A Multi-Omics Approach to Disclose Metabolic Pathways Impacting Intestinal Permeability in Obese Patients Undergoing Very Low Calorie Ketogenic Diet
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Experimental Design and Recruited Participants
2.2. DNA Extraction from Fecal Samples
2.3. Sequencing Quality Check and Metataxonomic Bioinformatics Pipeline
2.4. Metabolic Pathway Prediction
2.5. NCBI Bioproject
2.6. Fecal and Urinary GC-MS Metabolite Profiles
2.7. Statistical Analyses
3. Results
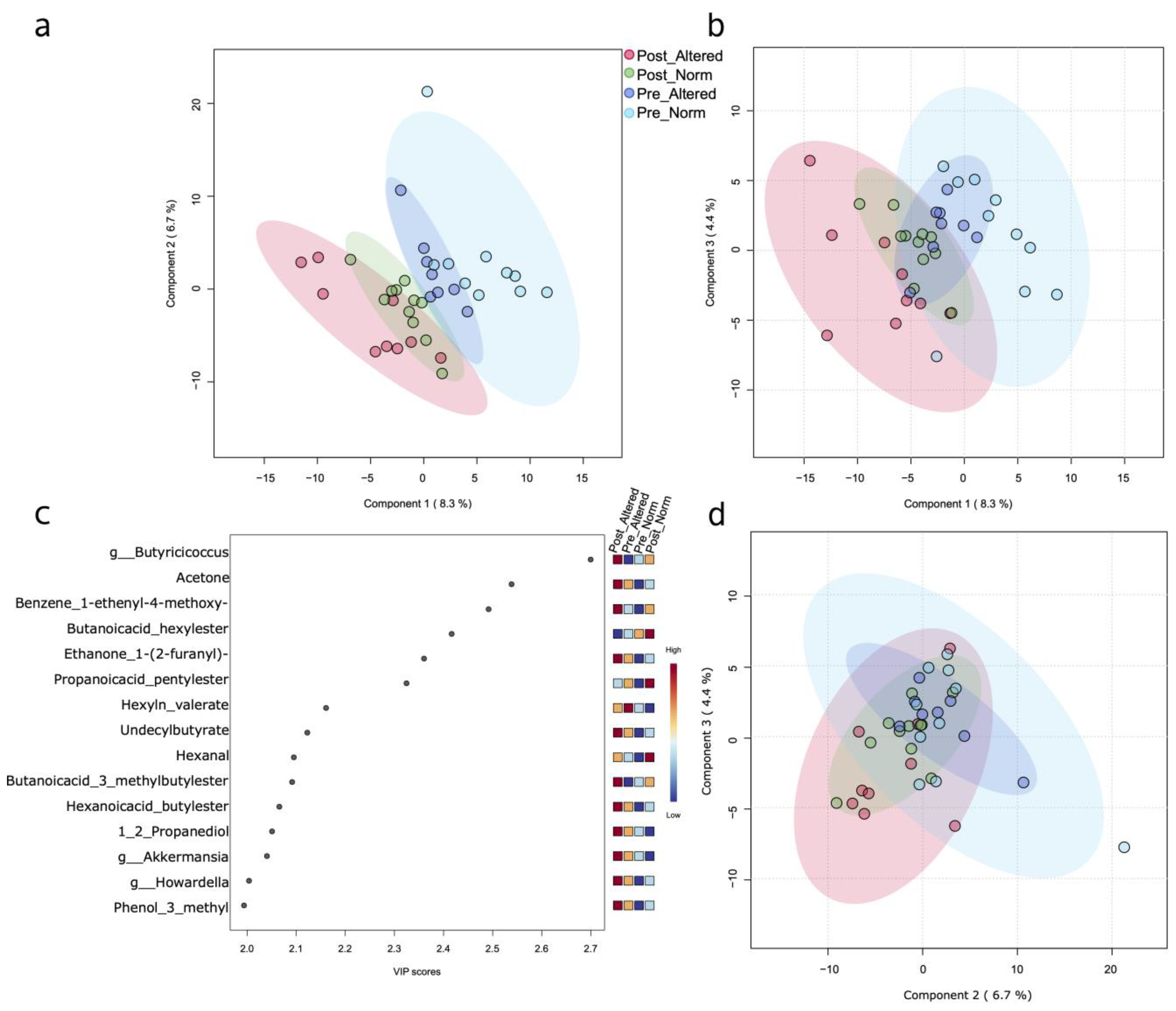
3.1. Multivariate Analyses Based on Single and Mixed Data Matrices
3.2. Biochemical Pathway Prediction Based on 16S Taxa Abundances
3.3. 16S Metabolic Predicted Pathways vs. Untargeted Metabolomics VOCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Lorenzo, A.; Romano, L.; Di Renzo, L.; Di Lorenzo, N.; Cenname, G.; Gualtieri, P. Obesity: A Preventable, Treatable, but Relapsing Disease. Nutrition 2020, 71, 110615. [Google Scholar] [CrossRef]
- Kim, J.; Nam, J.-H. Insight into the Relationship between Obesity-Induced Low-Level Chronic Inflammation and COVID-19 Infection. Int. J. Obes. 2020, 44, 1541–1542. [Google Scholar] [CrossRef] [PubMed]
- Hersoug, L.-G.; Møller, P.; Loft, S. Role of Microbiota-Derived Lipopolysaccharide in Adipose Tissue Inflammation, Adipocyte Size and Pyroptosis during Obesity. Nutr. Res. Rev. 2018, 31, 153–163. [Google Scholar] [CrossRef]
- Whelan, K.; Bancil, A.S.; Lindsay, J.O.; Chassaing, B. Ultra-Processed Foods and Food Additives in Gut Health and Disease. Nat. Rev. Gastroenterol. Hepatol. 2024, 21, 406–427. [Google Scholar] [CrossRef] [PubMed]
- Linsalata, M.; Russo, F.; Riezzo, G.; D’Attoma, B.; Prospero, L.; Orlando, A.; Ignazzi, A.; Di Chito, M.; Sila, A.; De Nucci, S.; et al. The Effects of a Very-Low-Calorie Ketogenic Diet on the Intestinal Barrier Integrity and Function in Patients with Obesity: A Pilot Study. Nutrients 2023, 15, 2561. [Google Scholar] [CrossRef]
- Celano, G.; Calabrese, F.M.; Riezzo, G.; D’Attoma, B.; Ignazzi, A.; Di Chito, M.; Sila, A.; De Nucci, S.; Rinaldi, R.; Linsalata, M.; et al. Effects of a Very-Low-Calorie Ketogenic Diet on the Fecal and Urinary Volatilome in an Obese Patient Cohort: A Preliminary Investigation. Nutrients 2023, 15, 3752. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Calabrese, F.M.; Celano, G.; Riezzo, G.; D’Attoma, B.; Ignazzi, A.; Di Chito, M.; Sila, A.; De Nucci, S.; Rinaldi, R.; Linsalata, M.; et al. Metabolomic Profiling of Obese Patients with Altered Intestinal Permeability Undergoing a Very Low-Calorie Ketogenic Diet. Nutrients 2023, 15, 5026. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Houtman, T.A.; Eckermann, H.A.; Smidt, H.; de Weerth, C. Gut Microbiota and BMI throughout Childhood: The Role of Firmicutes, Bacteroidetes, and Short-Chain Fatty Acid Producers. Sci. Rep. 2022, 12, 3140. [Google Scholar] [CrossRef]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-Analyses of Human Gut Microbes Associated with Obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and Disease Markers Correlate with Gut Microbiome Composition across Thousands of People. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef]
- Sze, M.A.; Schloss, P.D. Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. mBio 2016, 7, e01018-16. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary Intervention Impact on Gut Microbial Gene Richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Puljiz, Z.; Kumric, M.; Vrdoljak, J.; Martinovic, D.; Ticinovic Kurir, T.; Krnic, M.O.; Urlic, H.; Puljiz, Z.; Zucko, J.; Dumanic, P.; et al. Obesity, Gut Microbiota, and Metabolome: From Pathophysiology to Nutritional Interventions. Nutrients 2023, 15, 2236. [Google Scholar] [CrossRef]
- Lakshmanan, A.P.; Al Zaidan, S.; Bangarusamy, D.K.; Al-Shamari, S.; Elhag, W.; Terranegra, A. Increased Relative Abundance of Ruminoccocus Is Associated with Reduced Cardiovascular Risk in an Obese Population. Front. Nutr. 2022, 9, 849005. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Li, H.; Dai, Y.; Chen, D.; Wang, M.; Jiang, X.; Huang, Z.; Yu, H.; Huang, J.; et al. Altered Fecal Microbiota Composition in Older Adults with Frailty. Front. Cell Infect. Microbiol. 2021, 11, 696186. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-Producing Clostridium Cluster XIVa Species Specifically Colonize Mucins in an in Vitro Gut Model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef]
- Seo, B.; Jeon, K.; Moon, S.; Lee, K.; Kim, W.-K.; Jeong, H.; Cha, K.H.; Lim, M.Y.; Kang, W.; Kweon, M.-N.; et al. Roseburia spp. Abundance Associates with Alcohol Consumption in Humans and Its Administration Ameliorates Alcoholic Fatty Liver in Mice. Cell Host Microbe 2020, 27, 25–40.e6. [Google Scholar] [CrossRef]
