Vaginal Microbiome in Pregnant Women with and without Short Cervix
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection and DNA Extraction
2.3. Library Preparation and Sequencing
2.4. Amplicon Sequence Variance Assemblage
2.5. 16 S Metabarcoding Statistical Analysis
3. Results
3.1. Population Characteristics
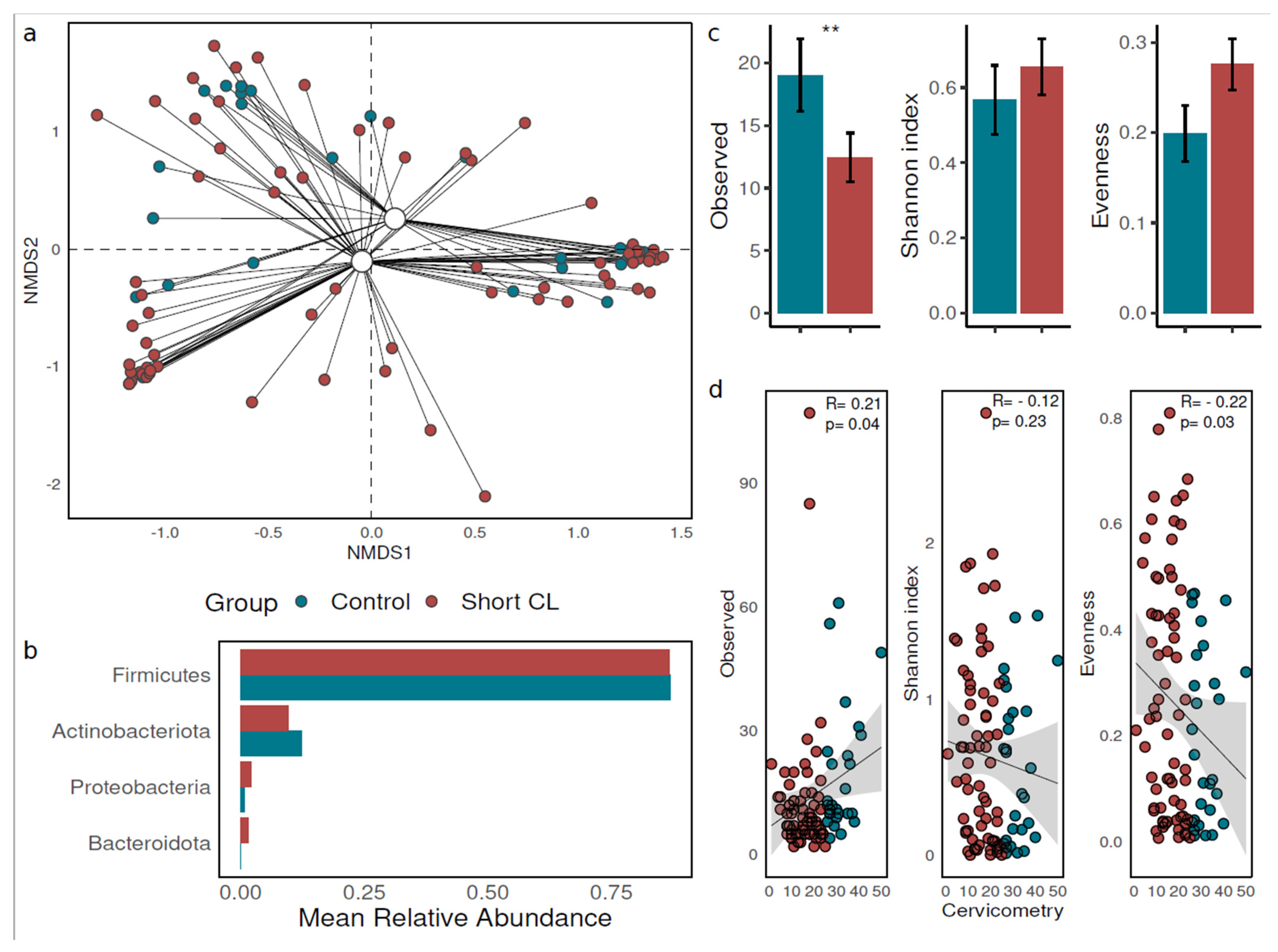
3.2. Vaginal Microbial Community Composition and Diversity
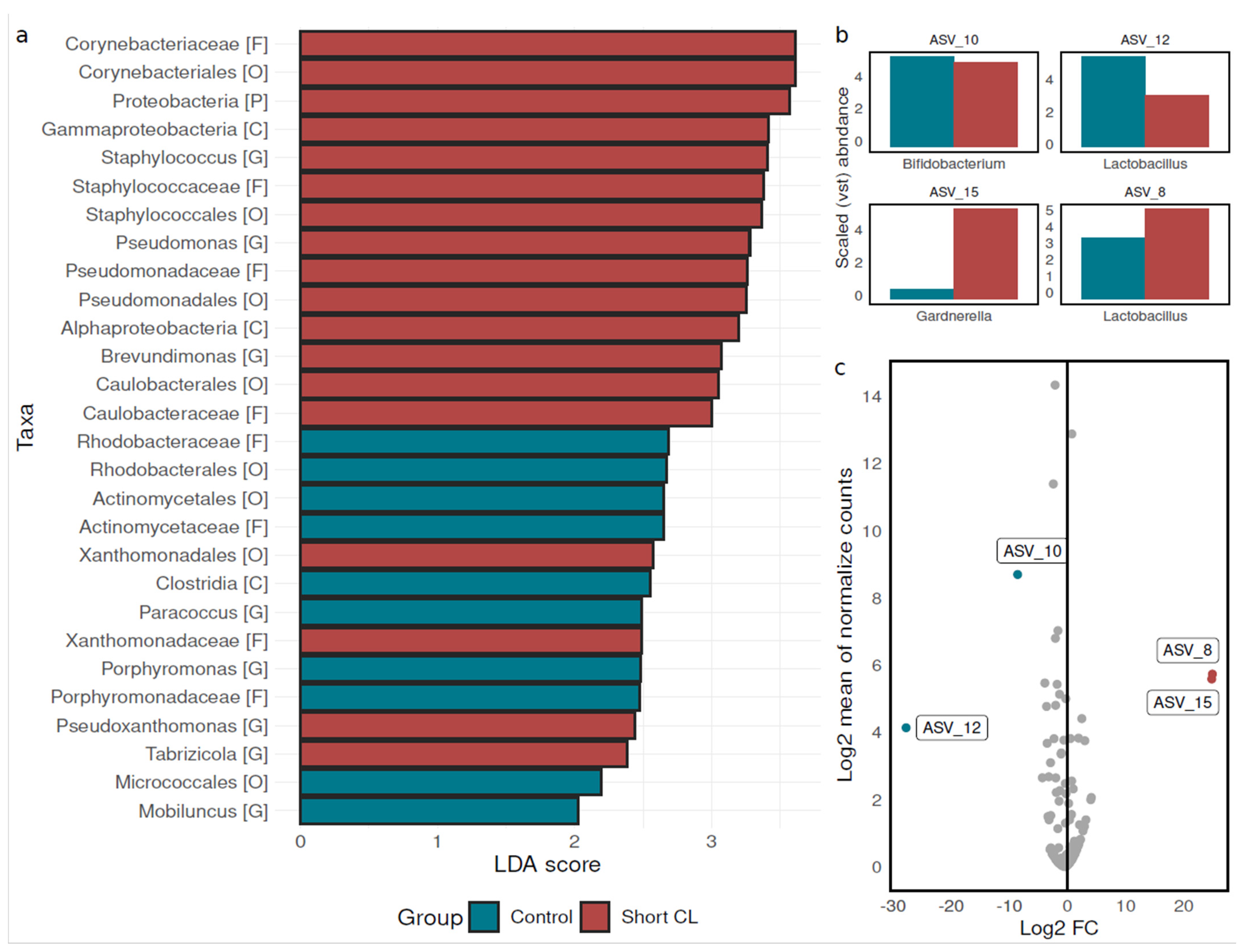
3.3. Bacterial Biomarkers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The Vaginal Microbiome and Preterm Birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Singh, M.P.; Goyal, K. Diversity of Vaginal Microbiome in Pregnancy: Deciphering the Obscurity. Front. Public Health 2020, 8, 326. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, M.L.; Goldkamp, J.M.; Boerrigter, A.; Jakes, C.; Pyon, R.; Vricella, L.K.; Gross, G.A.; Aurora, R. Cervicovaginal Microbiome in Twin vs Singleton Gestations. Am. J. Obstet. Gynecol. MFM 2022, 4, 100579. [Google Scholar] [CrossRef]
- Romero, R.; Hassan, S.S.; Gajer, P.; Tarca, A.L.; Fadrosh, D.W.; Nikita, L.; Galuppi, M.; Lamont, R.F.; Chaemsaithong, P.; Miranda, J.; et al. Correction to: The Composition and Stability of the Vaginal Microbiota of Normal Pregnant Women Is Different from That of Non-Pregnant Women. Microbiome 2014, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Walther-António, M.R.S.; Jeraldo, P.; Berg Miller, M.E.; Yeoman, C.J.; Nelson, K.E.; Wilson, B.A.; White, B.A.; Chia, N.; Creedon, D.J. Pregnancy’s Stronghold on the Vaginal Microbiome. PLoS ONE 2014, 9, e98514. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.C.; Chaban, B.; Bocking, A.; Rocco, M.; Yang, S.; Hill, J.E.; Money, D.M.; Hemmingsen, S.; Reid, G.; Dumonceaux, T.; et al. The Vaginal Microbiome of Pregnant Women Is Less Rich and Diverse, with Lower Prevalence of Mollicutes, Compared to Non-Pregnant Women. Sci. Rep. 2017, 7, 9212. [Google Scholar] [CrossRef]
