Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
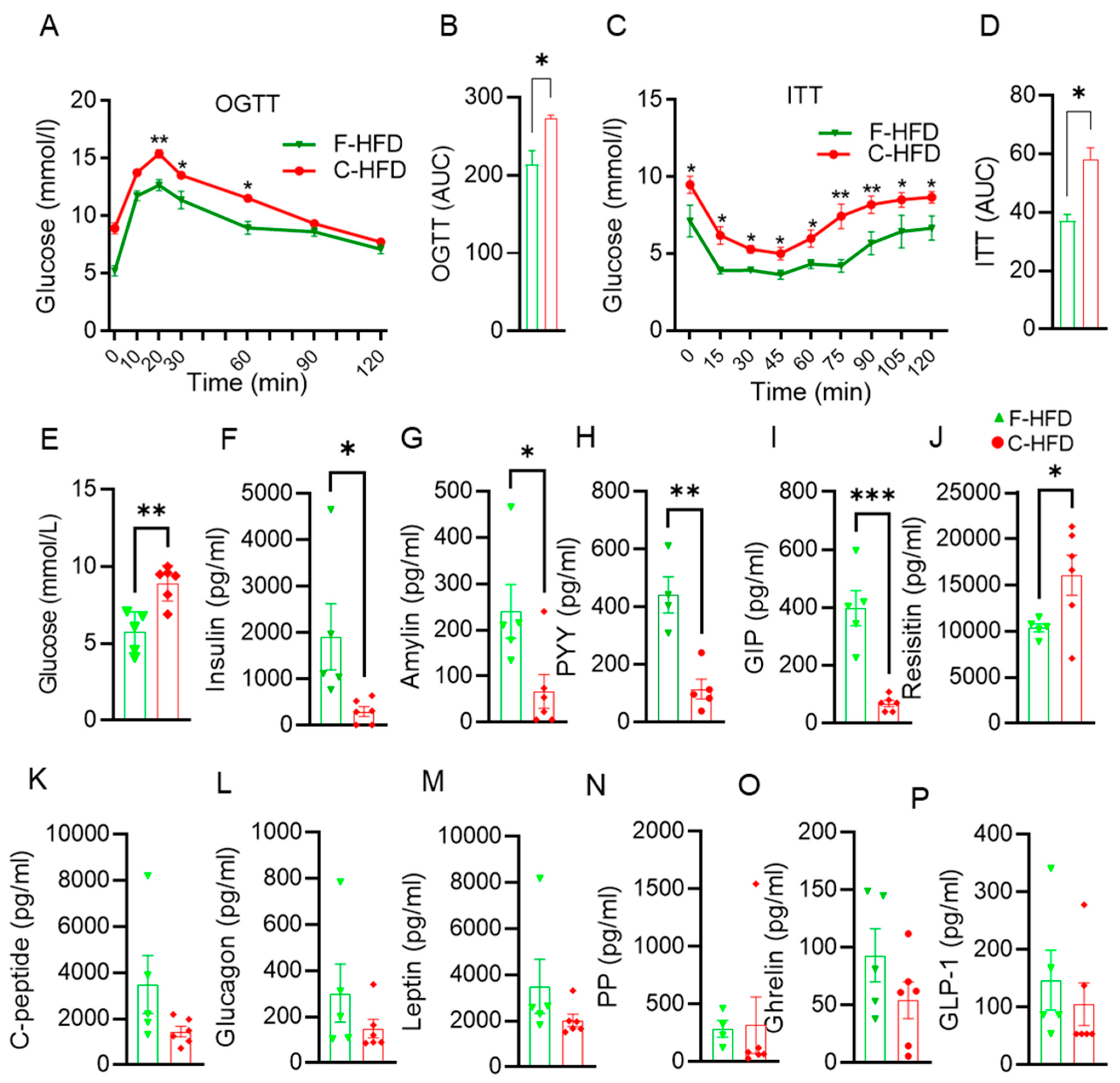
2.2. Oral Glucose Tolerance Test (OGTT)
2.3. Insulin Tolerance Test (ITT)
2.4. Plasma Measurements
2.5. Histological Analysis
2.6. Gene Expression Analysis by qRT-PCR
2.7. Microbiome Sequencing
2.8. 16S rRNA Microbiome Analysis and Bioinformatics Statistics
2.9. Biodiversity Analysis
2.10. Identification of Biomarker Microbiome
2.11. Microbial Metagenomic Functional Predictions
2.12. Prediction of Biomarker Microbiome-Epigenome Interactions
3. Results
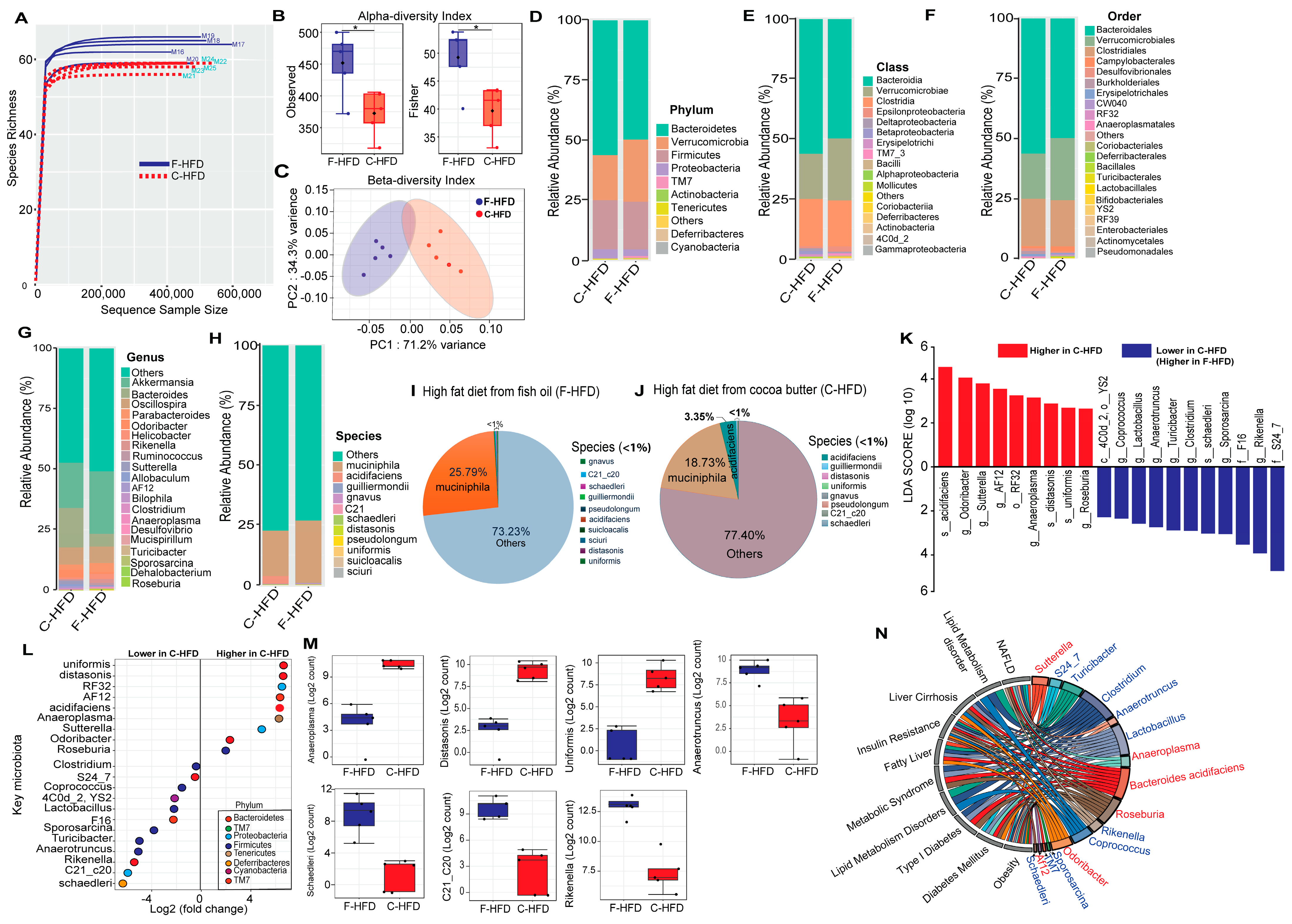
3.1. C-HFD and F-HFD Have Differential Effects on the Gut Microbiome in Mice
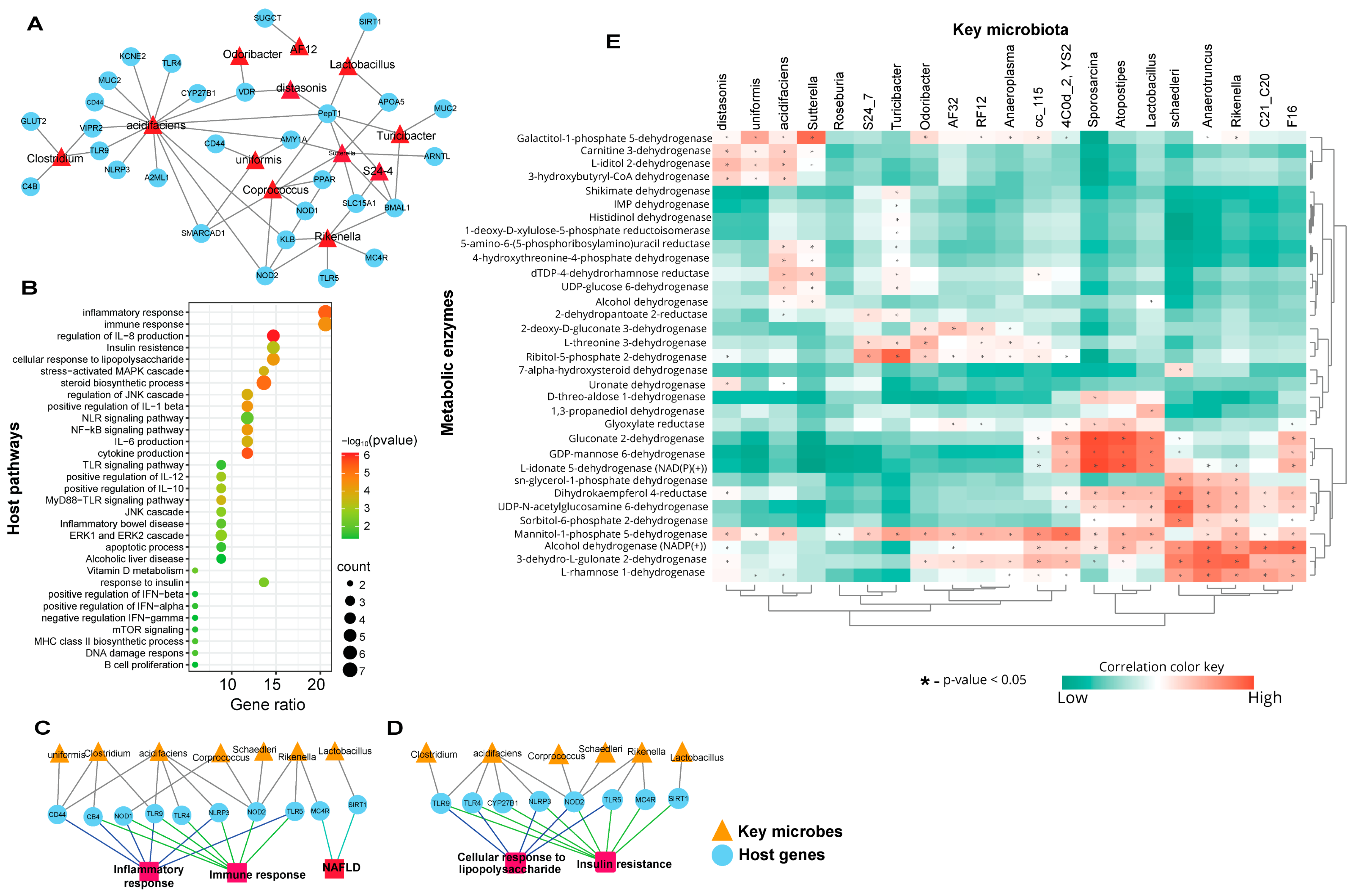
3.2. Biomarker Gut Microbiota Associate with Host Genes Involved in Immune-Metabolic Regulation
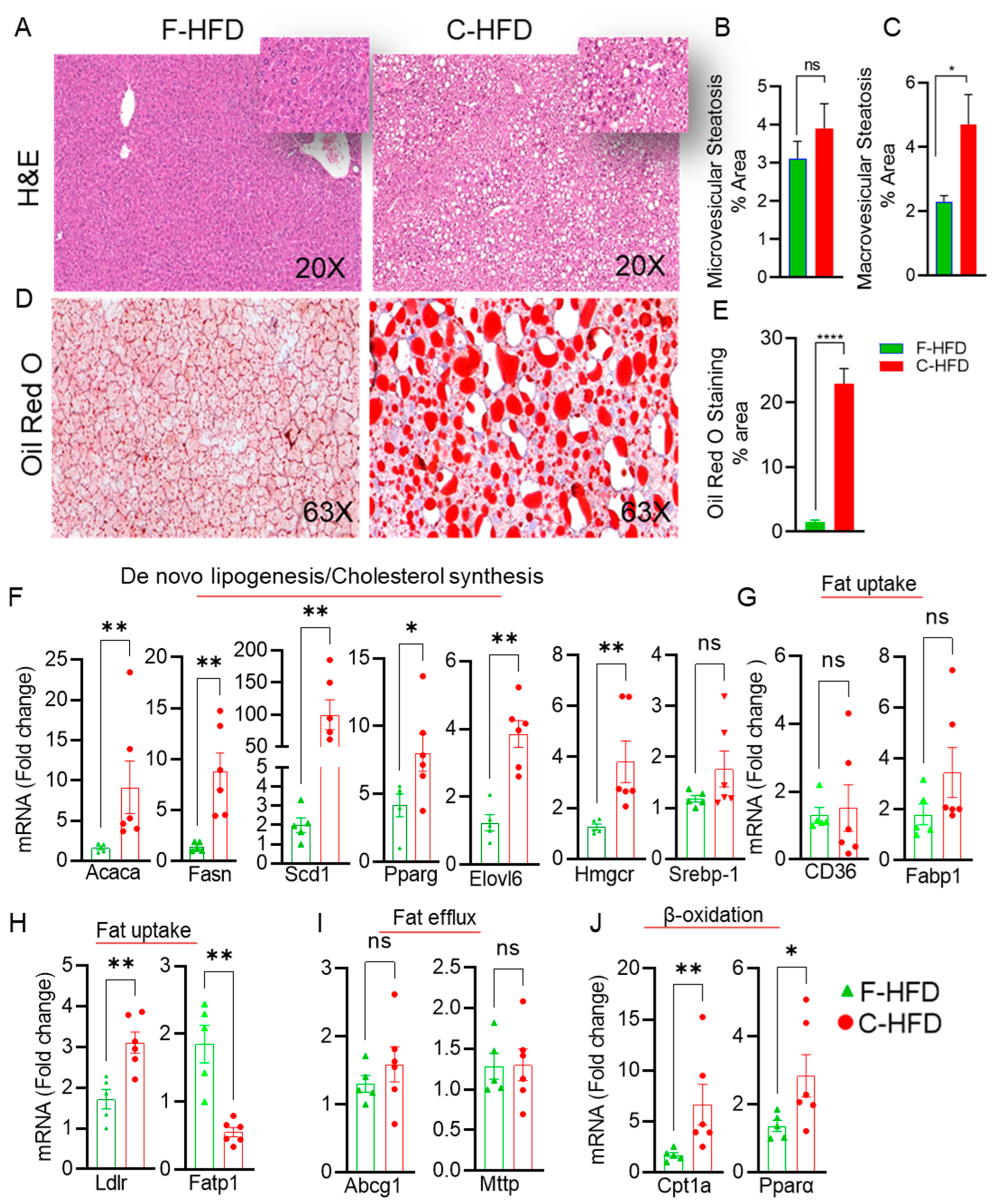
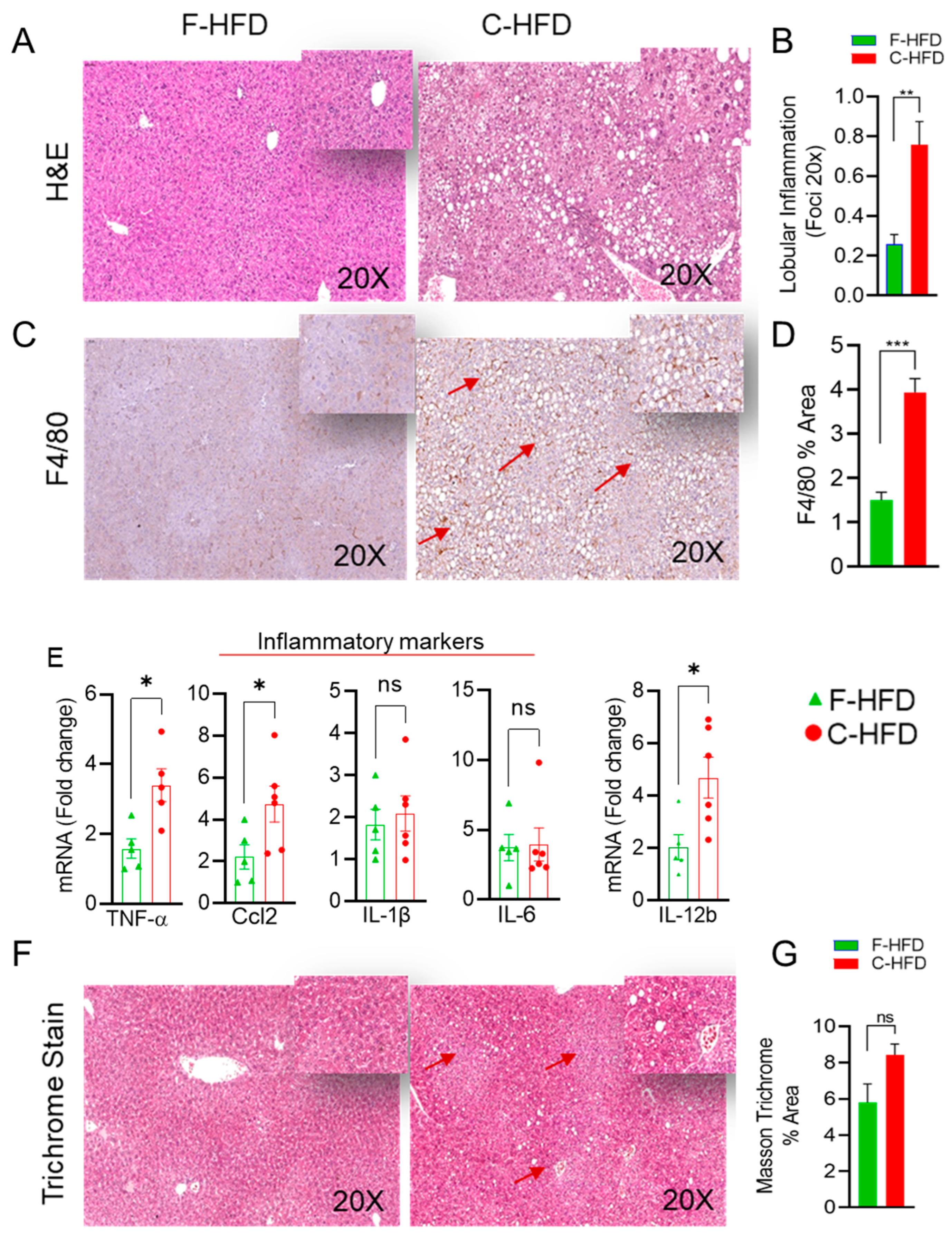
3.3. Mice Fed with C-HFD Display Liver Steatosis and Inflammation
3.4. Impaired Glucose Tolerance in C-HFD Mice, Compared with F-HFD Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, P. Influence of foods and nutrition on the gut microbiome and implications for intestinal health. Int. J. Mol. Sci. 2022, 23, 9588. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tan, Y.; Cheng, H.; Zhang, D.; Feng, W.; Peng, C. Functions of gut microbiota metabolites, current status and future perspectives. Aging Dis. 2022, 13, 1106. [Google Scholar] [CrossRef] [PubMed]
- Cano, P.G.; Santacruz, A.; Trejo, F.M.; Sanz, Y. Bifidobacterium cect 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet-fed mice. Obesity 2013, 21, 2310–2321. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, Y.; Kanazawa, A.; Aida, M.; Yoshida, Y.; Yamashiro, Y.; Watada, H. Association of gut microbiota and inflammatory markers in obese patients with type 2 diabetes mellitus: Post hoc analysis of a synbiotic interventional study. Biosci. Microbiota Food Health 2022, 41, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Martínez-López, Y.E.; Esquivel-Hernández, D.A.; Sánchez-Castañeda, J.P.; Neri-Rosario, D.; Guardado-Mendoza, R.; Resendis-Antonio, O. Type 2 diabetes, gut microbiome, and systems biology: A novel perspective for a new era. Gut Microbes 2022, 14, 2111952. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Wang, Q.; Yi, S.; Liu, X.; Jin, H.; Xu, J.; Wen, G.; Zhu, J.; Tuo, B. The source of the fat significantly affects the results of high-fat diet intervention. Sci. Rep. 2022, 12, 4315. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, X.-J. Effects of a high fat diet on intestinal microbiota and gastrointestinal diseases. World J. Gastroenterol. 2016, 22, 8905. [Google Scholar] [CrossRef]