- Brüssow, H.; Parkinson, S.J. You Are What You Eat. Nat. Biotechnol. 2014, 32, 243–245. [Google Scholar] [CrossRef]
- Gagliardi, A.; Totino, V.; Cacciotti, F.; Iebba, V.; Neroni, B.; Bonfiglio, G.; Trancassini, M.; Passariello, C.; Pantanella, F.; Schippa, S. Rebuilding the Gut Microbiota Ecosystem. Int. J. Environ. Res. Public Health 2018, 15, 1679. [Google Scholar] [CrossRef]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of High-Fat Diet on Gut Microbiota: A Driving Force for Chronic Disease Risk. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 515–520. [Google Scholar] [CrossRef]
- Pinto, A.; Bonucci, A.; Maggi, E.; Corsi, M.; Businaro, R. Anti-Oxidant and Anti-Inflammatory Activity of Ketogenic Diet: New Perspectives for Neuroprotection in Alzheimer’s Disease. Antioxidants 2018, 7, 63. [Google Scholar] [CrossRef]
- Waldram, A.; Holmes, E.; Wang, Y.; Rantalainen, M.; Wilson, I.D.; Tuohy, K.M.; McCartney, A.L.; Gibson, G.R.; Nicholson, J.K. Top-Down Systems Biology Modeling of Host Metabotype−Microbiome Associations in Obese Rodents. J. Proteome Res. 2009, 8, 2361–2375. [Google Scholar] [CrossRef]
- Belda, E.; Voland, L.; Tremaroli, V.; Falony, G.; Adriouch, S.; Assmann, K.E.; Prifti, E.; Aron-Wisnewsky, J.; Debédat, J.; Le Roy, T.; et al. Impairment of Gut Microbial Biotin Metabolism and Host Biotin Status in Severe Obesity: Effect of Biotin and Prebiotic Supplementation on Improved Metabolism. Gut 2022, 71, 2463–2480. [Google Scholar] [CrossRef]
- Denou, E.; Lolmède, K.; Garidou, L.; Pomie, C.; Chabo, C.; Lau, T.C.; Fullerton, M.D.; Nigro, G.; Zakaroff-Girard, A.; Luche, E.; et al. Defective NOD2 Peptidoglycan Sensing Promotes Diet-induced Inflammation, Dysbiosis, and Insulin Resistance. EMBO Mol. Med. 2015, 7, 259–274. [Google Scholar] [CrossRef]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Valiuskyte, K.; Dewhirst, F.E.; Nichols, F.C.; Davey, M.E. Synthesis of Sphingolipids Impacts Survival of Porphyromonas Gingivalis and the Presentation of Surface Polysaccharides. Front. Microbiol. 2016, 7, 1919. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Oh, S.F.; Olszak, T.; Neves, J.F.; Avci, F.Y.; Erturk-Hasdemir, D.; Lu, X.; Zeissig, S.; Blumberg, R.S.; Kasper, D.L. Sphingolipids from a Symbiotic Microbe Regulate Homeostasis of Host Intestinal Natural Killer T Cells. Cell 2014, 156, 123–133. [Google Scholar] [CrossRef]