- Aagaard, K.; Riehle, K.; Ma, J.; Segata, N.; Mistretta, T.A.; Coarfa, C.; Raza, S.; Rosenbaum, S.; van den Veyver, I.; Milosavljevic, A.; et al. A Metagenomic Approach to Characterization of the Vaginal Microbiome Signature in Pregnancy. PLoS ONE 2012, 7, e36466. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.G.; Parikh, H.I.; Brooks, J.P.; Edwards, D.J.; Tom, J.; Edupuganti, L.; Huang, B.; Girerd, P.H.; Bokhari, Y.A.; Bradley, S.P.; et al. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019, 25, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.K.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal Microbiome of Reproductive-Age Women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shawa, G.; et al. Temporal and Spatial Variation of the Human Microbiota during Pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065. [Google Scholar] [CrossRef]
- Ocaña, V.S.; Elena, B.; De Holgado, A.A.P.R.; Nader-Macias, M.E. Surface Characteristics of Lactobacilli Isolated from Human Vagina. J. Gen. Appl. Microbiol. 1999, 45, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Anahtar, M.N.; Byrne, E.H.; Doherty, K.E.; Bowman, B.A.; Yamamoto, H.S.; Soumillon, M.; Padavattan, N.; Ismail, N.; Moodley, A.; Sabatini, M.E.; et al. Cervicovaginal Bacteria Are a Major Modulator of Host Inflammatory Responses in the Female Genital Tract. Immunity 2015, 42, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Torcia, M.G. Interplay among Vaginal Microbiome, Immune Response and Sexually Transmitted Viral Infections. Int. J. Mol. Sci. 2019, 20, 266. [Google Scholar] [CrossRef] [PubMed]
