Experimental Protocols Used to Mimic Gastrointestinal Protein Digestion: A Systematic Review
Abstract
1. Introduction
2. Material and Methods
2.1. Search Strategies
2.2. Eligibility Criteria
2.3. Data Extraction Process
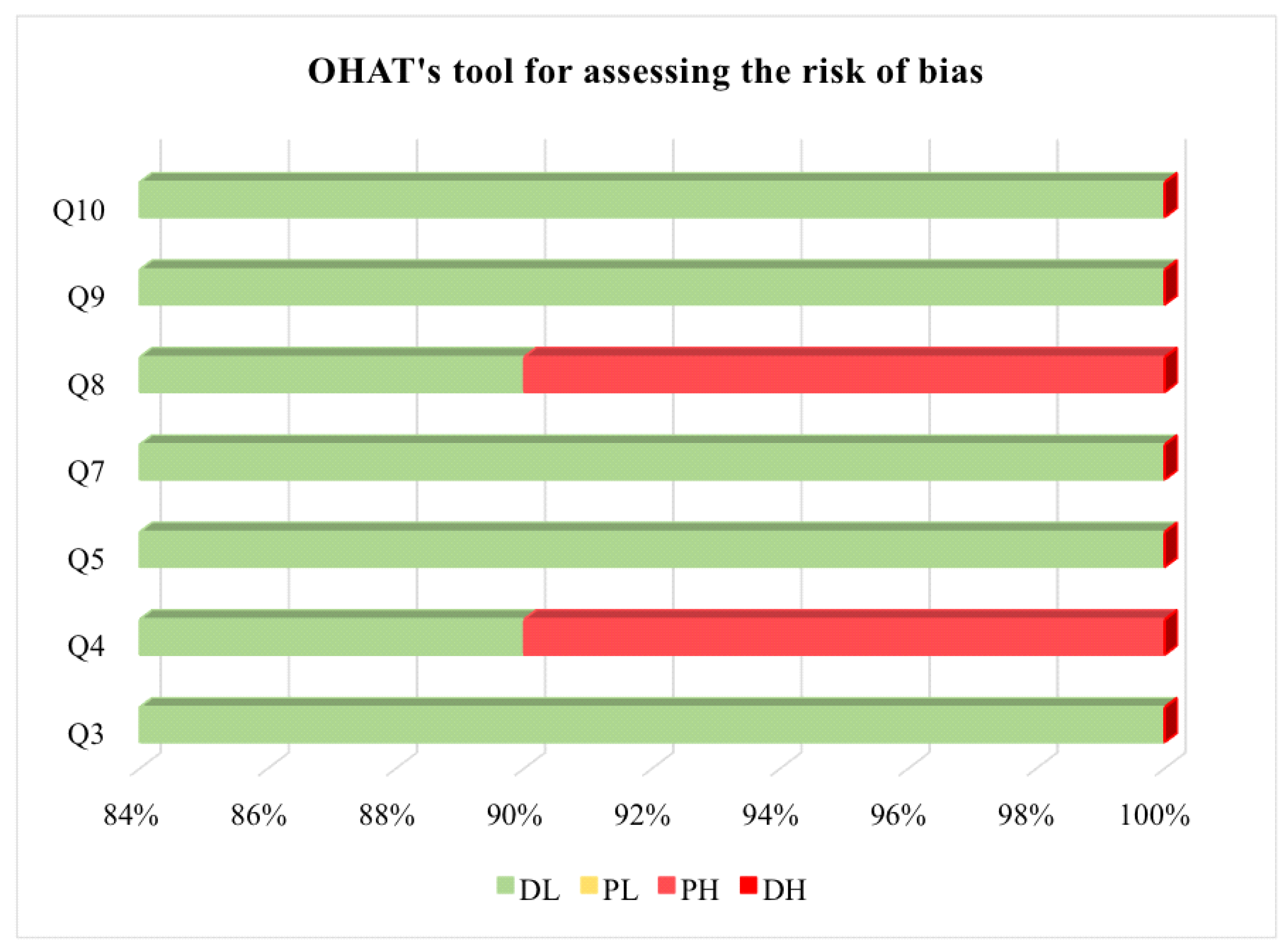
2.4. Bias Risk Assessment and Study Quality Assessment
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Santos-Hernández, M.; Alfieri, F.; Gallo, V.; Miralles, B.; Masi, P.; Romano, A.; Ferranti, P.; Recio, I. Compared digestibility of plant protein isolates by using the INFOGEST digestion protocol. Food Res. Int. 2020, 137, 109708. [Google Scholar] [CrossRef]
- Daroit, D.J.; Brandelli, A. In vivo bioactivities of food protein-derived peptides—A current review. Curr. Opin. Food Sci. 2021, 39, 120–129. [Google Scholar] [CrossRef]
- Rivero-Pino, F. Bioactive food-derived peptides for functional nutrition: Effect of forti cation, processing and storage on peptide stability and bioactivity within food matrices. Food Chem. 2022, 406, 1–10. [Google Scholar] [CrossRef]
- Boutrou, R.; Gaudichon, C.; Dupont, D.; Jardin, J.; Airinei, G.; Marsset-Baglieri, A.; Benamouzig, R.; Tomé, D.; Leonil, J. Sequential release of milk protein–derived bioactive peptides in the jejunum in healthy humans. Am. J. Clin. Nutr. 2013, 97, 1314–1323. [Google Scholar] [CrossRef]
- Sullivan, L.M.; Kehoe, J.J.; Barry, L.; Buckley, M.J.M.; Shanahan, F.; Mok, K.H.; Brodkorb, A. Gastric digestion of a-lactalbumin in adult human subjects using capsule endoscopy and nasogastric tube sampling. Br. J. Nutr. 2014, 112, 638–646. [Google Scholar] [CrossRef]
- Brodkorb, A.; Egger, L.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Orlien, V.; Aalaei, K.; Poojary, M.M.; Nielsen, D.S.; Ahrné, L.; Carrascal, J.R. Effect of processing on in vitro digestibility (IVPD) of food proteins. Crit. Rev. Food Sci. Nutr. 2021, 63, 2790–2839. [Google Scholar] [CrossRef]
- Sá, A.G.A.; Moreno, Y.M.F.; Carciofi, B.A.M. Food processing for the improvement of plant proteins digestibility. Crit. Rev. Food Sci. Nutr. 2019, 60, 3367–3386. [Google Scholar] [CrossRef]
- Akobeng, A.K. Principles of evidence based medicine. Arch. Dis. Child. 2005, 90, 837–840. [Google Scholar] [CrossRef]
- Sampaio, R.F.; Mancini, M.C. Estudos de revisão sistemática: Um guia para síntese criteriosa da evidência científica. Rev. Bras. Fisioter. 2007, 11, 83–89. [Google Scholar] [CrossRef]
- Moher, D.; Shamseer, L.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015, 4, 1–9. [Google Scholar] [CrossRef]
- Luz, A.B.S.; Costa, R.O.A.; Medeiros, G.C.B.S.; Piuvezam, G.; Passos, T.S.; Morais, A.H.A. What are the digestion and absorption models used to reproduce gastrointestinal protein processes? A protocol for systematic review. Medicine 2021, 100, 1–5. [Google Scholar]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Ismail, M.; Alsalahi, A.; Imam, M.U.; Ooi, D.J.; Khaza’ai, H.; Aljaberi, M.A.; Shamsudin, M.N.; Idrus, Z. Safety and Neuroprotective Efficacy of Palm Oil and Tocotrienol-Rich Fraction from Palm Oil: A Systematic Review. Nutrients 2020, 12, 521. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. OHAT Risk of Bias Rating Tool for Human and Animal Studies Organization of This Document Indirectness, Timing, and Other Factors Related to Risk of Bias. 2015; pp. 1–37. Available online: https://ntp.niehs.nih.gov/sites/default/files/ntp/ohat/pubs/riskofbiastool_508.pdf (accessed on 13 December 2022).
- Hooijmans, C.R.; Rovers, M.M.; Vries, R.B.M.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Br. Med. J. 2021, 372, 1–9. [Google Scholar]