- Meijnikman, A.S.; Aydin, O.; Prodan, A.; Tremaroli, V.; Herrema, H.; Levin, E.; Acherman, Y.; Bruin, S.; Gerdes, V.e.; Backhed, F.; et al. Distinct Differences in Gut Microbial Composition and Functional Potential from Lean to Morbidly Obese Subjects. J. Intern. Med. 2020, 288, 699–710. [Google Scholar] [CrossRef]
- Kushkevych, I.; Cejnar, J.; Treml, J.; Dordević, D.; Kollar, P.; Vítězová, M. Recent Advances in Metabolic Pathways of Sulfate Reduction in Intestinal Bacteria. Cells 2020, 9, 698. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Kollar, P.; Vítězová, M.; Drago, L. Hydrogen Sulfide as a Toxic Product in the Small–Large Intestine Axis and Its Role in IBD Development. J. Clin. Med. 2019, 8, 1054. [Google Scholar] [CrossRef]
- Figliuolo, V.R.; dos Santos, L.M.; Abalo, A.; Nanini, H.; Santos, A.; Brittes, N.M.; Bernardazzi, C.; de Souza, H.S.P.; Vieira, L.Q.; Coutinho-Silva, R.; et al. Sulfate-Reducing Bacteria Stimulate Gut Immune Responses and Contribute to Inflammation in Experimental Colitis. Life Sci. 2017, 189, 29–38. [Google Scholar] [CrossRef]
- Sankaranarayanan, K.; Antaris, X.X.; Palanski, B.A.; El Gamal, A.; Kao, C.M.; Fitch, W.L.; Fischer, C.R.; Khosla, C. Tunable Enzymatic Synthesis of the Immunomodulator Lipid IVA To Enable Structure-Activity Analysis. J. Am. Chem. Soc. 2019, 141, 9474–9478. [Google Scholar] [CrossRef]
- Yuan, X.; Chen, R.; McCormick, K.L.; Zhang, Y.; Lin, X.; Yang, X. The Role of the Gut Microbiota on the Metabolic Status of Obese Children. Microb. Cell Factories 2021, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Shmagel, A.; Demmer, R.; Knights, D.; Butler, M.; Langsetmo, L.; Lane, N.E.; Ensrud, K. The Effects of Glucosamine and Chondroitin Sulfate on Gut Microbial Composition: A Systematic Review of Evidence from Animal and Human Studies. Nutrients 2019, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Kobayashi, T.; Moroi, S.; Kotake, H.; Ikoma, T.; Saeki, H.; Ura, K.; Takagi, Y. Anti-Obesity Effects of Chondroitin Sulfate Oligosaccharides from the Skate Raja pulchra. Carbohydr. Polym. 2019, 214, 303–310. [Google Scholar] [CrossRef]