- Trakman, G.L.; Fehily, S.; Basnayake, C.; Hamilton, A.L.; Russell, E.; Wilson-O’Brien, A.; Kamm, M.A. Diet and gut microbiome in gastrointestinal disease. J. Gastroenterol. Hepatol. 2022, 37, 237–245. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.-H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front. Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Cho, K.Y. Association of gut microbiota with obesity in children and adolescents. Clin. Exp. Pediatr. 2023, 66, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and firmicutes/bacteroidetes ratio in an adult ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Chanda, D.; De, D. Meta-analysis reveals obesity associated gut microbial alteration patterns and reproducible contributors of functional shift. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kübeck, R.; Bonet-Ripoll, C.; Hoffmann, C.; Walker, A.; Müller, V.M.; Schüppel, V.L.; Lagkouvardos, I.; Scholz, B.; Engel, K.-H.; Daniel, H. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016, 5, 1162–1174. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Zhao, F.; Song, S.; Li, Y.; Xu, X.; Zhou, G.; Li, C. Fish oil, lard and soybean oil differentially shape gut microbiota of middle-aged rats. Sci. Rep. 2017, 7, 826. [Google Scholar] [CrossRef] [PubMed]
- Quek, R.Y.C.; Peh, E.W.Y.; Henry, C.J. Effects of cocoa butter and cocoa butter equivalent in a chocolate confectionery on human blood triglycerides, glucose and insulin. Foods 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Patterson, W.L.; Georgel, P.T. Breaking the cycle: The role of omega-3 polyunsaturated fatty acids in inflammation-driven cancers. Biochem. Cell Biol. 2014, 92, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, C.; Parini, P.; Ostojic, J.; Cheung, L.; Hu, J.; Zadjali, F.; Tahir, F.; Brismar, K.; Norstedt, G.; Tollet-Egnell, P. Cocoa butter and safflower oil elicit different effects on hepatic gene expression and lipid metabolism in rats. Lipids 2009, 44, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Bäckhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef]
- Jennison, E.; Byrne, C.D. The role of the gut microbiome and diet in the pathogenesis of non-alcoholic fatty liver disease. Clin. Mol. Hepatol. 2021, 27, 22. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.; Ferreira, G.; Reis, F.; Viana, S. Impact of dietary sugars on gut microbiota and metabolic health. Diabetology 2022, 3, 549–560. [Google Scholar] [CrossRef]
- Konstantopoulos, P.; Doulamis, I.P.; Tzani, A.; Korou, M.L.; Agapitos, E.; Vlachos, I.S.; Pergialiotis, V.; Verikokos, C.; Mastorakos, G.; Katsilambros, N.L.; et al. Metabolic effects of crocus sativus and protective action against non-alcoholic fatty liver disease in diabetic rats. Biomed. Rep. 2017, 6, 513–518. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fengler, V.H.I.; Macheiner, T.; Kessler, S.M.; Czepukojc, B.; Gemperlein, K.; Müller, R.; Kiemer, A.K.; Magnes, C.; Haybaeck, J.; Lackner, C.; et al. Susceptibility of different mouse wild type strains to develop diet-induced nafld/afld-associated liver disease. PLoS ONE 2016, 11, e0155163. [Google Scholar] [CrossRef]
