Pediococcus pentosaceus MIANGUAN2 Alleviates Influenza Virus Infection by Modulating Gut Microbiota and Enhancing Short-Chain Fatty Acid Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria and Virus
2.2. Animals and Ethics Statement
2.3. Experimental Design and Sample Collection
2.4. Measurement of Lung Viral Titers
2.5. Quantitative Real-Time PCR
2.6. Histopathology Analysis of the Lung
2.7. Cytokine Measurement
2.8. Transcriptome Analysis of Lung Tissues
2.9. 16S rRNA Gene Sequencing Analysis of Gut Microbiota
2.10. Metabolome Analysis of Cecal Content
2.11. Statistical Analysis
3. Results
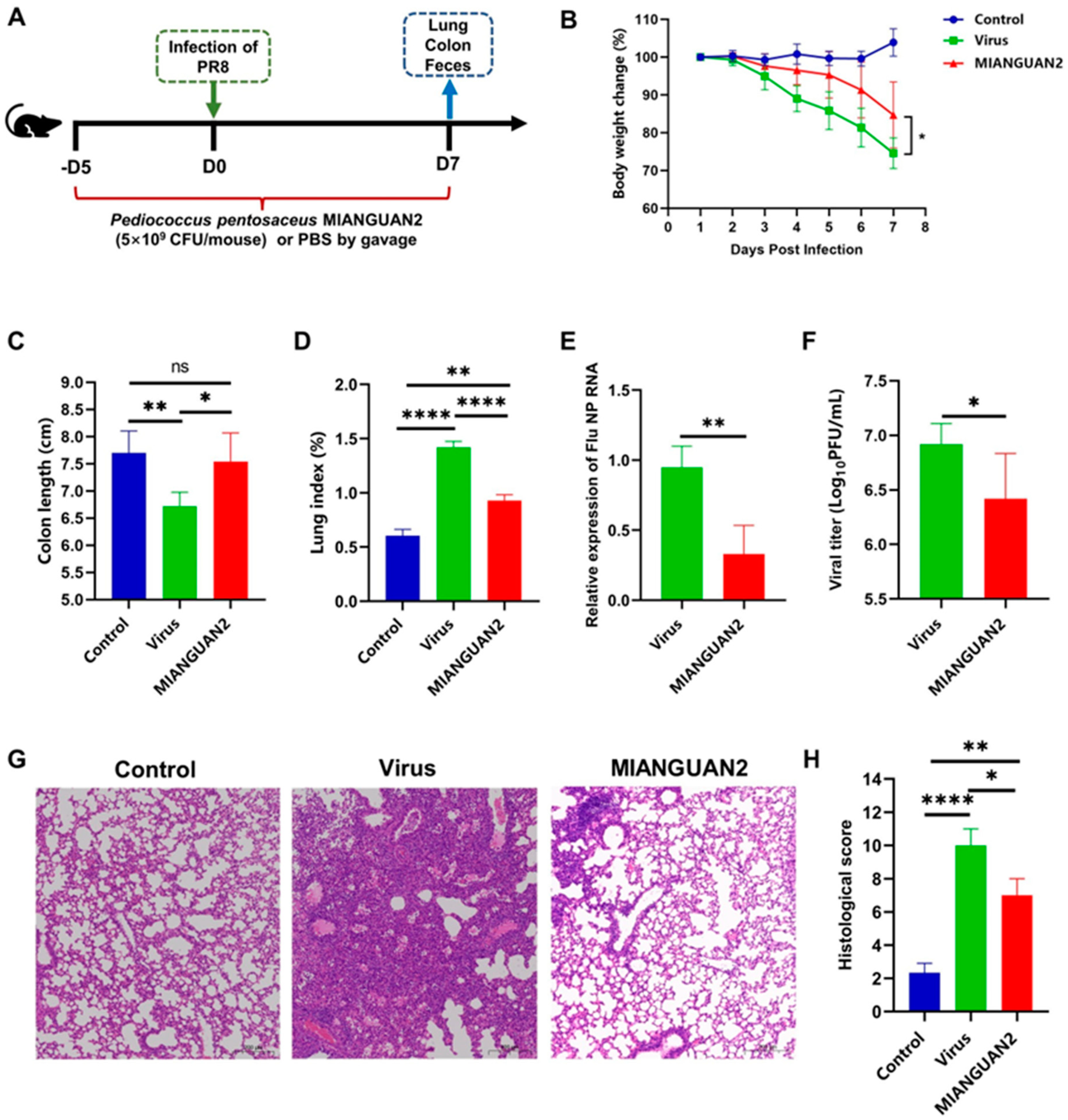
3.1. P. pentosaceus MIANGUAN2 Protects the Host against Influenza Virus Infection
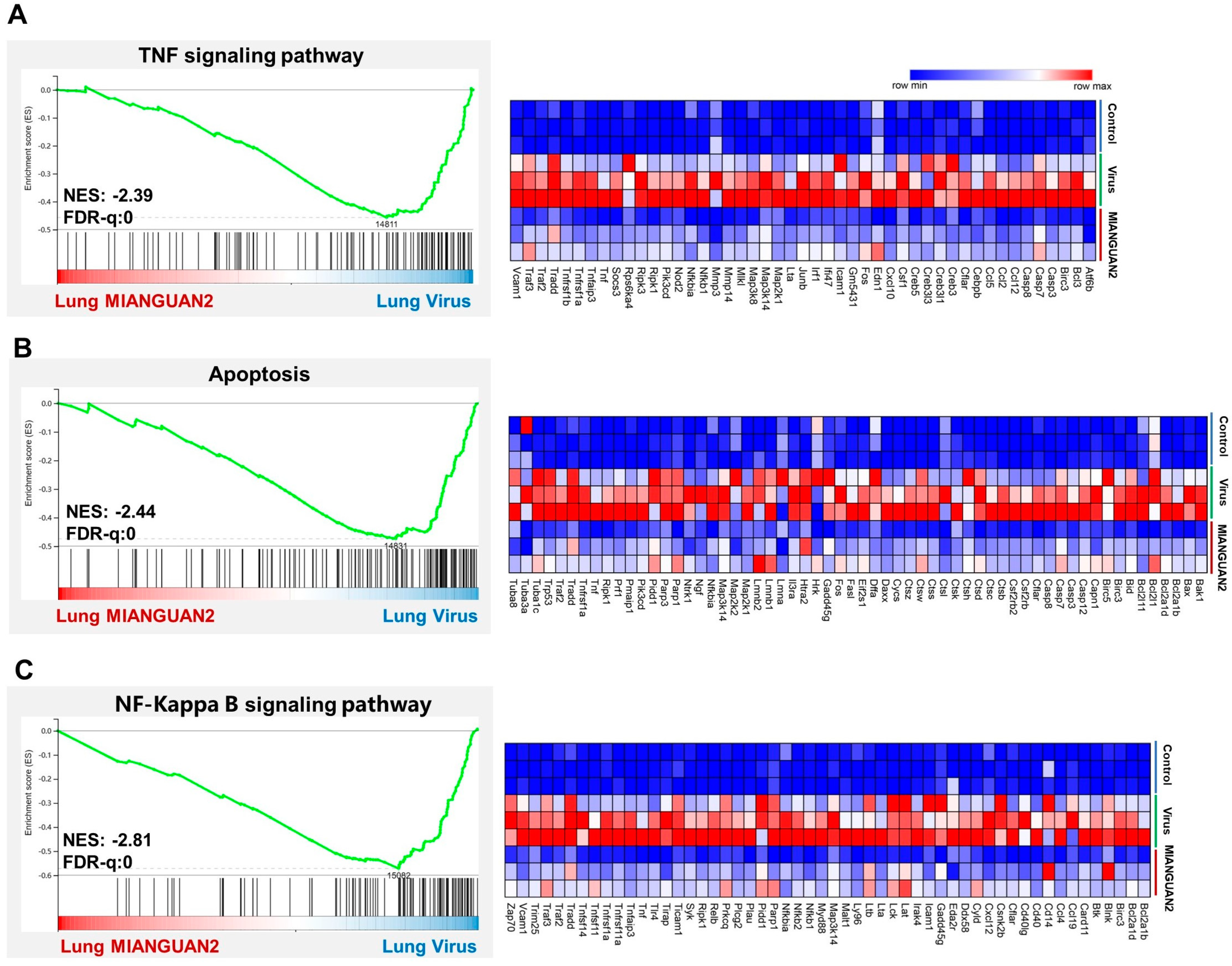
3.2. P. pentosaceus MIANGUAN2 Regulates Cytokine Production and Suppresses the Expression of Multiple Inflammatory Signaling Pathways in the Lung
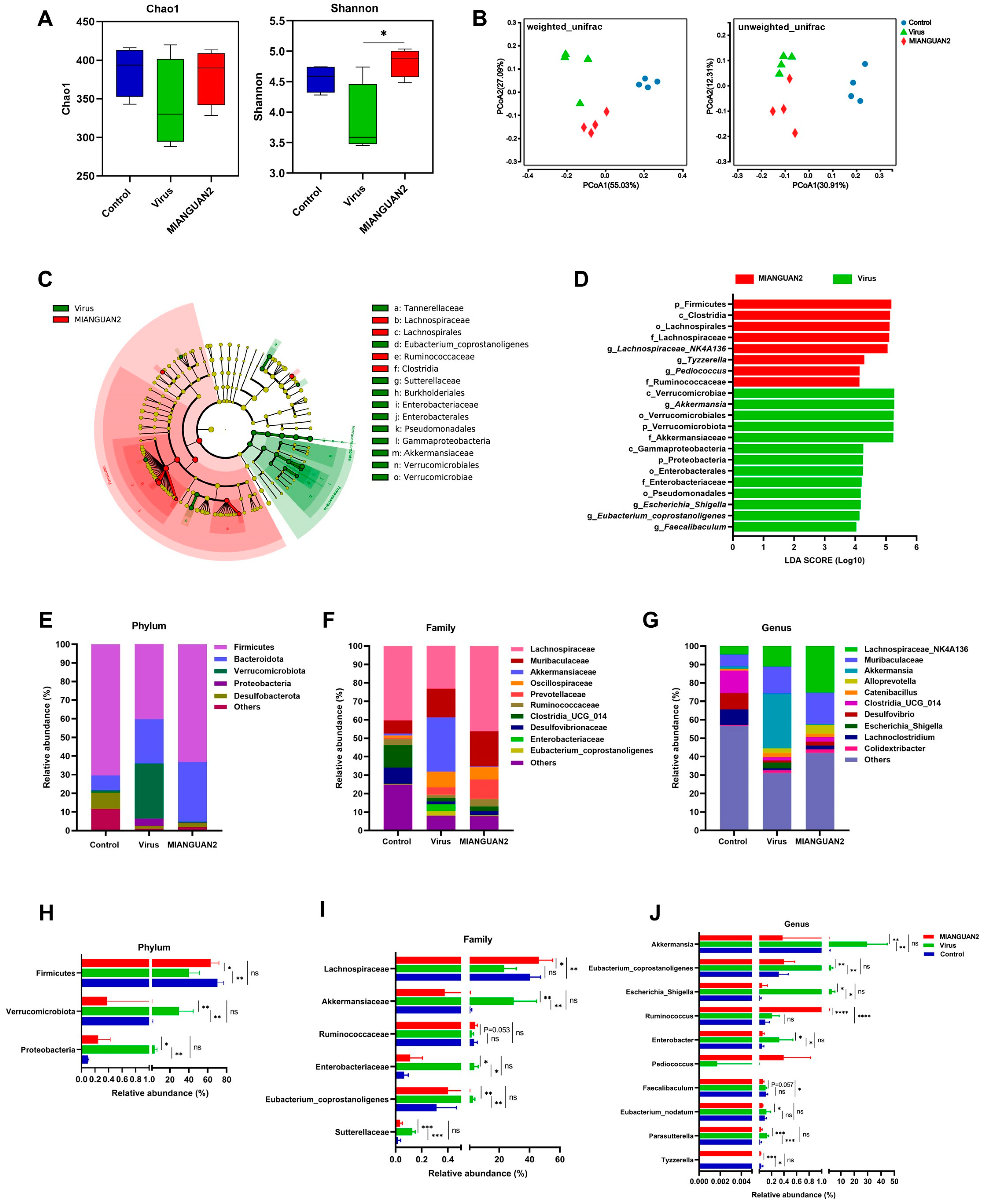
3.3. P. pentosaceus MIANGUAN2 Alters the Gut Microbiota Composition of Influenza Infected-Mice
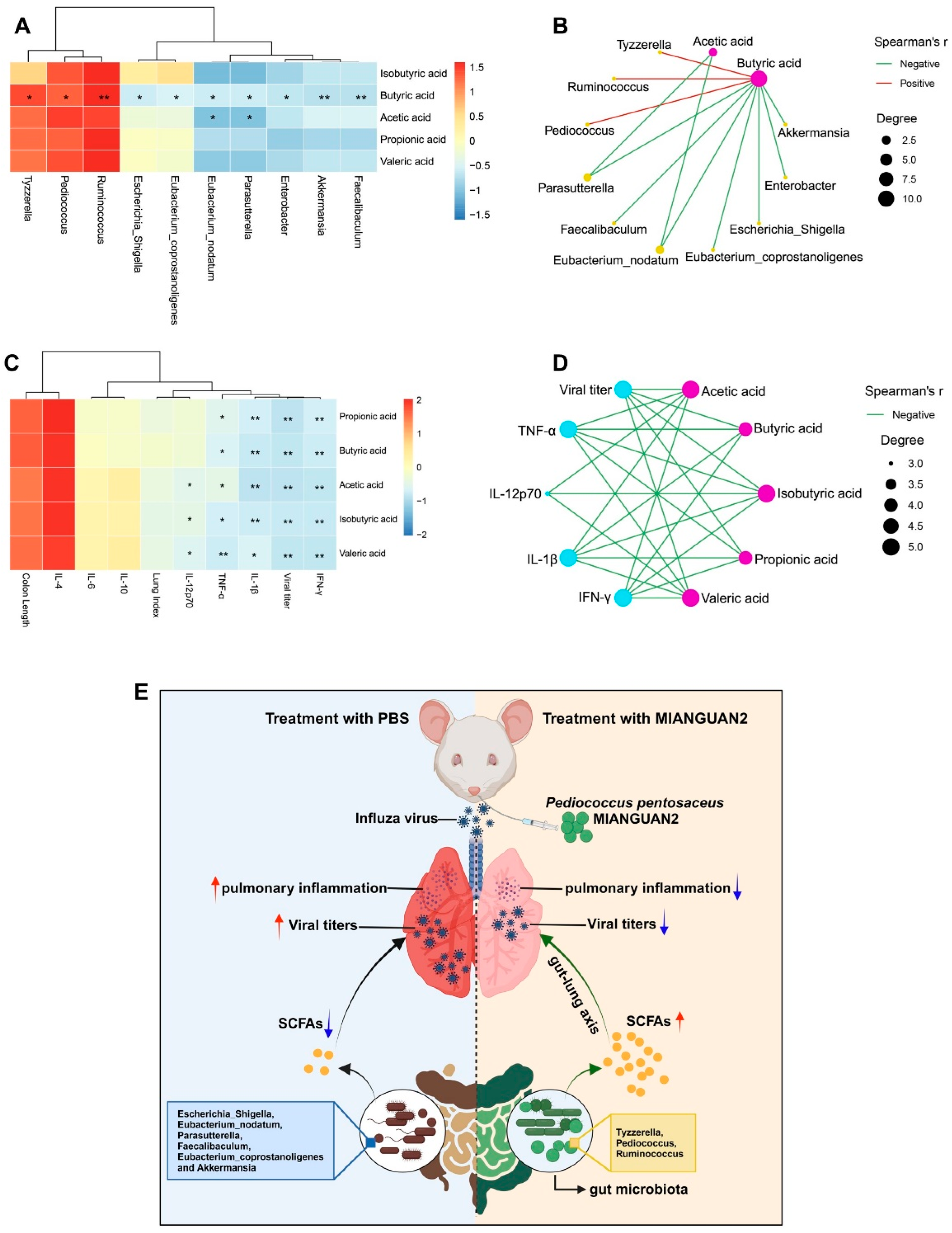
3.4. P. pentosaceus MIANGUAN2 Increases the SCFA Levels in the Feces of Influenza-Infected Mice
3.5. Correlations between SCFAs and the Gut Microbiota or Cytokine Profiles of Lung Tissues or Influenza Infection Phenotype
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keilman, L.J. Seasonal Influenza (Flu). Nurs. Clin. N. Am. 2019, 54, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- Buchy, P.; Badur, S. Who and when to vaccinate against influenza. Int. J. Infect. Dis. 2020, 93, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Sharma, S.D.; Kumar, A.; Ende, Z.; Mishina, M.; Wang, Y.; Falls, Z.; Samudrala, R.; Pohl, J.; Knight, P.R.; et al. Antiviral Approaches against Influenza Virus. Clin. Microbiol. Rev. 2023, 36, e0004022. [Google Scholar] [CrossRef]
