of Moderate-Intensity Continuous Training, but Not High-Intensity Interval Training, Increases Insulin-Stimulated Intestinal Glucose Uptake. J. Appl. Physiol. 2017, 122, 1188–1197. [Google Scholar] [CrossRef]
- Motiani, K.K.; Collado, M.C.; Eskelinen, J.-J.; Virtanen, K.A.; Löyttyniemi, E.; Salminen, S.; Nuutila, P.; Kalliokoski, K.K.; Hannukainen, J.C. Exercise Training Modulates Gut Microbiota Profile and Improves Endotoxemia. Med. Sci. Sports Exerc. 2020, 52, 94–104. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and Associated Dietary Extremes Impact on Gut Microbial Diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The Microbiome of Professional Athletes Differs from That of More Sedentary Subjects in Composition and Particularly at the Functional Metabolic Level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Jang, L.-G.; Choi, G.; Kim, S.-W.; Kim, B.-Y.; Lee, S.; Park, H. The Combination of Sport and Sport-Specific Diet Is Associated with Characteristics of Gut Microbiota: An Observational Study. J. Int. Soc. Sports Nutr. 2019, 16, 21. [Google Scholar] [CrossRef]
- Estaki, M.; Pither, J.; Baumeister, P.; Little, J.P.; Gill, S.K.; Ghosh, S.; Ahmadi-Vand, Z.; Marsden, K.R.; Gibson, D.L. Cardiorespiratory Fitness as a Predictor of Intestinal Microbial Diversity and Distinct Metagenomic Functions. Microbiome 2016, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Shahar, R.T.; Koren, O.; Matarasso, S.; Shochat, T.; Magzal, F.; Agmon, M. Attributes of Physical Activity and Gut Microbiome in Adults: A Systematic Review. Int. J. Sports Med. 2020, 41, 801–814. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, Y.; Wiklund, P.; Tan, X.; Wu, N.; Zhang, X.; Tikkanen, O.; Zhang, C.; Munukka, E.; Cheng, S. The Association between Cardiorespiratory Fitness and Gut Microbiota Composition in Premenopausal Women. Nutrients 2017, 9, 792. [Google Scholar] [CrossRef]
- Allen, J.M.; Mailing, L.J.; Niemiro, G.M.; Moore, R.; Cook, M.D.; White, B.A.; Holscher, H.D.; Woods, J.A. Exercise Alters Gut Microbiota Composition and Function in Lean and Obese Humans. Med. Sci. Sports Exerc. 2018, 50, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Szurkowska, J.; Wiącek, J.; Laparidis, K.; Karolkiewicz, J. A Comparative Study of Selected Gut Bacteria Abundance and Fecal pH in Bodybuilders Eating High-Protein Diet and More Sedentary Controls. Nutrients 2021, 13, 4093. [Google Scholar] [CrossRef]
- Naukkarinen, J.; Rissanen, A.; Kaprio, J.; Pietiläinen, K.H. Causes and Consequences of Obesity: The Contribution of Recent Twin Studies. Int. J. Obes. 2012, 36, 1017–1024. [Google Scholar] [CrossRef]