- Crowe, A.R.; Yue, W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: An integrated protocol. Bio-protocol 2019, 9, e3465. [Google Scholar] [CrossRef] [PubMed]
- Mehlem, A.; Hagberg, C.E.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red o for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef]
- Yen, K.; Le, T.T.; Bansal, A.; Narasimhan, S.D.; Cheng, J.X.; Tissenbaum, H.A. A comparative study of fat storage quantitation in nematode caenorhabditis elegans using label and label-free methods. PLoS ONE 2010, 5, e12810. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using qiime 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. Microbiomeanalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Ombrello, A.K. Dada2. In Encyclopedia of Medical Immunology: Immunodeficiency Diseases; MacKay, I., Rose, N.R., Eds.; Springer: New York, NY, USA, 2020; pp. 1–7. [Google Scholar]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16s rrna gene database and workbench compatible with arb. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. Unifrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The metacyc database of metabolic pathways and enzymes and the biocyc collection of pathway/genome databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, L.; Zeng, P.; Huang, C.; Li, J.; Geng, B.; Yang, J.; Kong, W.; Zhou, X.; Cui, Q. An analysis of human microbe-disease associations. Brief. Bioinform. 2017, 18, 85–97. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, W.; Wu, X.; Liang, X.; Cao, L.; Zhai, J.; Yang, Y.; Chen, Q.; Liu, H.; Zhang, J.; et al. Miaome: Human microbiome affect the host epigenome. Comput. Struct. Biotechnol. J. 2022, 20, 2455–2463. [Google Scholar] [CrossRef]
- Delpino, F.M.; Figueiredo, L.M.; da Silva, B.G.C. Effects of omega-3 supplementation on body weight and body fat mass: A systematic review. Clin. Nutr. ESPEN 2021, 44, 122–129. [Google Scholar] [CrossRef]
- Togo, J.; Hu, S.; Li, M.; Niu, C.; Speakman, J.R. Impact of dietary sucrose on adiposity and glucose homeostasis in c57bl/6j mice depends on mode of ingestion: Liquid or solid. Mol. Metab. 2019, 27, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, B.; Peng, B.; Wang, J.; Hu, Y.; Wang, R.; Wang, S. Different dose of sucrose consumption divergently influences gut microbiota and ppar-γ/mapk/nf-κb pathway in dss-induced colitis mice. Nutrients 2022, 14, 2765. [Google Scholar] [CrossRef] [PubMed]