- Hyman, R.W.; Fukushima, M.; Jiang, H.; Fung, E.; Rand, L.; Johnson, B.; Vo, K.C.; Caughey, A.B.; Hilton, J.F.; Davis, R.W.; et al. Diversity of the Vaginal Microbiome Correlates with Preterm Birth. Reprod. Sci. 2014, 21, 32–40. [Google Scholar] [CrossRef]
- Callahan, B.J.; DiGiulio, D.B.; Aliaga Goltsman, D.S.; Sun, C.L.; Costello, E.K.; Jeganathan, P.; Biggio, J.R.; Wong, R.J.; Druzin, M.L.; Shaw, G.M.; et al. Replication and Refinement of a Vaginal Microbial Signature of Preterm Birth in Two Racially Distinct Cohorts of US Women. Proc. Natl. Acad. Sci. USA 2017, 114, 9966–9971. [Google Scholar] [CrossRef]
- Iams, J.D.; Goldenberg, R.L.; Meis, P.J.; Mercer, B.M.; Moawad, A.; Das, A.; Thom, E.; McNellis, D.; Copper, R.L.; Johnson, F.; et al. The Length of the Cervix and the Risk of Spontaneous Premature Delivery. National Institute of Child Health and Human Development Maternal Fetal Medicine Unit Network. N. Engl. J. Med. 1996, 334, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, M.; Berghella, V. Cervical Length for the Prediction and Prevention of Preterm Birth. Expert Rev. Obstet. Gynecol. 2013, 8, 345–355. [Google Scholar] [CrossRef]
- Waldorf, K.M.A.; Singh, N.; Mohan, A.R.; Young, R.C.; Ngo, L.; Das, A.; Tsai, J.; Bansal, A.; Paolella, L.; Herbert, B.R.; et al. Uterine Overdistention Induces Preterm Labor Mediated by Inflammation: Observations in Pregnant Women and Nonhuman Primates. Am. J. Obstet. Gynecol. 2015, 213, 830.e1–830.e19. [Google Scholar] [CrossRef]
- Gerson, K.D.; McCarthy, C.; Elovitz, M.A.; Ravel, J.; Sammel, M.D.; Burris, H.H. Cervicovaginal Microbial Communities Deficient in Lactobacillus Species Are Associated with Second Trimester Short Cervix. Am. J. Obstet. Gynecol. 2020, 222, 491.e1–491.e8. [Google Scholar] [CrossRef]