- Smyk, J.M.; Szydłowska, N.; Szulc, W.; Majewska, A. Evolution of Influenza Viruses-Drug Resistance, Treatment Options, and Prospects. Int. J. Mol. Sci. 2022, 23, 12244. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Yildiz, S.; Mazel-Sanchez, B.; Kandasamy, M.; Manicassamy, B.; Schmolke, M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 2018, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Sencio, V.; Barthelemy, A.; Tavares, L.P.; Machado, M.G.; Soulard, D.; Cuinat, C.; Queiroz-Junior, C.M.; Noordine, M.L.; Salomé-Desnoulez, S.; Deryuter, L.; et al. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 2020, 30, 2934–2947.e2936. [Google Scholar] [CrossRef]
- Monto, A.S.; Gravenstein, S.; Elliott, M.; Colopy, M.; Schweinle, J. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 2000, 160, 3243–3247. [Google Scholar] [CrossRef]
- Sencio, V.; Gallerand, A.; Gomes Machado, M.; Deruyter, L.; Heumel, S.; Soulard, D.; Barthelemy, J.; Cuinat, C.; Vieira, A.T.; Barthelemy, A.; et al. Influenza Virus Infection Impairs the Gut’s Barrier Properties and Favors Secondary Enteric Bacterial Infection through Reduced Production of Short-Chain Fatty Acids. Infect. Immun. 2021, 89, e0073420. [Google Scholar] [CrossRef]