- Nie, X.; Chen, J.; Ma, X.; Ni, Y.; Shen, Y.; Yu, H.; Panagiotou, G.; Bao, Y. A Metagenome-Wide Association Study of Gut Microbiome and Visceral Fat Accumulation. Comput. Struct. Biotechnol. J. 2020, 18, 2596–2609. [Google Scholar] [CrossRef]
- Zemel, M.B.; Bruckbauer, A. Effects of a Leucine and Pyridoxine-Containing Nutraceutical on Fat Oxidation, and Oxidative and Inflammatory Stress in Overweight and Obese Subjects. Nutrients 2012, 4, 529–541. [Google Scholar] [CrossRef]
- Mukherjee, S.; Choi, M.; Yun, J.W. Trans-Anethole Induces Thermogenesis via Activating SERCA/SLN Axis in C2C12 Muscle Cells. Biotechnol. Bioprocess Eng. 2022, 27, 938–948. [Google Scholar] [CrossRef]
- Moreno-Navarrete, J.M.; Rodríguez, A.; Ortega, F.; Becerril, S.; Girones, J.; Sabater-Masdeu, M.; Latorre, J.; Ricart, W.; Frühbeck, G.; Fernández-Real, J.M. Heme Biosynthetic Pathway Is Functionally Linked to Adipogenesis via Mitochondrial Respiratory Activity. Obesity 2017, 25, 1723–1733. [Google Scholar] [CrossRef]

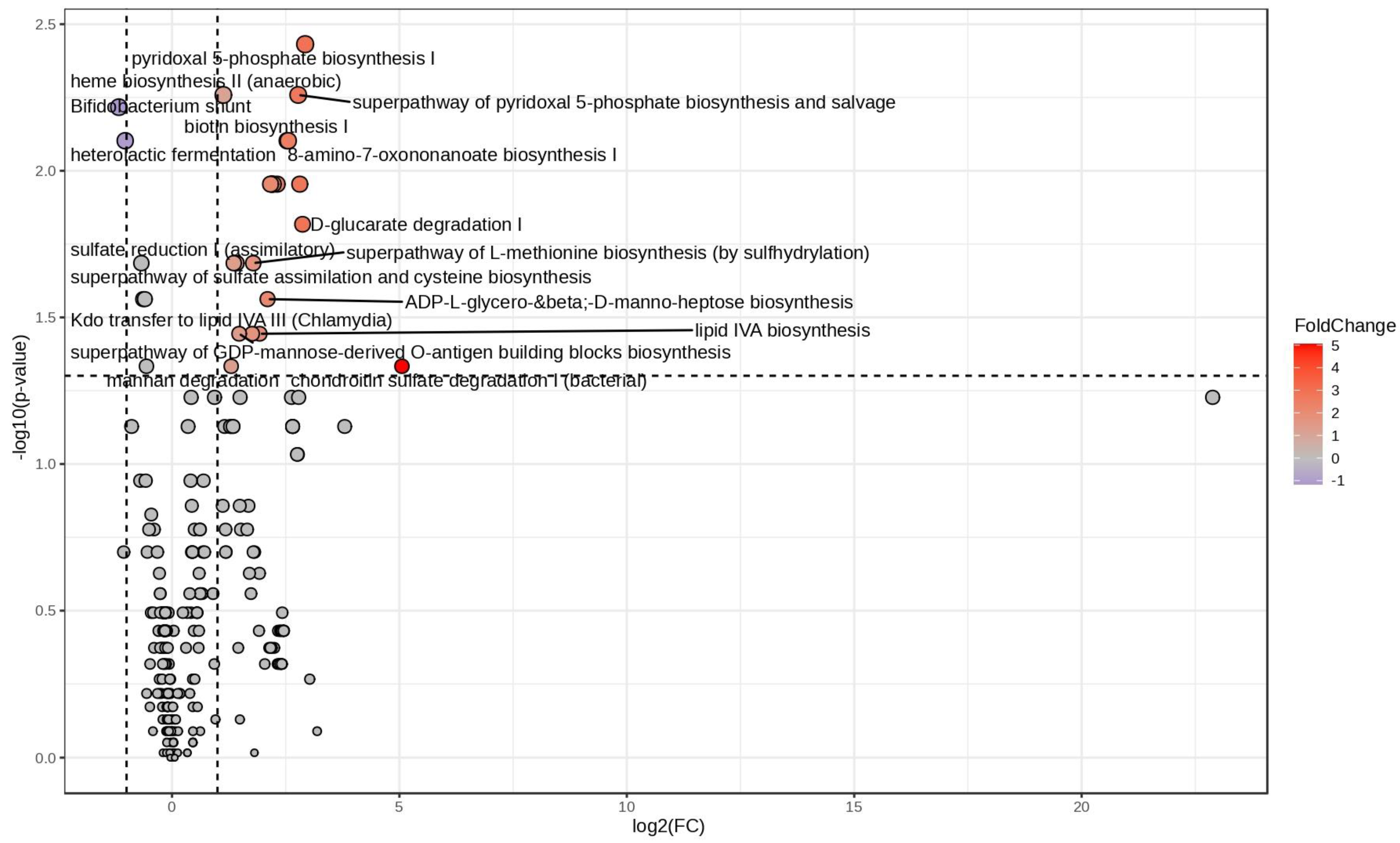

| Pathway | FC | log2(FC) | Corrected p-Value | −LOG10(p) |
|---|---|---|---|---|
| Chondroitin sulfate degradation I (bacterial) | 33.248 | 5.0552 | 0.046401 | 1.3335 |
| Pyridoxal 5-phosphate biosynthesis I | 7.6054 | 2.927 | 0.0037022 | 2.4315 |
| D-glucarate degradation I | 7.3074 | 2.8694 | 0.01522 | 1.8176 |
| Anhydromuropeptides recycling | 7.0071 | 2.8088 | 0.011107 | 1.9544 |
| Super-pathway of pyridoxal 5-phosphate biosynthesis and salvage | 6.8423 | 2.7745 | 0.0055121 | 2.2587 |
| 8-amino-7-oxononanoate biosynthesis I | 5.8769 | 2.555 | 0.007898 | 2.1025 |
| Biotin biosynthesis I | 5.7884 | 2.5332 | 0.007898 | 2.1025 |
| Palmitate biosynthesis II (bacteria and plants) | 4.9619 | 2.3109 | 0.011107 | 1.9544 |
| Oleate biosynthesis IV (anaerobic) | 4.6762 | 2.2253 | 0.011107 | 1.9544 |
| Super-pathway of fatty acid biosynthesis initiation (E. coli) | 4.596 | 2.2004 | 0.011107 | 1.9544 |
| Palmitoleate biosynthesis I (from (5Z)-dodec-5-enoate) | 4.5835 | 2.1965 | 0.011107 | 1.9544 |
| Stearate biosynthesis II (bacteria and plants) | 4.5657 | 2.1908 | 0.011107 | 1.9544 |
| mycolate biosynthesis | 4.4898 | 2.1666 | 0.011107 | 1.9544 |
| ADP-L-glycero-beta-D-manno-heptose biosynthesis | 4.2874 | 2.1001 | 0.027396 | 1.5623 |
| lipid IVA biosynthesis | 3.8011 | 1.9264 | 0.035952 | 1.4443 |