- Kindinger, L.M.; Bennett, P.R.; Lee, Y.S.; Marchesi, J.R.; Smith, A.; Cacciatore, S.; Holmes, E.; Nicholson, J.K.; Teoh, T.G.; MacIntyre, D.A. The Interaction between Vaginal Microbiota, Cervical Length, and Vaginal Progesterone Treatment for Preterm Birth Risk. Microbiome 2017, 5, 6. [Google Scholar] [CrossRef]
- Di Paola, M.; Seravalli, V.; Paccosi, S.; Linari, C.; Parenti, A.; De Filippo, C.; Tanturli, M.; Vitali, F.; Torcia, M.G.; Di Tommaso, M. Identification of Vaginal Microbial Communities Associated with Extreme Cervical Shortening in Pregnant Women. J. Clin. Med. 2020, 9, 3621. [Google Scholar] [CrossRef]
- Stout, M.J.; Zhou, Y.; Wylie, K.M.; Tarr, P.I.; Macones, G.A.; Tuuli, M.G. Early Pregnancy Vaginal Microbiome Trends and Preterm Birth. Am. J. Obstet. Gynecol. 2017, 217, 356.e1–356.e18. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Peraçoli, J.C.; De Carvalho, R.; Sérgio, C.; De Almeida, H.; Costa, M.; Gustavo, L.; Francisco, D.O.; Pereira, L.; Henri, D.S. Pre-Eclampsia/Eclampsia. Rev. Bras. Ginecol. Obstet. 2019, 41, 318–332. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Shetty, S.A.; Lahti, L. Microbiomeutilities: Utilities for Microbiome Analytics. R Package Version 1.00.16. 2022. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; Wagner, H.; Barbour, M.; Bedward, M.; Bolker, B.; et al. Vegan: Community Ecology Package. 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 15 September 2022).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Vandeputte, D.; De Commer, L.; Tito, R.Y.; Kathagen, G.; Sabino, J.; Vermeire, S.; Faust, K.; Raes, J. Temporal Variability in Quantitative Human Gut Microbiome Profiles and Implications for Clinical Research. Nat. Commun. 2021, 12, 6740. [Google Scholar] [CrossRef]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R Package Version 0.7.0. Computer Software. 2021. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 10 November 2022).