- Bradley, K.C.; Finsterbusch, K.; Schnepf, D.; Crotta, S.; Llorian, M.; Davidson, S.; Fuchs, S.Y.; Staeheli, P.; Wack, A. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep. 2019, 28, 245–256.e244. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.T. Probiotics. Am. J. Health Syst. Pharm. 2010, 67, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The Gut-Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 2015, 12 (Suppl. S2), S150–S156. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Huang, Y.; Liu, G.; Li, X.; Xiao, Y.; Liu, C.; Zhang, Y.; Li, J.; Xu, J.; Lu, S.; et al. A Novel Immunobiotics Bacteroides dorei Ameliorates Influenza Virus Infection in Mice. Front. Immunol. 2021, 12, 828887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hu, J.; Feng, J.W.; Hu, X.T.; Wang, T.; Gong, W.X.; Huang, K.; Guo, Y.X.; Zou, Z.; Lin, X.; et al. Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol. 2020, 21, 99. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Limaye, A.; Liu, J.R.; Wu, T.N. Potential probiotics for regulation of the gut-lung axis to prevent or alleviate influenza in vulnerable populations. J. Tradit. Complement. Med. 2023, 13, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ou, G.; Xu, H.; Wu, J.; Wang, S.; Chen, Y.; Deng, L.; Chen, X. The gut-lung axis in influenza A: The role of gut microbiota in immune balance. Front. Immunol. 2023, 14, 1147724. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Sun, X.; Zhao, Y.; Iv, C.; Sun, X.; Jin, M.; Zhang, Q. GlcNac produced by the gut microbiome enhances host influenza resistance by modulating NK cells. Gut Microbes 2023, 15, 2271620. [Google Scholar] [CrossRef]
- Steed, A.L.; Christophi, G.P.; Kaiko, G.E.; Sun, L.; Goodwin, V.M.; Jain, U.; Esaulova, E.; Artyomov, M.N.; Morales, D.J.; Holtzman, M.J.; et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 2017, 357, 498–502. [Google Scholar] [CrossRef]