- Bisinotto, M.S.; Silva, D.C.; Fino, L.C.; Simabuco, F.M.; Bezerra, R.M.N.; Antunes, A.E.C.; Pacheco, M.T.B. Bioaccessibility of cashew nut kernel our compounds released after simulated in vitro human gastrointestinal digestion. Food Res. Int. 2021, 139, 1–8. [Google Scholar]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Martini, S.; Conte, A.; Tagliazucchi, D. Comparative peptidomic profile and bioactivities of cooked beef, pork, T chicken and turkey meat after in vitro gastro-intestinal digestion. J. Proteomics. 2019, 208, 103500. [Google Scholar] [CrossRef]
- Luo, Q.; Li, X.; Zhang, Z.; Chen, A.; Li, S.; Shen, G.; Li, M.; Liu, X.; Yin, X.; Cheng, L.; et al. Extraction of Zanthoxylum seed protein and identi cation of its simulated digestion products. LWT-Food Sci. Technol. 2022, 161, 113412. [Google Scholar] [CrossRef]
- Abrahamse, E.; Thomassen, G.G.M.; Renes, I.B.; Wierenga, P.A.; Hettinga, K.A. Gastrointestinal Protein Hydrolysis Kinetics: Opportunities for Further Infant Formula Improvement. Nutrients 2022, 14, 1512. [Google Scholar] [CrossRef]
- Torcello-Gómez, A.; Dupont, D.; Jardin, J.; Briard-Bion, V.; Deglaire, A.; Risse, K.; Mechoulan, E.; Mackie, A. The pattern of peptides released from dairy and egg proteins is highly dependent on the simulated digestion scenario. Food Funct. 2020, 11, 5240–5256. [Google Scholar] [CrossRef]
- Ménard, O.; Bourlieu, C.; Oliveira, S.C.; Dellarosa, N.; Laghi, L.; Carrière, F.; Capozzi, F.; Dupont, D.; Deglaire, A. A first step towards a consensus static in vitro model for simulating full-term infant digestion. Food Chem. 2018, 240, 338–345. [Google Scholar] [CrossRef]
- Liu, Y.; Pischetsrieder, M. Identification and Relative Quantification of Bioactive Peptides Sequentially Released during Simulated Gastrointestinal Digestion of Commercial Kefir. J. Agric. Food Chem. 2017, 65, 1865–1873. [Google Scholar] [CrossRef]
- Versantvoort, C.H.; Oomen, A.G.; Van de Kamp, E.; Rompelberg, C.J.; Sips, A.J. Applicability of an in vitro digestion model in assessing the bioaccessibility of mycotoxins from food. Food Chem. Toxicol. 2005, 43, 31–40. [Google Scholar] [CrossRef]
- Sousa, R.; Portmann, R.; Dubois, S.; Recio, I.; Egger, L. Protein digestion of different protein sources using the INFOGEST static T digestion model. Food Res. Int. 2020, 130, 108996. [Google Scholar] [CrossRef]
- Torcello-Gómez, A.; Dupont, D.; Jardin, J.; Briard-Bion, V.; Deglaire, A.; Risse, K.; Mechoulan, E.; Mackie, A. Human gastrointestinal conditions affect in vitro digestibility of peanut and bread proteins. Food Funct. 2020, 11, 6921–6932. [Google Scholar] [CrossRef]
- Hall, A.E.; Moraru, C.I. Comparative effects of high pressure processing and heat treatment on in vitro digestibility of pea protein and starch. NPJ Sci. Food. 2022, 6, 2. [Google Scholar] [CrossRef]
- Minekus, M. The TNO Gastro-Intestinal Model (TIM). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Springer International Publishing: Cham, Switzerland, 2015; pp. 37–46. [Google Scholar]
- Denis, S.; Sayd, T.; Georges, A.; Chambon, C.; Chalancon, S.; Santé-Lhoutellier, V.; Blanquet-Diot, S. Digestion of cooked meat proteins is slightly affected by age as assessed using the dynamic gastrointestinal TIM model and mass spectrometry. Food Funct. 2016, 7, 2682–2691. [Google Scholar] [CrossRef]
- Ribnicky, D.M.; Roopchand, D.E.; Oren, A.; Grace, M.; Poulev, A.; Lila, M.A.; Havenaar, R.; Raskin, I. Effects of a high fat meal matrix and protein complexation on the bioaccessibility of blueberry anthocyanins using the TNO gastrointestinal model (TIM-1). Food Chem. 2014, 142, 349–357. [Google Scholar] [CrossRef]