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots (Version 0.1.7). Obtido Desde. 2018. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Tantengco, O.A.G.; Menon, R. Breaking Down the Barrier: The Role of Cervical Infection and Inflammation in Preterm Birth. Front. Glob. Women’s Health 2022, 2, 777643. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; Lamere, B.; Weihe, C.; Wandro, S.; Lindsay, K.L.; Wadhwa, P.D.; Mills, D.A.; Pride, D.T.; Fiehn, O.; Northen, T.; et al. Cervicovaginal Microbiome Composition Is Associated with Metabolic Profiles in Healthy Pregnancy. MBio 2020, 11, e01851-20. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.E. Progesterone and Preterm Birth. Int. J. Gynecol. Obstet. 2020, 150, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Donders, G.G.G.; Bellen, G.; Rezeberga, D. Aerobic Vaginitis in Pregnancy. BJOG 2011, 118, 1163–1170. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas Spp: Emerging Global Opportunistic Pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef]
- Lev-Sagie, A.; De Seta, F.; Verstraelen, H.; Ventolini, G.; Lonnee-Hoffmann, R.; Vieira-Baptista, P. The Vaginal Microbiome: II. Vaginal Dysbiotic Conditions. J. Low. Genit. Tract Dis. 2022, 26, 79–84. [Google Scholar] [CrossRef]
- Krauss-Silva, L.; Almada-Horta, A.; Alves, M.B.; Camacho, K.G.; Moreira, M.E.L.; Braga, A. Basic Vaginal pH, Bacterial Vaginosis and Aerobic Vaginitis: Prevalence in Early Pregnancy and Risk of Spontaneous Preterm Delivery, a Prospective Study in a Low Socioeconomic and Multiethnic South American Population. BMC Pregnancy Childbirth 2014, 14, 107. [Google Scholar] [CrossRef]
- Ncib, K.; Bahia, W.; Leban, N.; Mahdhi, A.; Trifa, F.; Mzoughi, R.; Haddad, A.; Jabeur, C.; Donders, G. Microbial Diversity and Pathogenic Properties of Microbiota Associated with Aerobic Vaginitis in Women with Recurrent Pregnancy Loss. Diagnostics 2022, 12, 2444. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, Y.; Gao, J.; Wu, M.; Li, C.; Wang, Z.; Huang, N.; Cui, L.; Du, M.; Ying, C. Characterization of Vaginal Microbiota in Women With Recurrent Spontaneous Abortion That Can Be Modified by Drug Treatment. Front. Cell. Infect. Microbiol. 2021, 11, 770. [Google Scholar] [CrossRef]
- Ma, X.; Wu, M.; Wang, C.; Li, H.; Fan, A.; Wang, Y.; Han, C.; Xue, F. The Pathogenesis of Prevalent Aerobic Bacteria in Aerobic Vaginitis and Adverse Pregnancy Outcomes: A Narrative Review. Reprod. Health 2022, 19, 21. [Google Scholar] [CrossRef]
- Witkin, S.S.; Moron, A.F.; Linhares, I.M.; Forney, L.J. Influence of Lactobacillus Crispatus, Lactobacillus Iners and Gardnerella Vaginalis on Bacterial Vaginal Composition in Pregnant Women. Arch. Gynecol. Obstet. 2021, 304, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Pacha-Herrera, D.; Erazo-Garcia, M.P.; Cueva, D.F.; Orellana, M.; Borja-Serrano, P.; Arboleda, C.; Tejera, E.; Machado, A. Clustering Analysis of the Multi-Microbial Consortium by Lactobacillus Species Against Vaginal Dysbiosis Among Ecuadorian Women. Front. Cell. Infect. Microbiol. 2022, 12, 428. [Google Scholar] [CrossRef] [PubMed]