- Alasmar, R.M.; Varadharajan, K.; Shanmugakonar, M.; Al-Naemi, H.A. Early-life sugar consumption affects the microbiome in juvenile mice. Mol. Nutr. Food Res. 2023, 67, 2200322. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Araki, Y.; Hanzawa, F.; Umeki, M.; Kojima, T.; Nishimura, N.; Ikeda, S.; Mochizuki, S.; Oda, H. High sucrose diet-induced dysbiosis of gut microbiota promotes fatty liver and hyperlipidemia in rats. J. Nutr. Biochem. 2021, 93, 108621. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; Ross, A.W.; Walker, A.W.; Morgan, P.J. Dietary uncoupling of gut microbiota and energy harvesting from obesity and glucose tolerance in mice. Cell Rep. 2017, 21, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; González, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974. [Google Scholar] [CrossRef] [PubMed]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from europe and rural africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Cronin, P.; Joyce, S.A.; O’Toole, P.W.; O’Connor, E.M. Dietary fibre modulates the gut microbiota. Nutrients 2021, 13, 1655. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kong, Q.; Li, X.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients 2020, 12, 3197. [Google Scholar] [CrossRef]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjære, E.; Midtbø, L.K.; et al. High-fat feeding rather than obesity drives taxonomical and functional changes in the gut microbiota in mice. Microbiome 2017, 5, 43. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xiao, Y.; Li, D.; Zhang, S.; Wu, Y.; Zhang, Q.; Bai, W. New insights into the mechanisms of high-fat diet mediated gut microbiota in chronic diseases. iMeta 2023, 2, e69. [Google Scholar] [CrossRef]
- De Bandt, J.P.; Waligora-Dupriet, A.J.; Butel, M.J. Intestinal microbiota in inflammation and insulin resistance: Relevance to humans. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Ju, X.; Chen, W.; Yuan, J.; Wang, Z.; Aluko, R.E.; He, R. Rice bran attenuated obesity via alleviating dyslipidemia, browning of white adipocytes and modulating gut microbiota in high-fat diet-induced obese mice. Food Funct. 2020, 11, 2406–2417. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Anderson, J.R.; Carroll, I.; Azcarate-Peril, M.A.; Rochette, A.D.; Heinberg, L.J.; Peat, C.; Steffen, K.; Manderino, L.M.; Mitchell, J.; Gunstad, J. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med. 2017, 38, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef]
- Chipashvili, O.; Utter, D.R.; Bedree, J.K.; Ma, Y.; Schulte, F.; Mascarin, G.; Alayyoubi, Y.; Chouhan, D.; Hardt, M.; Bidlack, F.; et al. Episymbiotic saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe 2021, 29, 1649–1662.e7. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef]