- Sanchón, J.; Fernández-Tomé, S.; Miralles, B.; Hernández-Ledesma, B.; Tomé, D.; Gaudichon, C.; Recio, I. Protein degradation and peptide release from milk proteins in human jejunum. Comparison with in vitro gastrointestinal simulation. Food Chem. 2018, 239, 486–494. [Google Scholar] [CrossRef]
- Corrochano, A.R.; Sariçay, Y.; Arranz, E.; Kelly, P.M.; Buckin, V.; Giblin, L. Comparison of antioxidant activities of bovine whey proteins before and after simulated gastrointestinal digestion. J. Dairy Sci. 2019, 102, 54–67. [Google Scholar] [CrossRef]
- Wang, X.; Ye, A.; Lin, Q.; Han, J.; Singh, H. Gastric digestion of milk protein ingredients: Study using an in vitro dynamic model. J. Dairy Sci. 2018, 101, 6842–6852. [Google Scholar] [CrossRef]
- Kong, F.; Singh, R.P. A Model Stomach System to Investigate Disintegration Kinetics of Solid Foods during Gastric Digestion. Food Eng. Phys. Prop. 2008, 73, E202–E210. [Google Scholar]
- Kiewiet, M.B.G.; Dekkers, R.; Ulfman, L.H.; Groeneveld, A.; Vos, P.; Faas, M.M. Immunomodulating protein aggregates in soy and whey hydrolysates and their resistance to digestion in an in vitro infant gastrointestinal model: New insights in the mechanism of immunomodulatory hydrolysates. Food Funct. 2018, 9, 604–613. [Google Scholar] [CrossRef]
- Nguyen, T.T.P.; Bhandari, B.; Cichero, J.; Prakash, S. Gastrointestinal digestion of dairy and soy proteins in infant formulas: An in vitro study. Food Res. Int. 2015, 76, 348–358. [Google Scholar] [CrossRef]
- Crowley, D.; O’Callaghan, Y.; McCarthy, A.; Connolly, A.; Piggott, C.O.; FitzGerald, R.J.; O’Brien, N.M. Immunomodulatory potential of a brewers’ spent grain protein hydrolysate incorporated into low-fat milk following in vitro gastrointestinal digestion. Int. J. Food Sci. Nutr. 2015, 66, 672–676. [Google Scholar] [CrossRef]
- McCarthy, A.L.; O’Callaghan, Y.C.; Connolly, A.; Piggott, C.O.; FitzGerald, R.J.; O’Brien, N.M. A study of the ability of bioactive extracts from brewers’ spent grain to enhance the antioxidant and immunomodulatory potential of food formulations following in vitro digestion. Int. J. Food Sci. Nutr. 2015, 66, 230–235. [Google Scholar] [CrossRef]
- Rios-Villa, K.A.; Bhattacharya, M.; La, E.H.; Barile, D.; Bornhorst, G.M. Interactions between whey proteins and cranberry juice after thermal or non-thermal processing during in vitro gastrointestinal digestion. Food Funct. 2020, 11, 7661–7680. [Google Scholar] [CrossRef]
- Bornhorst, G.M.; Singh, R.P. Kinetics of in Vitro Bread Bolus Digestion with Varying Oral and Gastric Digestion Parameters. Food Biophys. 2013, 8, 50–59. [Google Scholar] [CrossRef]
- Roman, M.J.; Burri, B.J.; Singh, R.P. Release and bioaccessibility of beta-carotene from fortified almond butter during in vitro digestion. J. Agric. Food Chem. 2012, 60, 9659–9666. [Google Scholar] [CrossRef] [PubMed]
- Vilcacundo, R.; Miralles, B.; Carrillo, W.; Hernández-Ledesma, B. In vitro chemopreventive properties of peptides released from quinoa (Chenopodium quinoa Willd.) protein under simulated gastrointestinal digestion. Food Res. Int. 2018, 105, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Picone, G.; Noni, I.; Ferranti, P.; Nicolai, M.A.; Alamprese, C.; Trimigno, A.; Brodkorb, A.; Portmann, R.; Pihianto, A.; El, S.N.; et al. Monitoring molecular composition and digestibility of ripened bresaola through a combined foodomics approach. Food Res. Int. 2019, 115, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Salelles, L.; Floury, J.; Le Feunteun, S. Pepsin activity as a function of pH and digestion time on caseins and egg white proteins under static in vitro conditions. Food Funct. 2021, 12, 12468–12478. [Google Scholar] [CrossRef] [PubMed]