- Solomon, T.P.J. Sources of Inter-Individual Variability in the Therapeutic Response of Blood Glucose Control to Exercise in Type 2 Diabetes: Going Beyond Exercise Dose. Front. Physiol. 2018, 9, 896. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of Mammals and Their Gut Microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Kevans, D.; Smith, M.I.; Guttman, D.S.; Griffiths, A.; Panaccione, R.; Otley, A.; et al. Association of Host Genome with Intestinal Microbial Composition in a Large Healthy Cohort. Nat. Genet. 2016, 48, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The Effect of Host Genetics on the Gut Microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human Genetic Variation and the Gut Microbiome in Disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Akkermans, A.D.L.; Vliet, W.M.A.-V.; Arjan, G.M.; de Visser, J.; de Vos, W.M. The Host Genotype Affects the Bacterial Community in the Human Gastronintestinal Tract. Microb. Ecol. Health Dis. 2001, 13, 129–134. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A Core Gut Microbiome in Obese and Lean Twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef]
- Vilchez-Vargas, R.; Skieceviciene, J.; Lehr, K.; Varkalaite, G.; Thon, C.; Urba, M.; Morkūnas, E.; Kucinskas, L.; Bauraite, K.; Schanze, D.; et al. Gut Microbial Similarity in Twins Is Driven by Shared Environment and Aging. EBioMedicine 2022, 79, 104011. [Google Scholar] [CrossRef]
- Heiskanen, M.A.; Honkala, S.M.; Hentilä, J.; Ojala, R.; Lautamäki, R.; Koskensalo, K.; Lietzén, M.S.; Saunavaara, V.; Saunavaara, J.; Helmiö, M.; et al. Systemic Cross-Talk between Brain, Gut, and Peripheral Tissues in Glucose Homeostasis: Effects of Exercise Training (CROSSYS). Exercise Training Intervention in Monozygotic Twins Discordant for Body Weight. BMC Sports Sci. Med. Rehabil. 2021, 13, 16. [Google Scholar] [CrossRef]
- Ojala, R.; Hentilä, J.; Lietzén, M.S.; Arponen, M.; Heiskanen, M.A.; Honkala, S.M.; Virtanen, H.; Koskensalo, K.; Lautamäki, R.; Löyttyniemi, E.; et al. Bone Marrow Metabolism Is Affected by Body Weight and Response to Exercise Training Varies According to Anatomical Location. Diabetes Obes. Metab. 2024, 26, 251–261. [Google Scholar] [CrossRef]
- Heinonen, S.; Saarinen, L.; Naukkarinen, J.; Rodríguez, A.; Frühbeck, G.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Vuolteenaho, K.; Moilanen, E.; et al. Adipocyte Morphology and Implications for Metabolic Derangements in Acquired Obesity. Int. J. Obes. 2014, 38, 1423–1431. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association 2. Classification and Diagnosis of Diabetes. Diabetes Care 2014, 38, S8–S16. [Google Scholar] [CrossRef]
- Voipio-Pulkki, L.-M.; Nuutila, P.; Knuuti, M.J.; Ruotsalainen, U.; Haaparanta, M.; Teräs, M.; Wegelius, U.; Koivisto, V.A. Heart and Skeletal Muscle Glucose Disposal in Type 2 Diabetic Patients as Determined by Positron Emission Tomography. J. Nucl. Med. 1993, 34, 2064–2067. [Google Scholar]