- Vieira, J.R.P.; Rezende, A.T.d.O.; Fernandes, M.R.; da Silva, N.A. Intestinal microbiota and active systemic lupus erythematosus: A systematic review. Adv. Rheumatol. 2021, 61, 42. [Google Scholar] [CrossRef]
- Le Roy, T.; Moens de Hase, E.; Van Hul, M.; Paquot, A.; Pelicaen, R.; Regnier, M.; Depommier, C.; Druart, C.; Everard, A.; Maiter, D.; et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 2022, 71, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.; Gholami, A.M.; Berry, D.; Desmarchelier, C.; Hahne, H.; Loh, G.; Mondot, S.; Lepage, P.; Rothballer, M.; Walker, A.; et al. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014, 8, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, K.; Itoh, K. Intestinal colonization by a lachnospiraceae bacterium contributes to the development of diabetes in obese mice. Microbes Environ. 2014, 29, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhang, L.; Gao, Y.; Wu, H.; Zhang, J. The dynamic effects of maternal high-calorie diet on glycolipid metabolism and gut microbiota from weaning to adulthood in offspring mice. Front. Nutr. 2022, 9, 941969. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, S.; Zhou, Y.; Yao, Z.; Zhang, D.; Cao, S.; Wei, Z.; Tan, B.; Li, Y.; Lian, Z.; et al. Evolutionary biologic changes of gut microbiota in an ‘adenoma-carcinoma sequence’ mouse colorectal cancer model induced by 1,2-dimethylhydrazine. Oncotarget 2016, 8, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Zhang, X. Bacterial atlas of mouse gut microbiota. Cell. Microbiol. 2022, 2022, 5968814. [Google Scholar] [CrossRef]
- Yan, G.; Li, S.; Wen, Y.; Luo, Y.; Huang, J.; Chen, B.; Lv, S.; Chen, L.; He, L.; He, M.; et al. Characteristics of intestinal microbiota in c57bl/6 mice with non-alcoholic fatty liver induced by high-fat diet. Front. Microbiol. 2022, 13, 1051200. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, K.T.; Enos, R.T.; Bader, J.E.; Sougiannis, A.T.; Carson, M.S.; Chatzistamou, I.; Carson, J.A.; Nagarkatti, P.S.; Nagarkatti, M.; Murphy, E.A. Prolonged high-fat-diet feeding promotes non-alcoholic fatty liver disease and alters gut microbiota in mice. World J. Hepatol. 2019, 11, 619–637. [Google Scholar] [CrossRef]
- Wang, M.; Li, L.; Qian, J.; Wang, N.; Bao, J.; Lu, J.; Chen, F.; Li, Y.; Zhang, Y.; Yan, F. Periodontitis salivary microbiota exacerbates nonalcoholic fatty liver disease in high-fat diet-induced obese mice. iScience 2023, 26, 106346. [Google Scholar] [CrossRef]
- Characteristics of intestinal bacteria with fatty liver diseases and cirrhosis. Ann. Hepatol. 2019, 18, 796–803. [CrossRef]
- Norheim, F.; Chella Krishnan, K.; Bjellaas, T.; Vergnes, L.; Pan, C.; Parks, B.W.; Meng, Y.; Lang, J.; Ward, J.A.; Reue, K.; et al. Genetic regulation of liver lipids in a mouse model of insulin resistance and hepatic steatosis. Mol. Syst. Biol. 2021, 17, e9684. [Google Scholar] [CrossRef]