- Jamnik, P.; Istenič, K.; Koštomaj, T.; Wulff, T.; Geirsdóttir, M.; Almgren, A.; Jónsdóttir, R.; Kristinsson, H.G.; Undeland, I. Bioactivity of Cod and Chicken Protein Hydrolysates before and after in vitro Gastrointestinal Digestion. Food Technol. Biotechnol. 2017, 55, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Tibäck, E.A.; Svelander, C.A.; Colle, I.J.P.; Altskär, A.I.; Alminger, M.A.G.; Hendrickx, M.E.G.; Ahrné, L.M.; Langton, M.I.B.C. Mechanical and thermal pre-treatments of crushed tomatoes: Effects on consistency and in vitro accessibility of lycopene. J. Food Sci. 2009, 74, E386–E395. [Google Scholar] [CrossRef] [PubMed]
- Vilcacundo, R.; Martínez-Villaluenga, C.; Miralles, B.; Hernández-Ledesma, B. Release of multifunctional peptides from kiwicha (Amaranthus caudatus) protein under in vitro gastrointestinal digestion. J. Sci. Food Agric. 2019, 99, 1225–1232. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef]
- Peredo-Lovillo, A.; Hernández-Mendoza, A.; Vallejo-Cordoba, B.; Romero-Luna, H.E. Conventional and in silico approaches to select promising food-derived bioactive peptides: A review. Food Chem. 2022, 13, 100183. [Google Scholar] [CrossRef]
- Heda, R.; Toro, F.; Tombazzi, C.R. Physiology, Pepsin. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023; pp. 1–7. [Google Scholar]
- Duijsens, D.; Pälchen, K.; Guevara-Zambrano, J.M.; Verkempinck, S.H.E.; Infantes-Garcia, M.R.; Hendrickx, M.E.; Van Loey, A.; Grauwet, T. Strategic choices for in vitro food digestion methodologies enabling food digestion design. Trends Food Sci. Technol. 2022, 126, 61–72. [Google Scholar] [CrossRef]
- Verhoeckx, K.; Bøgh, K.L.; Dupont, D.; Egger, L.; Gadermaier, G.; Larré, C.; Mackie, A.; Menard, O.; Adel-Patient, K.; Picariello, G.; et al. The relevance of a digestibility evaluation in the allergenicity risk assessment of novel proteins. Opinion of a joint initiative of COST action ImpARAS and COST action INFOGEST. Food Chem. Toxicol. 2019, 129, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.A.; Mariutti, L.R. Static and semi-dynamic in vitro digestion methods: State of the art and recent achievements towards standardization. Curr. Opin. Food Sci. 2021, 41, 260–273. [Google Scholar] [CrossRef]
- Egger, L.; Schlegel, P.; Baumann, C.; Stoffers, H.; Guggisberg, D.; Brügger, C.; Dürr, D.; Stoll, P.; Vergères, G.; Portmann, R. Physiological comparability of the harmonized INFOGEST in vitro digestion method to in vivo pig digestion. Food Res. Int. 2017, 102, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, A.; Kleiner, M. Dietary protein and the intestinal microbiota: An understudied relationship. iScience 2022, 25, 105313. [Google Scholar] [CrossRef] [PubMed]
- Shani-Levi, C.; Alvito, P.; Andrés, A.; Assunção, R.; Barberá, R.; Blanquet-Diot, S.; Bourlieu, C.; Brodkorb, A.; Cilla, A.; Deglaire, A.; et al. Extending in vitro digestion models to specific human populations: Perspectives, practical tools and bio-relevant information. Trends Food Sci. Technol. 2017, 60, 52–63. [Google Scholar] [CrossRef]
- Chen, J. Food oral processing—A review. Food Hydrocoll. 2009, 23, 1–25. [Google Scholar] [CrossRef]
- Guo, Q.; Ye, A.; Lad, M.; Ferrua, M.; Dalgleish, D.; Singh, H. Disintegration kinetics of food gels during gastric digestion and its role on gastric emptying: An in vitro analysis. Food Funct. 2015, 6, 756–764. [Google Scholar] [CrossRef]