- Niu, J.; Cui, M.; Yang, X.; Li, J.; Yao, Y.; Guo, Q.; Lu, A.; Qi, X.; Zhou, D.; Zhang, C.; et al. Microbiota-derived acetate enhances host antiviral response via NLRP3. Nat. Commun. 2023, 14, 642. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, M.; Yamashita, M.; Ariyoshi, T.; Eguchi, S.; Minemura, A.; Miura, D.; Higashi, S.; Oka, K.; Nonogaki, T.; Mori, T.; et al. Clostridium butyricum-induced ω-3 fatty acid 18-HEPE elicits anti-influenza virus pneumonia effects through interferon-λ upregulation. Cell Rep. 2022, 41, 111755. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Kim, T.Y.; Lee, S.H.; Seo, S.U.; Kweon, M.N. Newly isolated Lactobacillus paracasei strain modulates lung immunity and improves the capacity to cope with influenza virus infection. Microbiome 2023, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- de Azevedo, P.O.S.; Mendonça, C.M.N.; Seibert, L.; Domínguez, J.M.; Converti, A.; Gierus, M.; Oliveira, R.P.S. Bacteriocin-like inhibitory substance of Pediococcus pentosaceus as a biopreservative for Listeria sp. control in ready-to-eat pork ham. Braz. J. Microbiol. 2020, 51, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Gong, P.; Pan, J.; Wang, N.; Tong, J.; Wang, M.; Long, M.; Li, P.; He, J. Pediococcus pentosaceus xy46 Can Absorb Zearalenone and Alleviate its Toxicity to the Reproductive Systems of Male Mice. Microorganisms 2019, 7, 266. [Google Scholar] [CrossRef] [PubMed]
