Nrf2-Mediated Pathway Activated by Prunus spinosa L. (Rosaceae) Fruit Extract: Bioinformatics Analyses and Experimental Validation
Abstract
1. Introduction
2. Materials and Methods
2.1. Extract Preparation
2.2. Determination of the Phenolic Composition of P. spinosa Extract by HPLC-DAD-ESI-MSn
2.2.1. Chemicals and Materials
2.2.2. SPE Purification
2.2.3. HPLC-DAD-ESI-MSn Analysis
2.3. The miRNet Analysis
2.4. Chemprop Analysis
2.5. Cell Culture
2.6. Cell Viability
2.7. Fluorescence Analysis of Cells Treated with P. spinosa Extract
2.8. Measurement of Mitochondrial Membrane Potential (MMP)
2.9. Chloromethyl-2′,7′-dichlorodihydrofluorescein Diacetate (DCF) Fluorescence Assay
2.10. Cell Extract Preparation and Western Immunoblotting Analysis
2.11. Nrf2 Activity Assay
2.12. Statistical Analysis
3. Results
3.1. Phenolic Content of P. spinosa Extract
3.2. The miRNet Analysis of P. spinosa Extract
3.3. Chemprop Analysis of Phenolic Compounds of P. spinosa Extract
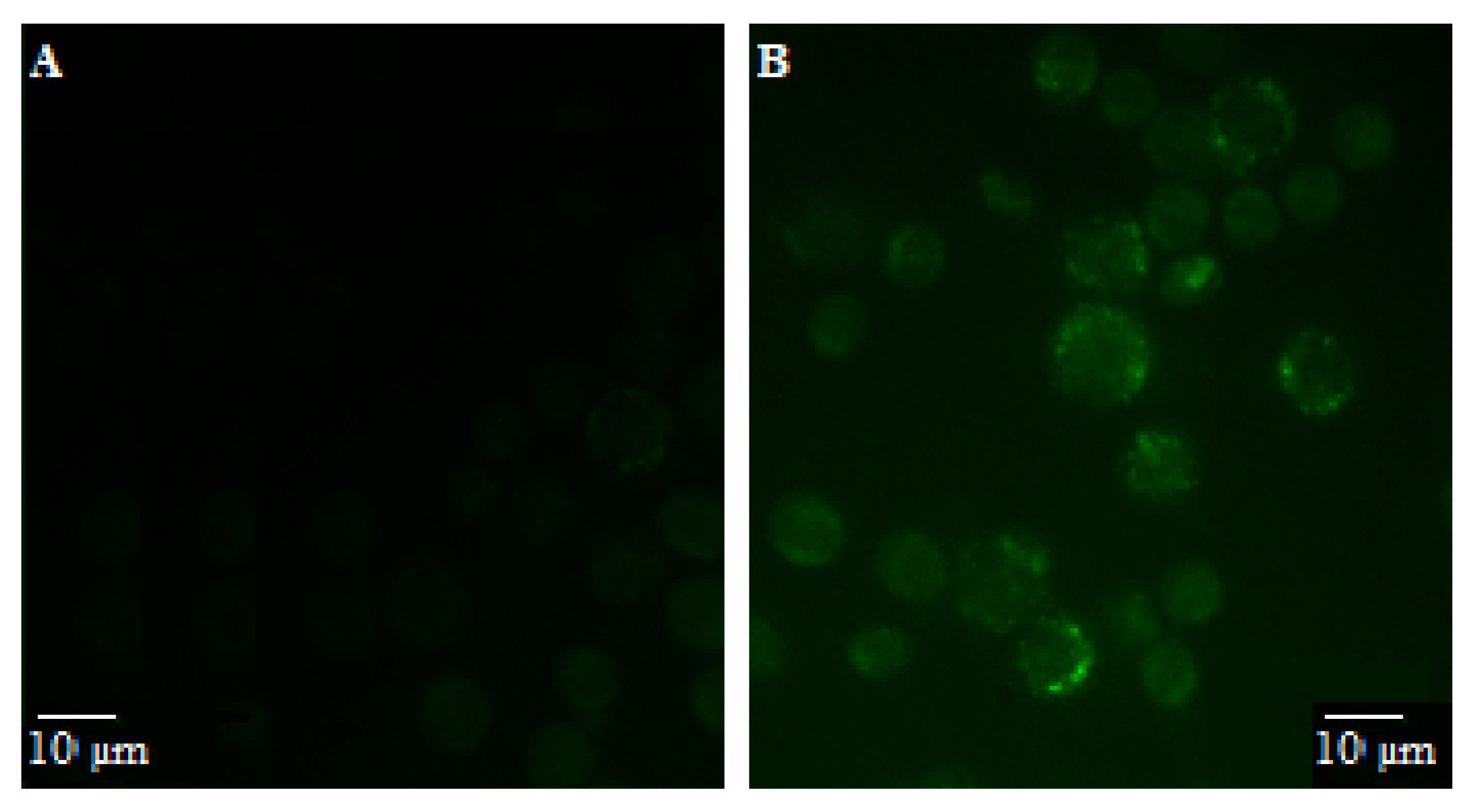
3.4. Internalization of P. spinosa Extract Quercetin Derivatives
3.5. Mitochondrial Membrane Potential (MMP) Evaluation
3.6. Evaluation of the Antioxidant Activity (DCF Analysis)
3.7. Nrf2 Modulation by P. spinosa Extract
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alkadi, H. A Review on Free Radicals and Antioxidants. Infect. Disord. Drug Targets 2020, 20, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.J. Free radicals and redox regulation in ageing. Free. Radic. Biol. Med. 2019, 134, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Gan, R.-Y.; Li, S.; Zhou, Y.; Li, A.-N.; Xu, D.-P.; Li, H.-B. Antioxidant Phytochemicals for the Prevention and Treatment of Chronic Diseases. Molecules 2015, 20, 21138–21156. [Google Scholar] [CrossRef]
- Rahman, M.; Rahaman, S.; Islam, R.; Rahman, F.; Mithi, F.M.; Alqahtani, T.; Almikhlafi, M.A.; Alghamdi, S.Q.; Alruwaili, A.S.; Hossain, S.; et al. Role of Phenolic Compounds in Human Disease: Current Knowledge and Future Prospects. Molecules 2021, 27, 233. [Google Scholar] [CrossRef]
- Popescu, I.; Caudullo, G. Prunus spinosa in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Houston Durrant, T., Mauri, A., Eds.; Publications Office of the European Union: Luxembourg, 2016; p. e018f4e. [Google Scholar]
- Albertini, M.C.; Fraternale, D.; Semprucci, F.; Cecchini, S.; Colomba, M.; Rocchi, M.B.; Sisti, D.; Di Giacomo, B.; Mari, M.; Sabatini, L.; et al. Bioeffects of Prunus spinosa L. fruit ethanol extract on reproduction and phenotypic plasticity of Trichoplax adhaerens Schulze, 1883 (Placozoa). PeerJ 2019, 7, e6789. [Google Scholar] [CrossRef]
- Marchelak, A.; Owczarek, A.; Matczak, M.; Pawlak, A.; Kolodziejczyk-Czepas, J.; Nowak, P.; Olszewska, M.A. Bioactivity Potential of Prunus spinosa L. Flower Extracts: Phytochemical Profiling, Cellular Safety, Pro-inflammatory Enzymes Inhibition and Protective Effects Against Oxidative Stress In Vitro. Front. Pharmacol. 2017, 8, 680. [Google Scholar] [CrossRef]