- Smeets, P.A.M.; Deng, R.; Eijnatten, E.J.M.; Mayar, M. Monitoring food digestion with magnetic resonance techniques. Proc. Nutr. Soc. 2021, 80, 148–158. [Google Scholar] [CrossRef]
- Barbé, F.; Ménard, O.; Le Gouar, Y.; Buffière, C.; Famelart, M.H.; Laroche, B.; Le Fenteun, S.; Dupont, D.; Rémond, D. The heat treatment and the gelation are strong determinants of the kinetics of milk proteins digestion and of the peripheral availability of amino acids. Food Chem. 2013, 136, 1203–1212. [Google Scholar] [CrossRef]
- Suárez, S.E.; Rabesona, H.; Ménard, O.; Jardin, J.; Anton, M.; Añón, M.C. Dynamic digestion of a high protein beverage based on amaranth: Structural changes and antihypertensive activity. Food Res Int. 2024, 187, 114416. [Google Scholar] [CrossRef] [PubMed]
- Passari, L.M.Z.G.; Soares, P.K.; Bruns, R.E. Estatística aplicada à química: Dez dúvidas comuns. Quim. Nova 2011, 34, 888–892. [Google Scholar] [CrossRef]
- Vitha, M.F.; Carr, P.W.; Mabbott, G.A. Appropriate Use of Blanks, Standards, and Controls W in Chemical Measurements. J. Chem. Educ. 2005, 82, 901–902. [Google Scholar] [CrossRef]
- Bohn, T.; Carriere, F.; Day, L.; Deglaire, A.; Egger, L.; Freitas, D.; Golding, M.; Le Feunteun, S.; Macierzanka, A.; Menard, O.; et al. Correlation between in vitro and in vivo data on food digestion. What can we predict with static in vitro digestion models? Crit. Rev. Food Sci. Nutr. 2018, 58, 2239–2261. [Google Scholar] [CrossRef] [PubMed]
- Hur, S.J.; Lim, B.O.; Decker, E.A.; McClements, D.J. In vitro human digestion models for food applications. Food Chem. 2011, 125, 1–12. [Google Scholar] [CrossRef]
- Egger, L.; Ménard, O.; Baumann, C.; Duerr, D.; Schlegel, P.; Stoll, P.; Vergères, G.; Dupont, D.; Portmann, R. Digestion of milk proteins: Comparing static and dynamic in vitro digestion systems with in vivo data. Food Res. Int. 2019, 118, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Egger, L.; Ménard, O.; Delgado-Andrade, C.; Alvito, P.; Assunção, R.; Balance, S.; Barberá, R.; Brodkorb, A.; Cattenoz, T.; Clemente, A.; et al. The harmonized INFOGEST in vitro digestion method: From knowledge to action. Food Res. Int. 2016, 88, 217–225. [Google Scholar] [CrossRef]
- Li, C.; Yu, W.; Wu, P.; Chen, X.D. Current in vitro digestion systems for understanding food digestion in human upper gastrointestinal tract. Trends Food Sci. Technol. 2020, 96, 114–126. [Google Scholar] [CrossRef]
- Mulet-Cabero, A.I.; Egger, L.; Portmann, R.; Ménard, O.; Marze, S.; Minekus, M.; Le Feunteun, S.; Sarkar, A.; Grundy, M.M.L.; Carrière, F.; et al. A standardised semi-dynamic in vitro digestion method suitable for food—An international consensus. Food Funct. 2020, 11, 1702–1720. [Google Scholar] [CrossRef]
- Mackie, A.; Macierzanka, A. Colloidal aspects of protein digestion. Curr. Opin. Colloid Interface Sci. 2010, 15, 102–108. [Google Scholar] [CrossRef]
- Macierzanka, A.; Sancho, A.I.; Mills, E.N.C.; Rigby, N.M.; MacKie, A.R. Emulsification alters simulated gastrointestinal proteolysis of β-casein and β-lactoglobulin. Soft Matter 2009, 5, 538–550. [Google Scholar] [CrossRef]
- Abdel-Aal, E.S.M. Effects of baking on protein digestibility of organic spelt products determined by two in vitro digestion methods. LWT-Food Sci. Technol. 2008, 41, 1282–1288. [Google Scholar] [CrossRef]
- Somaratne, G.; Ye, A.; Nau, F.; Ferrua, M.J.; Dupont, D.; Singh, R.P.; Singh, J. Egg white gel structure determines biochemical digestion with consequences on softening and mechanical disintegration during in vitro gastric digestion. Food Res. Int. 2020, 138, 109782. [Google Scholar] [CrossRef] [PubMed]