- Higashikawa, F.; Noda, M.; Awaya, T.; Danshiitsoodol, N.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Antiobesity effect of Pediococcus pentosaceus LP28 on overweight subjects: A randomized, double-blind, placebo-controlled clinical trial. Eur. J. Clin. Nutr. 2016, 70, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Ayyash, M.; Abu-Jdayil, B.; Olaimat, A.; Esposito, G.; Itsaranuwat, P.; Osaili, T.; Obaid, R.; Kizhakkayil, J.; Liu, S.Q. Physicochemical, bioactive and rheological properties of an exopolysaccharide produced by a probiotic Pediococcus pentosaceus M41. Carbohydr. Polym. 2020, 229, 115462. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Yang, L.; Wu, W.; Lv, L.; Jiang, X.; Wang, Q.; Wu, J.; Li, Y.; Ye, J.; Fang, D.; et al. Pediococcus pentosaceus LI05 alleviates DSS-induced colitis by modulating immunological profiles, the gut microbiota and short-chain fatty acid levels in a mouse model. Microb. Biotechnol. 2020, 13, 1228–1244. [Google Scholar] [CrossRef]
- Dong, F.; Xiao, F.; Li, X.; Li, Y.; Wang, X.; Yu, G.; Zhang, T.; Wang, Y. Pediococcus pentosaceus CECT 8330 protects DSS-induced colitis and regulates the intestinal microbiota and immune responses in mice. J. Transl. Med. 2022, 20, 33. [Google Scholar] [CrossRef]
- Le, B.; Kim, D.G.; Phuoc, N.N.; Linh, N.T.H.; Yang, S.H. Dietary supplementation with Pediococcus pentosaceus enhances the innate immune response in and promotes growth of Litopenaeus vannamei shrimp. J. Fish Dis. 2022, 45, 1343–1354. [Google Scholar] [CrossRef]
- Huang, J.B.; Wu, Y.C.; Chi, S.C. Dietary supplementation of Pediococcus pentosaceus enhances innate immunity, physiological health and resistance to Vibrio anguillarum in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2014, 39, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cao, Z.; Lu, S.; Wang, Z.; Ma, C.; Zhang, G.; Chen, M.; Yang, J.; Ren, Z.; Xu, J. Pediococcus pentosaceus MIANGUAN Enhances the Immune Response to Vaccination in Mice. Probiotics Antimicrob. Proteins 2024. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Toapanta, F.R.; Ross, T.M. Impaired immune responses in the lungs of aged mice following influenza infection. Respir. Res. 2009, 10, 112. [Google Scholar] [CrossRef] [PubMed]
- Mann, P.C.; Vahle, J.; Keenan, C.M.; Baker, J.F.; Bradley, A.E.; Goodman, D.G.; Harada, T.; Herbert, R.; Kaufmann, W.; Kellner, R.; et al. International harmonization of toxicologic pathology nomenclature: An overview and review of basic principles. Toxicol. Pathol. 2012, 40, 7s–13s. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Kolde, R. pheatmap: Pretty Heatmaps. 2015. Available online: https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf (accessed on 17 March 2024).
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Bai, Y.; Huang, W.; Jiang, X.; Xu, W.; Li, Y.; Wang, Y.; Huang, S.; Wu, K.; Hu, L.; Chen, C. Metabolomic interplay between gut microbiome and plasma metabolome in cardiac surgery-associated acute kidney injury. Rapid Commun. Mass Spectrom. 2023, 37, e9504. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, W. Create Elegant Data Visualisations Using the Grammar of Graphics. R Package ggplot2 Version 3.3.2. 2020. Available online: https://mpn.metworx.com/packages/ggplot2/3.3.2/index.html (accessed on 17 March 2024).
- Revelle, W. psych: Procedures for Psychological, Psychometric, and Personality Research; R Package Version 1.0-95. 2013. Available online: https://CRAN.R-project.org/package=psych (accessed on 17 March 2024).