- Weng, X.; Maxwell-Warburton, S.; Hasib, A.; Ma, L.; Kang, L. The membrane receptor cd44: Novel insights into metabolism. Trends Endocrinol. Metab. 2022, 33, 318–332. [Google Scholar] [CrossRef]
- Bauer, S.; Hezinger, L.; Rexhepi, F.; Ramanathan, S.; Kufer, T.A. Nod-like receptors—Emerging links to obesity and associated morbidities. Int. J. Mol. Sci. 2023, 24, 8595. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Suganami, T.; Nakagawa, N.; Tanaka, M.; Yamamoto, Y.; Kamei, Y.; Terai, S.; Sakaida, I.; Ogawa, Y. Melanocortin 4 receptor-deficient mice as a novel mouse model of nonalcoholic steatohepatitis. Am. J. Pathol. 2011, 179, 2454–2463. [Google Scholar] [CrossRef]
- Vachharajani, V.T.; Liu, T.; Wang, X.; Hoth, J.J.; Yoza, B.K.; McCall, C.E. Sirtuins link inflammation and metabolism. J. Immunol. Res. 2016, 2016, 8167273. [Google Scholar] [CrossRef]
- Fontes-Cal, T.C.M.; Mattos, R.T.; Medeiros, N.I.; Pinto, B.F.; Belchior-Bezerra, M.; Roque-Souza, B.; Dutra, W.O.; Ferrari, T.C.A.; Vidigal, P.V.T.; Faria, L.C.; et al. Crosstalk between plasma cytokines, inflammation, and liver damage as a new strategy to monitoring nafld progression. Front. Immunol. 2021, 12, 708959. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Ma, F.; Guan, M. Role of steroid hormones in the pathogenesis of nonalcoholic fatty liver disease. Metabolites 2021, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Poulsen, K.L.; Wu, L.; Liu, S.; Miyata, T.; Song, Q.; Wei, Q.; Zhao, C.; Lin, C.; Yang, J. Targeted therapeutics and novel signaling pathways in non-alcohol-associated fatty liver/steatohepatitis (nafl/nash). Signal Transduct. Target. Ther. 2022, 7, 287. [Google Scholar] [CrossRef]
- Cani, P.D.; Geurts, L.; Matamoros, S.; Plovier, H.; Duparc, T. Glucose metabolism: Focus on gut microbiota, the endocannabinoid system and beyond. Diabetes Metab. 2014, 40, 246–257. [Google Scholar] [CrossRef]
- Lee, J.; Song, X.; Hyun, B.; Jeon, C.O.; Hyun, S. Drosophila gut immune pathway suppresses host development-promoting effects of acetic acid bacteria. Mol. Cells 2023, 46, 637–653. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.N.; Bonham, K.S.; Ilott, N.E.; Britton, G.J.; Colmenero, P.; Bullers, S.J.; McIver, L.J.; Ma, S.; Nguyen, L.H.; Filer, A.; et al. Alterations in the gut microbiome implicate key taxa and metabolic pathways across inflammatory arthritis phenotypes. Sci. Transl. Med. 2023, 15, eabn4722. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Guo, W.; Xiao, D.; Guan, M.; Liao, T.; Peng, S.; Feng, A.; Wang, Z.; Yin, H.; Li, M.; et al. Microbiota-gut-brain axis drives overeating disorders. Cell Metab. 2023, 35, 2011–2027.e2017. [Google Scholar] [CrossRef] [PubMed]