- DeFronzo, R.A.; Tobin, J.D.; Andres, R. Glucose Clamp Technique: A Method for Quantifying Insulin Secretion and Resistance. Am. J. Physiol. 1979, 237, E214–E223. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. MicrobiomeAnalyst 2.0: Comprehensive Statistical, Functional and Integrative Analysis of Microbiome Data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef] [PubMed]
- Franquet, E.; Watts, G.; Kolodny, G.M.; Goldfine, A.B.; Patti, M.-E. PET-CT Reveals Increased Intestinal Glucose Uptake after Gastric Surgery. Surg. Obes. Relat. Dis. 2019, 15, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Ait-Omar, A.; Monteiro-Sepulveda, M.; Poitou, C.; Le Gall, M.; Cotillard, A.; Gilet, J.; Garbin, K.; Houllier, A.; Château, D.; Lacombe, A.; et al. GLUT2 Accumulation in Enterocyte Apical and Intracellular Membranes. Diabetes 2011, 60, 2598–2607. [Google Scholar] [CrossRef]
- Merigo, F.; Brandolese, A.; Facchin, S.; Missaggia, S.; Bernardi, P.; Boschi, F.; D’Incà, R.; Savarino, E.V.; Sbarbati, A.; Sturniolo, G.C. Glucose Transporter Expression in the Human Colon. World J. Gastroenterol. 2018, 24, 775–793. [Google Scholar] [CrossRef] [PubMed]
- Honka, H.; Mäkinen, J.; Hannukainen, J.C.; Tarkia, M.; Oikonen, V.; Teräs, M.; Fagerholm, V.; Ishizu, T.; Saraste, A.; Stark, C.; et al. Validation of [18F]Fluorodeoxyglucose and Positron Emission Tomography (PET) for the Measurement of Intestinal Metabolism in Pigs, and Evidence of Intestinal Insulin Resistance in Patients with Morbid Obesity. Diabetologia 2013, 56, 893–900. [Google Scholar] [CrossRef]
- Tobin, V.; Le Gall, M.; Fioramonti, X.; Stolarczyk, E.; Blazquez, A.G.; Klein, C.; Prigent, M.; Serradas, P.; Cuif, M.-H.; Magnan, C.; et al. Insulin Internalizes GLUT2 in the Enterocytes of Healthy but Not Insulin-Resistant Mice. Diabetes 2008, 57, 555–562. [Google Scholar] [CrossRef]
- Gontier, E.; Fourme, E.; Wartski, M.; Blondet, C.; Bonardel, G.; Le Stanc, E.; Mantzarides, M.; Foehrenbach, H.; Pecking, A.-P.; Alberini, J.-L. High and Typical 18F-FDG Bowel Uptake in Patients Treated with Metformin. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Klip, A.; Bock, K.D.; Bilan, P.J.; Richter, E.A. Transcellular Barriers to Glucose Delivery in the Body. Annu. Rev. Physiol. 2024, 86, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Kim, H.-N.; Chang, Y.; Yun, Y.; Ryu, S.; Shin, H.; Kim, H.-L. Gut Microbiota and Physiologic Bowel 18F-FDG Uptake. EJNMMI Res. 2017, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-S.; Lkhagva, E.; Jung, S.; Kim, H.-J.; Chung, H.-J.; Hong, S.-T. Fecal Microbiome Does Not Represent Whole Gut Microbiome. Cell. Microbiol. 2023, 2023, 6868417. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human Gut Microbes Associated with Obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children from Europe and Rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, H.; Ye, L.; Huang, L.; Du, J.; Liang, X.; Zhang, X.; Chen, J.; Jiang, Y.; Chen, L. Combined Analysis of the Microbiome and Metabolome to Reveal the Characteristics of Saliva from Different Diets: A Comparison among Vegans, Seafood-Based Omnivores, and Red Meat (Beef and Lamb) Omnivores. Front. Microbiol. 2024, 15, 1419686. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Hoffmann, C.; Sherrill-Mix, S.A.; Keilbaugh, S.A.; Hamady, M.; Chen, Y.-Y.; Knight, R.; Ahima, R.S.; Bushman, F.; Wu, G.D. High-Fat Diet Determines the Composition of the Murine Gut Microbiome Independently of Obesity. Gastroenterology 2009, 137, 1716–1724.e1-2. [Google Scholar] [CrossRef]
- Krajmalnik-Brown, R.; Ilhan, Z.-E.; Kang, D.-W.; DiBaise, J.K. Effects of Gut Microbes on Nutrient Absorption and Energy Regulation. Nutr. Clin. Pract. 2012, 27, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Hu, Y.; Bruner, D.W. Composition of Gut Microbiota and Its Association with Body Mass Index and Lifestyle Factors in a Cohort of 7–18 Years Old Children from the American Gut Project. Pediatr. Obes. 2019, 14, e12480. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, R.; Nistal, E.; Estébanez, B.; Porras, D.; Juárez-Fernández, M.; Martínez-Flórez, S.; García-Mediavilla, M.V.; de Paz, J.A.; González-Gallego, J.; Sánchez-Campos, S.; et al. Exercise Training Modulates the Gut Microbiota Profile and Impairs Inflammatory Signaling Pathways in Obese Children. Exp. Mol. Med. 2020, 52, 1048–1061. [Google Scholar] [CrossRef]
- Munukka, E.; Ahtiainen, J.P.; Puigbó, P.; Jalkanen, S.; Pahkala, K.; Keskitalo, A.; Kujala, U.M.; Pietilä, S.; Hollmén, M.; Elo, L.; et al. Six-Week Endurance Exercise Alters Gut Metagenome That Is Not Reflected in Systemic Metabolism in Over-Weight Women. Front. Microbiol. 2018, 9, 2323. [Google Scholar] [CrossRef] [PubMed]
- Mazzarelli, A.; Giancola, M.L.; Fontana, A.; Piselli, P.; Binda, E.; Trivieri, N.; Mencarelli, G.; Marchioni, L.; Vulcano, A.; De Giuli, C.; et al. Gut Microbiota Composition in COVID-19 Hospitalized Patients with Mild or Severe Symptoms. Front. Microbiol. 2022, 13, 1049215. [Google Scholar] [CrossRef]
- Jensen, E.A.; Young, J.A.; Jackson, Z.; Busken, J.; List, E.O.; Carroll, R.K.; Kopchick, J.J.; Murphy, E.R.; Berryman, D.E. Growth Hormone Deficiency and Excess Alter the Gut Microbiome in Adult Male Mice. Endocrinology 2020, 161, bqaa026. [Google Scholar] [CrossRef]
- Bizjak, D.A.; Tomschi, F.; Bales, G.; Nader, E.; Romana, M.; Connes, P.; Bloch, W.; Grau, M. Does Endurance Training Improve Red Blood Cell Aging and Hemorheology in Moderate-Trained Healthy Individuals? J. Sport. Health Sci. 2020, 9, 595–603. [Google Scholar] [CrossRef]
- Oliver, A.; Chase, A.B.; Weihe, C.; Orchanian, S.B.; Riedel, S.F.; Hendrickson, C.L.; Lay, M.; Sewall, J.M.; Martiny, J.B.H.; Whiteson, K. High-Fiber, Whole-Food Dietary Intervention Alters the Human Gut Microbiome but Not Fecal Short-Chain Fatty Acids. mSystems 2021, 6, e00115-21. [Google Scholar] [CrossRef]