- Varfolomeev, E.; Vucic, D. Intracellular regulation of TNF activity in health and disease. Cytokine 2018, 101, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Kiraz, Y.; Adan, A.; Kartal Yandim, M.; Baran, Y. Major apoptotic mechanisms and genes involved in apoptosis. Tumour. Biol. 2016, 37, 8471–8486. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Letai, A.; Sarosiek, K. Regulation of apoptosis in health and disease: The balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019, 20, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2014, 74, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Y.; Li, X.; Sun, B. Dynamic balancing of intestinal short-chain fatty acids: The crucial role of bacterial metabolism. Trends Food Sci. Technol. 2020, 100, 118–130. [Google Scholar] [CrossRef]
- Mahooti, M.; Miri, S.M.; Abdolalipour, E.; Ghaemi, A. The immunomodulatory effects of probiotics on respiratory viral infections: A hint for COVID-19 treatment? Microb. Pathog. 2020, 148, 104452. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; He, S.; Yue, K.; Mi, J.; Huang, Y.; Song, L.; Yang, T.; Ren, Z.; Ren, L.; Xu, J. Lactobacillus plantarum GUANKE modulate anti-viral function of dendritic cells in mice. Int. Immunopharmacol. 2024, 134, 112169. [Google Scholar] [CrossRef]
- Xing, J.-H.; Shi, C.-W.; Sun, M.-J.; Gu, W.; Zhang, R.-R.; Chen, H.-L.; Li, Y.; Wang, D.; Li, J.; Niu, T.-M.; et al. Lactiplantibacillus plantarum 0111 Protects against Influenza Virus by Modulating Intestinal Microbial-Mediated Immune Responses. Front. Microbiol. 2022, 13, 820484. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Kumova, O.K.; Fike, A.J.; Thayer, J.L.; Nguyen, L.T.; Mell, J.C.; Pascasio, J.; Stairiker, C.; Leon, L.G.; Katsikis, P.D.; et al. Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal Lactobacillus rhamnosus GG. PLoS Pathog. 2019, 15, e1008072. [Google Scholar] [CrossRef] [PubMed]
- Mahooti, M.; Abdolalipour, E.; Salehzadeh, A.; Mohebbi, S.R.; Gorji, A.; Ghaemi, A. Immunomodulatory and prophylactic effects of Bifidobacterium bifidum probiotic strain on influenza infection in mice. World J. Microbiol. Biotechnol. 2019, 35, 91. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.D.; Simmons, C.P.; Thanh, T.T.; Hien, V.M.; Smith, G.J.; Chau, T.N.; Hoang, D.M.; Chau, N.V.; Khanh, T.H.; Dong, V.C.; et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006, 12, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Rello, J.; Pop-Vicas, A. Clinical review: Primary influenza viral pneumonia. Crit. Care 2009, 13, 235. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Hung, I.F.; Li, I.W.; Lee, K.L.; Koo, C.K.; Yan, W.W.; Liu, R.; Ho, K.Y.; Chu, K.H.; Watt, C.L.; et al. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin. Infect. Dis. 2010, 50, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Al Rumaih, Z.; Kels, M.J.T.; Ng, E.; Kc, R.; Malley, R.; Chaudhri, G.; Karupiah, G. Therapeutic Targeting of Inflammation and Virus Simultaneously Ameliorates Influenza Pneumonia and Protects from Morbidity and Mortality. Viruses 2023, 15, 318. [Google Scholar] [CrossRef] [PubMed]
- Belisle, S.E.; Tisoncik, J.R.; Korth, M.J.; Carter, V.S.; Proll, S.C.; Swayne, D.E.; Pantin-Jackwood, M.; Tumpey, T.M.; Katze, M.G. Genomic profiling of tumor necrosis factor alpha (TNF-alpha) receptor and interleukin-1 receptor knockout mice reveals a link between TNF-alpha signaling and increased severity of 1918 pandemic influenza virus infection. J. Virol. 2010, 84, 12576–12588. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pires, B.R.B.; Silva, R.; Ferreira, G.M.; Abdelhay, E. NF-kappaB: Two Sides of the Same Coin. Genes 2018, 9, 24. [Google Scholar] [CrossRef]
- Ludwig, S.; Planz, O. Influenza viruses and the NF-kappaB signaling pathway—Towards a novel concept of antiviral therapy. Biol. Chem. 2008, 389, 1307–1312. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e1321. [Google Scholar] [CrossRef]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019, 59, 3227–3236. [Google Scholar] [CrossRef] [PubMed]