- Houghton, M.; Kerimi, A.; Mouly, V.; Tumova, S.; Williamson, G. Gut microbiome catabolites as novel modulators of muscle cell glucose metabolism. FASEB J. 2019, 33, 1887–1898. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Ando, Y.; Kimura, I.; Miyamoto, J. Involvement of gut microbial metabolites derived from diet on host energy homeostasis. Int. J. Mol. Sci. 2022, 23, 5562. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.; Rivera, L.; Furness, J.B.; Angus, P.W. The role of the gut microbiota in nafld. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 412–425. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.; Yun, S.Y.; Fu, Z.; Jang, J.M.; Back, M.J.; Kim, H.H.; Kim, D.K. Recombinant human hapln1 mitigates pulmonary emphysema by increasing tgf-β receptor i and sirtuins levels in human alveolar epithelial cells. Mol. Cells 2023, 46, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Gabbia, D.; De Martin, S. Targeting the adipose tissue-liver-gut microbiota crosstalk to cure masld. Biology 2023, 12, 1471. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Kwon, Y.W.; Kim, H.R.; Shin, N.; Son, H.J.; Cheong, C.S.; Kim, D.J.; Hwang, O. A novel pyrazolo[3,4-d]pyrimidine induces heme oxygenase-1 and exerts anti-inflammatory and neuroprotective effects. Mol. Cells 2022, 45, 134–147. [Google Scholar] [CrossRef]
- Han, H.; Wang, M.; Zhong, R.; Yi, B.; Schroyen, M.; Zhang, H. Depletion of gut microbiota inhibits hepatic lipid accumulation in high-fat diet-fed mice. Int. J. Mol. Sci. 2022, 23, 9350. [Google Scholar] [CrossRef]
- Cortez-Pinto, H.; Borralho, P.; Machado, J.; Lopes, M.T.; Gato, I.V.; Santos, A.M.; Guerreiro, A.S. Microbiota modulation with synbiotic decreases liver fibrosis in a high fat choline deficient diet mice model of non-alcoholic steatohepatitis (nash). GE-Port. J. Gastroenterol. 2016, 23, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, T.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut microbiota and nafld: Pathogenetic mechanisms, microbiota signatures, and therapeutic interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Yajima, M.; Karaki, S.I.; Tsuruta, T.; Kimura, S.; Nio-Kobayashi, J.; Kuwahara, A.; Yajima, T. Diversity of the intestinal microbiota differently affects non-neuronal and atropine-sensitive ileal contractile responses to short-chain fatty acids in mice. Biomed. Res. 2016, 37, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Ding, Z.; Ishaq, M.; Bacha, A.S.; Khan, I.; Hanif, A.; Li, W.; Guo, X. Understanding the effects of gut microbiota dysbiosis on nonalcoholic fatty liver disease and the possible probiotics role: Recent updates. Int. J. Biol. Sci. 2021, 17, 818–833. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019, 26, 222–235.e225. [Google Scholar] [CrossRef] [PubMed]