| Super-pathway of L-methionine biosynthesis (by sulfhydrylation) | 3.4462 | 1.785 | 0.02065 | 1.6851 |
| Kdo transfer to lipid IVA III (Chlamydia) | 3.3877 | 1.7603 | 0.035952 | 1.4443 |
| Super-pathway of GDP-mannose-derived O-antigen building blocks biosynthesis | 2.7817 | 1.476 | 0.035952 | 1.4443 |
| Sulfate reduction I (assimilatory) | 2.6706 | 1.4172 | 0.02065 | 1.6851 |
| Super-pathway of sulfate assimilation and cysteine biosynthesis | 2.5613 | 1.3569 | 0.02065 | 1.6851 |
| Mannan degradation | 2.4624 | 1.3001 | 0.046401 | 1.3335 |
| Heme biosynthesis II (anaerobic) | 2.1869 | 1.1289 | 0.0055121 | 2.2587 |
| Heterolactic fermentation | 0.48974 | −1.0299 | 0.007898 | 2.1025 |
| Bifidobacterium shunt | 0.44482 | −1.1687 | 0.0060678 | 2.217 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celano, G.; Calabrese, F.M.; Riezzo, G.; D’Attoma, B.; Ignazzi, A.; Di Chito, M.; Sila, A.; De Nucci, S.; Rinaldi, R.; Linsalata, M.; et al. A Multi-Omics Approach to Disclose Metabolic Pathways Impacting Intestinal Permeability in Obese Patients Undergoing Very Low Calorie Ketogenic Diet. Nutrients 2024, 16, 2079. https://doi.org/10.3390/nu16132079
Celano G, Calabrese FM, Riezzo G, D’Attoma B, Ignazzi A, Di Chito M, Sila A, De Nucci S, Rinaldi R, Linsalata M, et al. A Multi-Omics Approach to Disclose Metabolic Pathways Impacting Intestinal Permeability in Obese Patients Undergoing Very Low Calorie Ketogenic Diet. Nutrients. 2024; 16(13):2079. https://doi.org/10.3390/nu16132079
Chicago/Turabian StyleCelano, Giuseppe, Francesco Maria Calabrese, Giuseppe Riezzo, Benedetta D’Attoma, Antonia Ignazzi, Martina Di Chito, Annamaria Sila, Sara De Nucci, Roberta Rinaldi, Michele Linsalata, and et al. 2024. "A Multi-Omics Approach to Disclose Metabolic Pathways Impacting Intestinal Permeability in Obese Patients Undergoing Very Low Calorie Ketogenic Diet" Nutrients 16, no. 13: 2079. https://doi.org/10.3390/nu16132079
APA StyleCelano, G., Calabrese, F. M., Riezzo, G., D’Attoma, B., Ignazzi, A., Di Chito, M., Sila, A., De Nucci, S., Rinaldi, R., Linsalata, M., Apa, C. A., Mancini, L., De Angelis, M., Giannelli, G., De Pergola, G., & Russo, F. (2024). A Multi-Omics Approach to Disclose Metabolic Pathways Impacting Intestinal Permeability in Obese Patients Undergoing Very Low Calorie Ketogenic Diet. Nutrients, 16(13), 2079. https://doi.org/10.3390/nu16132079