- Nicolò, S.; Tanturli, M.; Mattiuz, G.; Antonelli, A.; Baccani, I.; Bonaiuto, C.; Baldi, S.; Nannini, G.; Menicatti, M.; Bartolucci, G.; et al. Vaginal Lactobacilli and Vaginal Dysbiosis-Associated Bacteria Differently Affect Cervical Epithelial and Immune Homeostasis and Anti-Viral Defenses. Int. J. Mol. Sci. 2021, 22, 6487. [Google Scholar] [CrossRef] [PubMed]
- Pausan, M.-R.; Kolovetsiou-Kreiner, V.; Richter, G.L.; Madl, T.; Giselbrecht, E.; Obermayer-Pietsch, B.; Weiss, E.-C.; Jantscher-Krenn, E.; Moissl-Eichinger, C. Human Milk Oligosaccharides Modulate the Risk for Preterm Birth in a Microbiome-Dependent and -Independent Manner. mSystems 2020, 5, e00334-20. [Google Scholar] [CrossRef]
- Castanheira, C.P.; Sallas, M.L.; Nunes, R.A.L.; Lorenzi, N.P.C.; Termini, L. Microbiome and Cervical Cancer. Pathobiology 2021, 88, 187–197. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, W.; Yuan, Y.; Zhu, W.; Shang, A. Vaginal Microecological Characteristics of Women in Different Physiological and Pathological Period. Front. Cell. Infect. Microbiol. 2022, 12, 1071. [Google Scholar] [CrossRef]
- Sarmento, S.G.P.; Moron, A.F.; Forney, L.J.; Hatanaka, A.R.; Carvalho, F.H.C.; França, M.S.; Hamamoto, T.K.; Mattar, R.; Linhares, I.M.; Minis, E.; et al. An Exploratory Study of Associations with Spontaneous Preterm Birth in Primigravid Pregnant Women with a Normal Cervical Length. J. Matern. Fetal. Neonatal Med. 2022, 35, 5383–5388. [Google Scholar] [CrossRef]
| All Women | Cases with Cervical Length ≤ 25 mm | Controls with Cervical Length > 25 mm | p-Value | |
|---|---|---|---|---|
| Number of enrolled pregnant women | 97 | 68 | 29 | / |
| Ethnicity n (%) Caucasian Asian North-African | 95 (98%) 1 (1%) 1 (1%) | 66 (97%) 1 (1.4%) 1(1.4%) | 29 (100%) 0 0 | / |
| Body mass index mean ± SD | 23.6 ± 5.4 | 23.0 ± 4.97 | 24.9 ± 6.1 | 0.16 |
| Smoking * n/nTot (%) | 8/91 (8.8%) | 7/68 (10.3%) | 1/23 (4.3%) | 0.67 |
| Age at sampling (years) mean ± SD | 33.8 ± 6.4 | 33.7 ± 6.4 | 34.0 ± 6.4 | 0.993 |
| Gestational age at sampling (weeks) mean ± SD | 28.4 ± 2.6 | 28.2 ± 2.5 | 28.6 ± 3.0 | 0.775 |
| Length of the cervix at sampling (mm) mean ± SD | 20.0 ± 9.5 | 15.1 ± 5.8 | 31.6 ± 5.7 | <0.001 *** |
| Gestational diabetes mellitus n (%) | 27 (27.8%) | 17 (25%) | 10 (14.7%) | 0.352 |
| Gestational age at delivery (weeks) mean ± SD | 37.6 ± 2.9 | 37.5 ± 2.7 | 37.6 ± 3.3 | 0.145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silvano, A.; Meriggi, N.; Renzi, S.; Seravalli, V.; Torcia, M.G.; Cavalieri, D.; Di Tommaso, M. Vaginal Microbiome in Pregnant Women with and without Short Cervix. Nutrients 2023, 15, 2173. https://doi.org/10.3390/nu15092173
Silvano A, Meriggi N, Renzi S, Seravalli V, Torcia MG, Cavalieri D, Di Tommaso M. Vaginal Microbiome in Pregnant Women with and without Short Cervix. Nutrients. 2023; 15(9):2173. https://doi.org/10.3390/nu15092173
Chicago/Turabian StyleSilvano, Angela, Niccolò Meriggi, Sonia Renzi, Viola Seravalli, Maria Gabriella Torcia, Duccio Cavalieri, and Mariarosaria Di Tommaso. 2023. "Vaginal Microbiome in Pregnant Women with and without Short Cervix" Nutrients 15, no. 9: 2173. https://doi.org/10.3390/nu15092173
APA StyleSilvano, A., Meriggi, N., Renzi, S., Seravalli, V., Torcia, M. G., Cavalieri, D., & Di Tommaso, M. (2023). Vaginal Microbiome in Pregnant Women with and without Short Cervix. Nutrients, 15(9), 2173. https://doi.org/10.3390/nu15092173