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut through Type I Interferons. PLoS Pathog. 2016, 12, e1005572. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhao, Y.; Yang, Y.; Gong, W.; Sun, X.; Yang, L.; Zhang, Q.; Jin, M. Akkermansia muciniphila Improves Host Defense against Influenza Virus Infection. Front. Microbiol. 2020, 11, 586476. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Liu, Y.; Wei, S.; Zong, X.; Zhou, G.; Lu, Z.; Wang, F.; Wang, Y.; Jin, M. Dynamic changes of inulin utilization associated with longitudinal development of gut microbiota. Int. J. Biol. Macromol. 2023, 229, 952–963. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl. Environ. Microbiol. 1996, 62, 1589–1592. [Google Scholar] [CrossRef] [PubMed]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2020, 19, 77–94. [Google Scholar] [CrossRef]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014, 5, e00889. [Google Scholar] [CrossRef]
- Ratajczak, W.; Rył, A.; Mizerski, A.; Walczakiewicz, K.; Sipak, O.; Laszczyńska, M. Immunomodulatory potential of gut microbiome-derived shortchain fatty acids (SCFAs). Acta Biochim. Pol. 2019, 66, 1–12. [Google Scholar] [CrossRef]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221. [Google Scholar] [CrossRef] [PubMed]
- Ashique, S.; De Rubis, G.; Sirohi, E.; Mishra, N.; Rihan, M.; Garg, A.; Reyes, R.J.; Manandhar, B.; Bhatt, S.; Jha, N.K.; et al. Short Chain Fatty Acids: Fundamental mediators of the gut-lung axis and their involvement in pulmonary diseases. Chem. Biol. Interact. 2022, 368, 110231. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Mohanty, S.; Sharma, S.; Tripathi, P. Possible role of gut microbes and host’s immune response in gut-lung homeostasis. Front. Immunol. 2022, 13, 954339. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, P.; Santhakumar, P.; Hu, Q.; Djiadeu, P.; Wolever, T.M.S.; Palaniyar, N.; Grasemann, H. Short-chain fatty acids affect cystic fibrosis airway inflammation and bacterial growth. Eur. Respir. J. 2015, 46, 1033. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Pattaroni, C.; Lopez-Mejia, I.C.; Riva, E.; Pernot, J.; Ubags, N.; Fajas, L.; Nicod, L.P.; Marsland, B.J. Dietary Fiber Confers Protection against Flu by Shaping Ly6c− Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity 2018, 48, 992–1005.e1008. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kamiya, S.; Narasaki, S.; Sumii, A.; Tsutsumi, Y.M.; Machida, K.; Hara, K.; Izumi-Mishima, Y.; Tsutsumi, R.; Sakaue, H. Partially Hydrolyzed Guar Gum Intake Supports the Gut Microbiota and Attenuates Inflammation during Influenza H1N1 Virus Infection in Mice. Nutrients 2023, 15, 4252. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Song, L.; Chen, M.; Huang, Y.; Wang, Z.; Ren, Z.; Xu, J. Pediococcus pentosaceus MIANGUAN2 Alleviates Influenza Virus Infection by Modulating Gut Microbiota and Enhancing Short-Chain Fatty Acid Production. Nutrients 2024, 16, 1923. https://doi.org/10.3390/nu16121923
Chen Y, Song L, Chen M, Huang Y, Wang Z, Ren Z, Xu J. Pediococcus pentosaceus MIANGUAN2 Alleviates Influenza Virus Infection by Modulating Gut Microbiota and Enhancing Short-Chain Fatty Acid Production. Nutrients. 2024; 16(12):1923. https://doi.org/10.3390/nu16121923
Chicago/Turabian StyleChen, Yulu, Liqiong Song, Mengshan Chen, Yuanming Huang, Zhihuan Wang, Zhihong Ren, and Jianguo Xu. 2024. "Pediococcus pentosaceus MIANGUAN2 Alleviates Influenza Virus Infection by Modulating Gut Microbiota and Enhancing Short-Chain Fatty Acid Production" Nutrients 16, no. 12: 1923. https://doi.org/10.3390/nu16121923
APA StyleChen, Y., Song, L., Chen, M., Huang, Y., Wang, Z., Ren, Z., & Xu, J. (2024). Pediococcus pentosaceus MIANGUAN2 Alleviates Influenza Virus Infection by Modulating Gut Microbiota and Enhancing Short-Chain Fatty Acid Production. Nutrients, 16(12), 1923. https://doi.org/10.3390/nu16121923