- Floury, J.; Bianchi, T.; Thévenot, J.; Dupont, D.; Jamme, F.; Lutton, E.; Panouillé, M.; Boué, F.; Le Feunteun, S. Exploring the breakdown of dairy protein gels during in vitro gastric digestion using time-lapse synchrotron deep-UV fluorescence microscopy. Food Chem. 2018, 239, 898–910. [Google Scholar] [CrossRef] [PubMed]
- Mulet-Cabero, A.I.; Torcello-Gómez, A.; Saha, S.; Mackie, A.R.; Wilde, P.J.; Brodkorb, A. Impact of caseins and whey proteins ratio and lipid content on in vitro digestion and ex vivo absorption. Food Chem. 2020, 319, 126514. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.R.; Fox, P.F.; Flynn, A.; Kindstedt, P.S. Susceptibility of β-Lactoglobulin and Sodium Caseinate to Proteolysis by Pepsin and Trypsin. J. Dairy Sci. 1995, 78, 2336–2344. [Google Scholar] [CrossRef] [PubMed]
- Dupont, D.; Mandalari, G.; Molle, D.; Jardin, J.; Léonil, J.; Faulks, R.M.; Wickham, M.S.J.; Mills, E.N.C.; Mackie, A.R. Comparative resistance of food proteins to adult and infant in vitro digestion models. Mol. Nutr. Food Res. 2010, 54, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Lorieau, L.; Le Gouar, Y.; Henry, G.; Mao, T.T.; Ligneul, A.; Hazart, E.; Dupont, D.; Floury, J. Whey-based cheese provides more postprandial plasma leucine than casein-based cheese: A pig study. Food Chem. 2019, 277, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Shani-Levi, C.; Goldstein, N.; Portmann, R.; Lesmes, U. Emulsion and protein degradation in the elderly: Qualitative insights from a study coupling a dynamic in vitro digestion model with proteomic analyses. Food Hydrocoll. 2017, 69, 393–401. [Google Scholar] [CrossRef]
- Giromini, C.; Cheli, F.; Rebucci, R.; Baldi, A. Invited review: Dairy proteins and bioactive peptides: Modeling digestion and the intestinal barrier. J. Dairy Sci. 2019, 102, 929–942. [Google Scholar] [CrossRef]
- Wang, H.; Brown, P.C.; Chow, E.C.Y.; Ewart, L.; Ferguson, S.S.; Fitzpatrick, S.; Freedman, B.S.; Guo, G.L.; Hedrich, W.; Heyward, S.; et al. 3D cell culture models: Drug pharmacokinetics, safety assessment, and regulatory consideration. Clin. Transl. Sci. 2021, 14, 1659–1680. [Google Scholar] [CrossRef] [PubMed]
- Cavalheiro, M.; Barros, A.P.D.N.; Louback, R.A.; Rossi, M.I.D. Modelos tridimensionais de cultura de células: Aproximando o in vitro do in vivo. INCQS-FIOCRUZ 2018, 6, 72–83. [Google Scholar]
- Funata, M.; Nio, Y.; Erion, D.M.; Thompson, W.L.; Takebe, T. The promise of human organoids in the digestive system. Cell Death Differ. 2021, 28, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Dedhia, P.H.; Bertaux-Skeirik, N.; Zavros, Y.; Spence, J.R. Organoid Models of Human Gastrointestinal Development and Disease. Gastroenterology 2016, 150, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Prior, N.; Inacio, P.; Huch, M. Liver organoids: From basic research to therapeutic applications. Gut 2019, 68, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhuo, Q.; Ye, Z.; Xu, X.; Ji, S. Organoid model: A new hope for pancreatic cancer treatment? BBA-Rev. Cancer 2021, 1875, 188466. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Chen, Y.G. Generation of 3D human gastrointestinal organoids: Principle and applications. Cell Regeneration 2020, 9, 6. [Google Scholar] [CrossRef]
- Costa, R.; Luz, A.; Medeiros, G.; Piuvezam, G.; Morais, A. What Are the Digestion and Absorption Models Used to Reproduce Gastrointestinal Protein Processes? A Protocol for Systematic Review. PROSPERO 2020 CRD42020198709. Available online: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42020198709 (accessed on 10 April 2024).