- Xu, Y.; Zhong, F.; Zheng, X.; Lai, H.-Y.; Wu, C.; Huang, C. Disparity of Gut Microbiota Composition Among Elite Athletes and Young Adults With Different Physical Activity Independent of Dietary Status: A Matching Study. Front. Nutr. 2022, 9, 843076. [Google Scholar] [CrossRef]
- Mahdieh, M.S.; Maryam, J.; Bita, B.; Neda, F.; Motahare, M.; Mahboobeh, B.; LeBris S, Q.; Kalani Behrooz, S. A Pilot Study on the Relationship between Lactobacillus, Bifidibactrium Counts and Inflammatory Factors Following Exercise Training. Arch. Physiol. Biochem. 2023, 129, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; Bishop, N.C.; Oliveira, M.; Tauler, P. Daily Probiotic’s (Lactobacillus Casei Shirota) Reduction of Infection Incidence in Athletes. Int. J. Sport. Nutr. Exerc. Metab. 2011, 21, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-C.; Lee, M.-C.; Lee, C.-C.; Ng, K.-S.; Hsu, Y.-J.; Tsai, T.-Y.; Young, S.-L.; Lin, J.-S.; Huang, C.-C. Effect of Lactobacillus Plantarum TWK10 on Exercise Physiological Adaptation, Performance, and Body Composition in Healthy Humans. Nutrients 2019, 11, 2836. [Google Scholar] [CrossRef]
- Hołubiuk, Ł.; Imiela, J. Diet and Helicobacter Pylori Infection. Prz. Gastroenterol. 2016, 11, 150–154. [Google Scholar] [CrossRef]
- Lai, Z.-L.; Tseng, C.-H.; Ho, H.J.; Cheung, C.K.Y.; Lin, J.-Y.; Chen, Y.-J.; Cheng, F.-C.; Hsu, Y.-C.; Lin, J.-T.; El-Omar, E.M.; et al. Fecal Microbiota Transplantation Confers Beneficial Metabolic Effects of Diet and Exercise on Diet-Induced Obese Mice. Sci. Rep. 2018, 8, 15625. [Google Scholar] [CrossRef]
- Muñoz, M.; Guerrero-Araya, E.; Cortés-Tapia, C.; Plaza-Garrido, A.; Lawley, T.D.; Paredes-Sabja, D. Comprehensive Genome Analyses of Sellimonas Intestinalis, a Potential Biomarker of Homeostasis Gut Recovery. Microb. Genom. 2020, 6, mgen000476. [Google Scholar] [CrossRef]
- Lun, H.; Yang, W.; Zhao, S.; Jiang, M.; Xu, M.; Liu, F.; Wang, Y. Altered Gut Microbiota and Microbial Biomarkers Associated with Chronic Kidney Disease. Microbiologyopen 2018, 8, e00678. [Google Scholar] [CrossRef] [PubMed]
- Caudet, J.; Trelis, M.; Cifre, S.; Soriano, J.M.; Rico, H.; Merino-Torres, J.F. Interplay between Intestinal Bacterial Communities and Unicellular Parasites in a Morbidly Obese Population: A Neglected Trinomial. Nutrients 2022, 14, 3211. [Google Scholar] [CrossRef]
- Huang, W.-C.; Tung, C.-L.; Yang, Y.-C.S.H.; Lin, I.-H.; Ng, X.E.; Tung, Y.-T. Endurance Exercise Ameliorates Western Diet–Induced Atherosclerosis through Modulation of Microbiota and Its Metabolites. Sci. Rep. 2022, 12, 3612. [Google Scholar] [CrossRef]