- Foley, K.P.; Zlitni, S.; Duggan, B.M.; Barra, N.G.; Anhê, F.F.; Cavallari, J.F.; Henriksbo, B.D.; Chen, C.Y.; Huang, M.; Lau, T.C.; et al. Gut microbiota impairs insulin clearance in obese mice. Mol. Metab. 2020, 42, 101067. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.D.; Hughes, H.; Kendall, E.; Baron, A.D.; Anderson, C.M. Antiobesity effects of the β-cell hormone amylin in diet-induced obese rats: Effects on food intake, body weight, composition, energy expenditure, and gene expression. Endocrinology 2006, 147, 5855–5864. [Google Scholar] [CrossRef] [PubMed]
- Boey, D.; Lin, S.; Karl, T.; Baldock, P.; Lee, N.; Enriquez, R.; Couzens, M.; Slack, K.; Dallmann, R.; Sainsbury, A.; et al. Peptide yy ablation in mice leads to the development of hyperinsulinaemia and obesity. Diabetologia 2006, 49, 1360–1370. [Google Scholar] [CrossRef]
- El, K.; Gray, S.M.; Capozzi, M.E.; Knuth, E.R.; Jin, E.; Svendsen, B.; Clifford, A.; Brown, J.L.; Encisco, S.E.; Chazotte, B.M.; et al. Gip mediates the incretin effect and glucose tolerance by dual actions on α cells and β cells. Sci. Adv. 2021, 7, eabf1948. [Google Scholar] [CrossRef]
- Li, F.-P.; He, J.; Li, Z.-Z.; Luo, Z.-F.; Yan, L.; Li, Y. Effects of resistin expression on glucose metabolism and hepatic insulin resistance. Endocrine 2009, 35, 243–251. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochumon, S.; Malik, M.Z.; Sindhu, S.; Arefanian, H.; Jacob, T.; Bahman, F.; Nizam, R.; Hasan, A.; Thomas, R.; Al-Rashed, F.; et al. Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice. Nutrients 2024, 16, 1929. https://doi.org/10.3390/nu16121929
Kochumon S, Malik MZ, Sindhu S, Arefanian H, Jacob T, Bahman F, Nizam R, Hasan A, Thomas R, Al-Rashed F, et al. Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice. Nutrients. 2024; 16(12):1929. https://doi.org/10.3390/nu16121929
Chicago/Turabian StyleKochumon, Shihab, Md. Zubbair Malik, Sardar Sindhu, Hossein Arefanian, Texy Jacob, Fatemah Bahman, Rasheeba Nizam, Amal Hasan, Reeby Thomas, Fatema Al-Rashed, and et al. 2024. "Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice" Nutrients 16, no. 12: 1929. https://doi.org/10.3390/nu16121929
APA StyleKochumon, S., Malik, M. Z., Sindhu, S., Arefanian, H., Jacob, T., Bahman, F., Nizam, R., Hasan, A., Thomas, R., Al-Rashed, F., Shenouda, S., Wilson, A., Albeloushi, S., Almansour, N., Alhamar, G., Al Madhoun, A., Alzaid, F., Thanaraj, T. A., Koistinen, H. A., ... Ahmad, R. (2024). Gut Dysbiosis Shaped by Cocoa Butter-Based Sucrose-Free HFD Leads to Steatohepatitis, and Insulin Resistance in Mice. Nutrients, 16(12), 1929. https://doi.org/10.3390/nu16121929