| Database | Search Strategies |
|---|---|
| PubMed and Web of Science 1 | digestion AND gastrointestinal tract AND protein hydrolysate AND (“in vitro” OR “rat” OR “mice” OR “animal”) |
| Science direct 2 | digestion AND gastrointestinal tract AND “digestion process” (“protein hydrolysate” OR peptide) AND “in vitro” AND animal |
| EMBASE and Biblioteca Virtual em Saúde | digestion OR digestibility AND gastric AND (peptides OR proteins) AND (“in vitro” OR animal) |
| Scopus 1 | “digestion process” AND “gastrointestinal tract” AND (peptides OR proteins) AND (“in vitro” OR mice) |
| PICOS | Inclusion | Exclusion |
|---|---|---|
| Population | Original articles resulting from in vivo studies performed with rats and mice of both sexes and different ages (puppies, young, adults, or elderly) without water or diet restriction, cell studies, and in vitro (gastrointestinal simulating fluids). | Articles with digestion models with other animals, or studies with computer simulations (in silico) |
| Interventions | Studies in which the intervention group has been submitted to the administration of peptides, proteins, or gastrointestinal simulating fluids to reproduce digestive processes of peptides and/or proteins | Studies that mimic the digestion process with the application of non-protein molecules or these molecules associated with peptides and/or proteins |
| Controls | Studies that present the control group composed of animals, cells, or in vitro experiments without administration of peptides or proteins. | Studies without a control group |
| Outcomes | Studies describing the model used to mimic protein digestion. | Studies that do not describe the protocol to simulate gastrointestinal conditions or studies that do not present schedule, experiment duration, frequency, administered dosages, concentration, and temperature, or studies that do not mimic digestion in humans |
| Study design | In vitro and in vivo experimental studies | Other types of studies |
| Other (article type) | Published original research articles | Other types of studies |
| Reference | Samples Used | Enzymes Used | pH | Temperature (Experimental/Interruption) | Experiment Duration (Each Step) | Protocol Adopted 1 |
|---|---|---|---|---|---|---|
| [19] | Defatted Cashew Flour (DCF) | α-amylase, pepsin, and porcine pancreatin | 7; 3 and 7 | 37 °C/90 °C | Oral phase: 10 min. Gastric phase: 120 min. Intestinal phase: 120 min. | [20] |
| [21] | Beef, chicken, pork, and turkey | Porcine α-amylase, pepsin, pancreatin | 2 (pepsin); the others were not reported | 37 °C/Not reported | Oral phase: 5 min., Gastric phase: 120 min., Intestinal phase: 120 min. | INFOGEST [20] |
| [22] | Z. bungeanum seed protein | Pepsin, trypsin solution | 3 and 7 | 37 °C/60 °C | Gastric phase: 120 min., Intestinal phase: 120 min. | [20] |
| [23] | Human milk (HM) or infant formula (IF) | α-amylase, lipase, porcine pepsin, porcine pancreatin | 6.3 (salivary fluid with amylase); 5.8 (gastric fluid with lipase and pepsin); 7.0 (intestinal fluid) | 37 °C/Not reported | Gastric phase: 120 min., Intestinal phase: 180 min. | Semi-dynamic in vitro simulation of the infant gastrointestinal tract |
| [24] | Peanut Ara h 1; wheat gliadin for bread; bread or peanuts | α-amylase from human saliva (for solid samples: peanuts and bread), porcine pepsin, porcine trypsin, bovine chymotrypsin, pancreatin from porcine pancreas (solid samples). | 7 (oral phase—infant and adult fasting); 5.3 (gastric phase); 6.6 (intestinal phase)—infant/3 (gastric phase); 7 (intestinal phase)—fed adult; 1.2 (gastric phase), 7 (intestinal phase)—fasting adult | 37 °C/Not reported | Oral phase (for solid samples: peanuts and bread): 2 min., Gastric phase (infant digestion and fed adult-static): 60 min., Intestinal phase (infant digestion and fed adult-static): 60 min. | The oral phase for the three protocols (infant, adult fed, and adult fasting) was based on the INFOGEST protocol. The gastric and intestinal phases for the infant protocol were based on Ménard et al. [25]. Static in vitro digestion for fed adults was based on the INFOGEST protocol |
| [26] | Kefir | α-amylase and mucin, pepsin, pancreatin | 2–3 (gastric), 7 (intestinal) | 37 °C/Not reported | Oral phase: 5 min., Gastric phase: 120 min., Intestinal phase: 120 min. | [27] |
| [28] | Three isolated proteins (zein, whey protein, and collagen) and five foods (peanuts, sorghum flour, wheat bran cereals, pigeon peas, and black beans) | Amylase, pepsin, pancreatin | 7; 3 and 7 | 37 °C/Not reported | Oral phase: 2 min., Gastric phase: 120 min., Intestinal phase: 120 min. | INFOGEST [20]—Static digestion |