- Karl, J.P.; Margolis, L.M.; Madslien, E.H.; Murphy, N.E.; Castellani, J.W.; Gundersen, Y.; Hoke, A.V.; Levangie, M.W.; Kumar, R.; Chakraborty, N.; et al. Changes in Intestinal Microbiota Composition and Metabolism Coincide with Increased Intestinal Permeability in Young Adults under Prolonged Physiological Stress. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G559–G571. [Google Scholar] [CrossRef]
- Corvec, S. Clinical and Biological Features of Cutibacterium (Formerly Propionibacterium) Avidum, an Underrecognized Microorganism. Clin. Microbiol. Rev. 2018, 31, e00064-17. [Google Scholar] [CrossRef] [PubMed]

| Heavier | Leaner | p-Value | |||||
|---|---|---|---|---|---|---|---|
| Pre | Post | Pre | Post | Baseline | Time | Time × Group | |
| n | 12 | 11 | 12 | 10 | |||
| Male/female | 4/8 | 4/7 | 4/8 | 4/6 | |||
| Age, years | 40.4 (37.5; 43.4) | 40.4 (37.5; 43.4) | |||||
| Weight, kg | 108.7 (91.8; 125.7) | 108.0 (93.1; 122.9) | 86.4 (72.4; 100.3) | 86.9 (72.6; 101.2) | 0.001 * | 0.95 | 0.37 |
| BMI, kg/m2 | 36.7 (32.2; 41.1) | 36.4 (32.4; 40.4) | 29.1 (25.1; 33.1) | 29.3 (25.3; 33.2) | 0.001 * | 0.92 | 0.41 |
| Waist circumference, cm | 117.7 (106.3; 129.2) | 115.0 (106.8; 123.1) | 96.5 (84.7; 108.3) | 94.4 (82.3; 106.6) | 0.001 * | 0.17 | 0.74 |
| Fat free mass, kg | 35.9 (31.0; 40.7) | 36.2 (32.1; 40.3) | 33.1 (29.0; 37.2) | 33.9 (30.6; 37.2) | 0.003 *† | 0.14 | 0.10 |
| Fat mass, kg | 45.5 (33.8; 57.3) | 44.5 (34.6; 54.4) | 27.8 (17.9; 37.7) | 26.9 (14.4; 39.4) | 0.001 *† | 0.70 † | 0.97 † |
| Visceral fat mass, kg | 5.9 (4.5; 7.3) | 5.5 (4.4; 6.5) | 3.1 (2.0; 4.3) | 3.2 (2.0; 4.4) | 0.002 *† | 0.07 | 0.29 |
| Fat percentage, % | 40.6 (35.5; 45.7) | 40.0 (35.9; 44.1) | 30.4 (24.0; 36.9) | 29.5 (20.3; 38.7) | 0.001 * | 0.37 | 0.72 |
| VO2peak, mL·kg−1·min−1 | 25.6 (22.7; 28.5) | 28.3 (26.1; 30.6) | 32.4 (27.3; 37.4) | 35.1 (29.9; 40.2) | 0.003 * | 0.001 * | 0.94 |
| Triglycerides, mmol/L | 1.4 (0.9; 1.9) | 1.2 (0.9; 1.5) | 0.8 (0.6; 1.0) | 0.8 (0.6; 1.0) | 0.040 * | 0.54 † | 0.49 † |
| Ffa, mmol/L | 0.59 (0.51; 0.67) | 0.68 (0.45; 0.91) | 0.52 (0.34; 0.69) | 0.54 (0.14; 0.94) | 0.29 | 0.63 | 0.63 |
| CRP, mg/L | 2.8 (1.4; 4.2) | 3.7 (1.3; 6.2) | 2.1 (0.3; 3.9) | 1.2 (−0.6; 3.1) | 0.050 * | 0.69 † | 0.50 † |
| M-value, μmol/kg × min | 23.1 (16.3; 30.0) | 31.4 (20.4; 42.3) | 37.6 (26.7; 48.5) | 46.9 (31.7; 62.1) | 0.007 * | 0.022 * | 0.82 |
| Heavier | Leaner | p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pre | Mid | Post | Pre | Mid | Post | Baseline | Time | Time × Group | ||
| n | 10 | 9 | 10 | 10 | 9 | 9 | ||||
| Male/female | 4/8 | 4/7 | 4/7 | 4/8 | 4/6 | 4/6 | ||||
| Pielou Evenness | 0.73 (0.70; 0.76) | 0.73 (0.70; 0.76) | 0.69 (0.65; 0.73) | 0.75 (0.72; 0.78) | 0.76 (0.74; 0.78) | 0.72 (0.69; 0.75) | 0.33 | 0.019 * | 0.78 | |
| Chao 1 | 353.8 (312.3; 395.4) | 350.0 (311.3; 388.6) | 357.0 (301.7; 412.3) | 339.1 (277.8; 400.4) | 394.3 (358.3; 430.4) | 414.3 (361.7; 466.9) | 0.60 | 0.40 | 0.36 | |