| [29] | Bovine milk proteins β-lactoglobulin (BLG) and β-casein (BCS), whole fresh cow’s milk, and large free-range hard-boiled eggs | Pepsin from porcine gastric mucosa, trypsin from porcine pancreas and bovine chymotrypsin (intestinal digestion of isolated proteins), pancreatin from the porcine pancreas for intestinal meal phase and infant and adult fed models | 5.3—gastric and 6.6—intestinal (infant); 3 and 7 (adult fed); 1.2 and 7 (fasting adult) | 37 °C/Not reported | Gastric phase: 60 min., Intestinal phase: 60 min. | [25] (infant) and INFOGEST—[20] (adults) |
| [30] | Pea Protein Concentrate (PPC) 5% or 15% (static and dynamic) | α-amylase, pepsin and pancreatin (static)/amylase, porcine pancreatic lipase, porcine gastric mucosal pepsin and pancreatin (dynamics) | 7; 3 and 7 (static)/No reported (dynamics) | 37 °C/freezing with liquid nitrogen (static), 37 °C/Not reported (dynamics) | Oral phase: 2 min. Gastric phase: 120 min. Intestinal phase: 120 min. (static)/Total digestion time: 240 min. (dynamics) | INFOGEST [20]—Static digestion + Minekus [31]; Denis et al. [32]; Ribnicky et al. [33]—Dynamic digestion |
| [34] | Casein and whey protein powders | Pepsin from porcine gastric mucosa, pancreatin from porcine pancreas | No reported | 37 °C/Quick freezing with liquid nitrogen | Gastric phase: 20 and 120 min., Intestinal phase: 60 min. and 120 min. | [20] |
| [35] | Whey Protein Isolate (WPI) | Fresh stock solution of porcine pepsin, pancreatin | 3 and 7 | 37 °C/Not reported | Gastric phase: 120 min., Intestinal phase: 120 min. | INFOGEST [20] |
| [36] | Milk protein dissolved in water | Pepsin | 1.5 | 37 °C/90 °C | Total digestion time: 220 min. | Simulated gastric fluid was prepared according to [20], and dynamic digestion was performed according to Kong and Singh [37] |
| [38] | Soy and whey protein hydrolysates | Pepsin, lipase, pancreatin | 4.39 (stomach) and 6.8 (intestinal) | 37 °C/Not reported | Gastric phase: 40 min., Intestinal phase: 120 min. | Nguyen et al. [39] (infant digestion) |
| [40] | Milk supplemented with Brewers’ spent grain (BSG) | Pepsin, pancreatin | 2 and 7.4 | 37 °C/Not reported | Gastric phase: 60 min., Intestinal phase: 120 min. | McCarthy et al. [41] |
| [42] | Cranberry juice with whey protein isolate | Pepsin, pancreatin | 7 (spittle), 2–3 (stomach) and 7 (intestine) | 37 °C/Not reported | Oral phase: 30 s, Gastric phase: 120 min., Intestinal phase: 120 min. | Bornhorst and Singh [43] and Roman et al. [44] |
| [45] | Quinoa Protein Concentrate (QPC) | Porcine pepsin, porcine pancreatin | No reported (gastric)/7 (intestinal) | 37 °C/Quick freezing with liquid nitrogen | Gastric phase: 120 min., Intestinal phase: 120 min. | [20] |
| [46] | Bresaola ground | α-amylase, pepsin, pancreatin | 7; 3 and 7 | Not reported | Oral phase: 2 min., Gastric phase: 120 min., Intestinal phase: 120 min. | INFOGEST [20] Static digestion |
| [47] | Casein Aggregates (CAs) and Egg White Proteins (EWPs) | Porcine pepsin | 4 | 37 °C/Not reported | Gastric phase: 120 min. | INFOGEST [20]—Static digestion |
| [48] | Cod and chicken protein hydrolysates | No reported | No reported | Not reported | No reported | Tibäck et al. [49] (static digestion) |
| [50] | Kiwicha Protein Concentrate (KPC) | Porcine pepsin, porcine pancreatin | No reported | Not reported | Gastric phase: 120 min., Intestinal phase: 120 min. | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luz, A.B.S.; de Medeiros, A.F.; de Medeiros, G.C.B.S.; Piuvezam, G.; Passos, T.S.; Morais, A.H.d.A. Experimental Protocols Used to Mimic Gastrointestinal Protein Digestion: A Systematic Review. Nutrients 2024, 16, 2398. https://doi.org/10.3390/nu16152398
Luz ABS, de Medeiros AF, de Medeiros GCBS, Piuvezam G, Passos TS, Morais AHdA. Experimental Protocols Used to Mimic Gastrointestinal Protein Digestion: A Systematic Review. Nutrients. 2024; 16(15):2398. https://doi.org/10.3390/nu16152398
Chicago/Turabian StyleLuz, Anna Beatriz Santana, Amanda Fernandes de Medeiros, Gidyenne Christine Bandeira Silva de Medeiros, Grasiela Piuvezam, Thaís Souza Passos, and Ana Heloneida de Araújo Morais. 2024. "Experimental Protocols Used to Mimic Gastrointestinal Protein Digestion: A Systematic Review" Nutrients 16, no. 15: 2398. https://doi.org/10.3390/nu16152398
APA StyleLuz, A. B. S., de Medeiros, A. F., de Medeiros, G. C. B. S., Piuvezam, G., Passos, T. S., & Morais, A. H. d. A. (2024). Experimental Protocols Used to Mimic Gastrointestinal Protein Digestion: A Systematic Review. Nutrients, 16(15), 2398. https://doi.org/10.3390/nu16152398