| Dominance | 0.04 (0.03; 0.06) | 0.04 (0.03; 0.05) | 0.05 (0.03; 0.08) | 0.03 (0.02; 0.04) | 0.03 (0.02; 0.03) | 0.04 (0.02; 0.05) | 0.25 | 0.14 | 1.00 | |
| Observed otus | 347.9 (307.4; 388.4) | 343.4 (306.4; 380.4) | 345.0 (290.9; 399.1) | 334.5 (273.2; 395.8) | 387.1 (352.0; 422.3) | 401.6 (349.3; 453.8) | 0.63 | 0.53 | 0.37 | |
| Shannon | 6.1 (5.8; 6.4) | 6.1 (5.8; 6.4) | 5.8 (5.3; 6.2) | 6.2 (5.8; 6.7) | 6.5 (6.3; 6.8) | 6.2 (5.9; 6.6) | 0.64 | 0.07 | 0.58 | |
| Simpson | 1.0 (0.9; 1.0) | 1.0 (1.0; 1.0) | 1.0 (0.9; 1.0) | 1.0 (1.0; 1.0) | 1.0 (1.0; 1.0) | 1.0 (1.0; 1.0) | 0.31 | 0.23 | 0.83 | |
| Energy, kcal | 2111.2 (1781.8; 2440.7) | 2180.6 (1814.5; 2546.6) | 2184.0 (1941.7; 2426.4) | 1960.6 (1627.1; 2294.1) | 2390.7 (1908.5; 2872.8) | 2111.9 (1774.7; 2449.1) | 0.29 † | 0.18 | 0.58 | |
| Fat, g | 95.1 (74.2; 116) | 92.1 (71.6; 112.6) | 88.1 (70.3; 106.0) | 87.7 (68.2; 107.2) | 97.4 (73.5; 121.3) | 93.8 (72.6; 115.1) | 0.32 | 0.82 | 0.39 | |
| Carbohydrate, g | 205.6 (161.8; 249.3) | 212.6 (155.8; 269.4) | 233.0 (193.8; 272.2) | 179.69 (146.5; 212.9) | 256.6 (196.8; 316.3) | 209.8 (164.7; 255.0) | 0.31 | 0.07 † | 0.28 † | |
| Pre->Mid p = 0.030 †* | ||||||||||
| Protein, g | 85.0 (71.48; 98.6) | 95.3 (74.2; 116.3) | 94.9 (80.7; 109.0) | 92.3 (75.3; 109.2) | 100.7 (79.7; 121.8) | 92.9 (77.4; 108.4) | 0.43 † | 0.38 † | 0.78 † | |
| Dietary fiber, g | 19.3 (16.0; 22.5) | 20.6 (15.4; 25.8) | 22.9 (19.0; 26.9) | 16.4 (12.4; 20.3) | 20.5 (13.9; 27.0) | 20.8 (15.7; 25.8) | 0.053 * | 0.054 † | 0.80 † | |
| Fatty acids, g | 87.9 (68.1; 107.6) | 86.1 (66.3; 105.9) | 82.5 (65.3; 99.8) | 80.9 (62.7; 99.2) | 89.7 (66.4; 112.9) | 86.7 (66.2; 107.3) | 0.43 † | 0.89 † | 0.28 † | |
| Saturated fatty acid, g | 34.7 (26.3; 43.0) | 35.3 (24.4; 46.1) | 32.9 (25.4; 46.1) | 33.9 (24.5; 43.4) | 36.7 (25.3; 48.1) | 35.2 (27.2; 43.3) | 0.70 † | 0.93 † | 0.59 † | |
| Cholesterol, g | 280.1 (198.5; 361.6) | 320.5 (144.6; 496.5) | 255.5 (151.4; 359.6) | 286.4 (165.4; 407.4) | 358.2 (223.5; 492.9) | 281 (205.0; 357.0) | 0.72 | 0.25 | 0.50 | |
| Sterol, g | 307.5 (248.3; 366.7) | 290.8 (233.1; 348.5) | 281.7 (225.9; 337.5) | 252.8 (202.3; 303.2) | 302.0 (235.5; 368.5) | 311.3 (245.5; 377.1) | 0.10 † | 0.69 † | 0.06 † | |
| Organic acids, g | 4.0 (2.7; 5.3) | 3.9 (2.6; 5.1) | 4.5 (3.6; 5.4) | 2.9 (2.0; 3.8) | 4.2 (3.1; 5.4) | 3.6 (2.7; 4.4) | 0.06 | 0.55 | 0.07 | |
| Sugar, g | 92.24 (67.6; 116.9) | 90.9 (63.4; 118.4) | 98.1 (70.4; 125.8) | 69.1 (49.5; 88.6) | 113.1 (84.7; 141.4) | 95.1 (66.5; 123.7) | 0.15 | 0.07 | 0.043 | |
| Pre->Mid p = 0.004 †* | ||||||||||
| Fructose, g | 10.7 (8.1; 13.4) | 10.7 (7.9; 13.5) | 14.2 (10.6; 17.8) | 10.2 (3.8; 16.6) | 12.1 (7.9; 16.3) | 14.2 (8.6; 19.8) | 0.34 † | 0.051 *† | 0.50 † | |
| Glucose, g | 12.4 (9.4; 15.5) | 11.5 (8.1; 14.9) | 15.4 (11.9; 18.9) | 12.9 (6.5; 19.3) | 16.4 (10.1; 22.7) | 16.9 (9.7; 24.0) | 0.89 | 0.024 *† | 0.41 † | |
| Sucrose, g | 53.8 (33.7; 74.0) | 48.4 (22.7; 74.1) | 49.6 (31.2; 67.9) | 34.1 (23.2; 45.0) | 65.4 (42.4; 88.3) | 50.1 (33.4; 66.8) | 0.13 | 0.16 † | 0.046 *† | |
| Pre->Mid p = 0.005 | Mid->Post p = 0.016 | |||||||||
| Soluble fiber, g | 7.1 (5.9; 8.4) | 8.9 (6.5; 11.2) | 8.8 (6.8; 10.7) | 6.7 (4.4; 9.0) | 7.5 (4.2; 10.8) | 8.1 (5.4; 10.9) | 0.06 † | 0.18 † | 0.95 † | |
| Insoluble fiber, g | 12.1 (9.6; 14.6) | 11.5 (8.1; 14.9) | 14.1 (7.7; 16.8) | 9.7 (7.8; 11.6) | 12.4 (8.0; 16.8) | 12.6 (9.8; 15.6) | 0.034 *† | 0.09 † | 0.48 † | |
| Soluble polysaccharide, g | 4.5 (3.6; 5.4) | 3.7 (2.7; 4.6) | 4.9 (3.9; 5.9) | 3.7 (2.5; 4.9) | 4.2 (3.2; 5.2) | 4.4 (3.2; 5.5) | 0.030 *† | 0.16 † | 0.16 † | |
| Salt, g | 7.5 (5.8; 9.3) | 7.9 (6.3; 9.5) | 8.7 (7.4; 10.1) | 7.0 (5.6; 8.3) | 9.9 (5.8; 14.0) | 8.1 (4.5; 11.8) | 0.62 † | 0.07 † | 0.78 † | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lietzén, M.S.; Guzzardi, M.A.; Ojala, R.; Hentilä, J.; Heiskanen, M.A.; Honkala, S.M.; Lautamäki, R.; Löyttyniemi, E.; Kirjavainen, A.K.; Rajander, J.; et al. Regular Exercise Training Induces More Changes on Intestinal Glucose Uptake from Blood and Microbiota Composition in Leaner Compared to Heavier Individuals in Monozygotic Twins Discordant for BMI. Nutrients 2024, 16, 3554. https://doi.org/10.3390/nu16203554
Lietzén MS, Guzzardi MA, Ojala R, Hentilä J, Heiskanen MA, Honkala SM, Lautamäki R, Löyttyniemi E, Kirjavainen AK, Rajander J, et al. Regular Exercise Training Induces More Changes on Intestinal Glucose Uptake from Blood and Microbiota Composition in Leaner Compared to Heavier Individuals in Monozygotic Twins Discordant for BMI. Nutrients. 2024; 16(20):3554. https://doi.org/10.3390/nu16203554
Chicago/Turabian StyleLietzén, Martin S., Maria Angela Guzzardi, Ronja Ojala, Jaakko Hentilä, Marja A. Heiskanen, Sanna M. Honkala, Riikka Lautamäki, Eliisa Löyttyniemi, Anna K. Kirjavainen, Johan Rajander, and et al. 2024. "Regular Exercise Training Induces More Changes on Intestinal Glucose Uptake from Blood and Microbiota Composition in Leaner Compared to Heavier Individuals in Monozygotic Twins Discordant for BMI" Nutrients 16, no. 20: 3554. https://doi.org/10.3390/nu16203554
APA StyleLietzén, M. S., Guzzardi, M. A., Ojala, R., Hentilä, J., Heiskanen, M. A., Honkala, S. M., Lautamäki, R., Löyttyniemi, E., Kirjavainen, A. K., Rajander, J., Malm, T., Lahti, L., Rinne, J. O., Pietiläinen, K. H., Iozzo, P., & Hannukainen, J. C. (2024). Regular Exercise Training Induces More Changes on Intestinal Glucose Uptake from Blood and Microbiota Composition in Leaner Compared to Heavier Individuals in Monozygotic Twins Discordant for BMI. Nutrients, 16(20), 3554. https://doi.org/10.3390/nu16203554