- Daglia, M. Polyphenols as antimicrobial agents. Curr. Opin. Biotechnol. 2012, 23, 174–181. [Google Scholar] [CrossRef]
- Sabatini, L.; Fraternale, D.; Di Giacomo, B.; Mari, M.; Albertini, M.C.; Gordillo, B.; Rocchi, M.B.L.; Sisti, D.; Coppari, S.; Semprucci, F.; et al. Chemical composition, antioxidant, antimicrobial and anti-inflammatory activity of Prunus spinosa L. fruit ethanol extract. J. Funct. Foods 2020, 67, 103885. [Google Scholar] [CrossRef]
- Coppari, S.; Colomba, M.; Fraternale, D.; Brinkmann, V.; Romeo, M.; Rocchi, M.B.L.; Di Giacomo, B.; Mari, M.; Guidi, L.; Ramakrishna, S.; et al. Antioxidant and Anti-Inflammaging Ability of Prune (Prunus Spinosa L.) Extract Result in Improved Wound Healing Efficacy. Antioxidants 2021, 10, 374. [Google Scholar] [CrossRef]
- Tiboni, M.; Coppari, S.; Casettari, L.; Guescini, M.; Colomba, M.; Fraternale, D.; Gorassini, A.; Verardo, G.; Ramakrishna, S.; Guidi, L.; et al. Prunus spinosa Extract Loaded in Biomimetic Nanoparticles Evokes In Vitro Anti-Inflammatory and Wound Healing Activities. Nanomaterials 2021, 11, 36. [Google Scholar] [CrossRef]
- Kapeta, S.; Chondrogianni, N.; Gonos, E.S. Nuclear Erythroid Factor 2-mediated Proteasome Activation Delays Senescence in Human Fibroblasts. J. Biol. Chem. 2010, 285, 8171–8184. [Google Scholar] [CrossRef]
- Tossetta, G.; Marzioni, D. Targeting the NRF2/KEAP1 pathway in cervical and endometrial cancers. Eur. J. Pharmacol. 2023, 941, 175503. [Google Scholar] [CrossRef]
- Marzioni, D.; Mazzucchelli, R.; Fantone, S.; Tossetta, G. NRF2 modulation in TRAMP mice: An in vivo model of prostate cancer. Mol. Biol. Rep. 2023, 50, 873–881. [Google Scholar] [CrossRef]
- Ghareghomi, S.; Habibi-Rezaei, M.; Arese, M.; Saso, L.; Moosavi-Movahedi, A.A. Nrf2 Modulation in Breast Cancer. Biomedicines 2022, 10, 2668. [Google Scholar] [CrossRef]
- Shaw, P.; Chattopadhyay, A. Nrf2–ARE signaling in cellular protection: Mechanism of action and the regulatory mechanisms. J. Cell. Physiol. 2020, 235, 3119–3130. [Google Scholar] [CrossRef]
- Suraweera, T.L.; Rupasinghe, H.P.V.; Dellaire, G.; Xu, Z. Regulation of Nrf2/ARE Pathway by Dietary Flavonoids: A Friend or Foe for Cancer Management? Antioxidants 2020, 9, 973. [Google Scholar] [CrossRef]
- Bungau, S.; Abdel-Daim, M.M.; Tit, D.M.; Ghanem, E.; Sato, S.; Maruyama-Inoue, M.; Yamane, S.; Kadonosono, K. Health Benefits of Polyphenols and Carotenoids in Age-Related Eye Diseases. Oxidative Med. Cell. Longev. 2019, 2019, 9783429. [Google Scholar] [CrossRef]
- Rafiei, H.; Omidian, K.; Bandy, B. Dietary Polyphenols Protect Against Oleic Acid-Induced Steatosis in an in Vitro Model of NAFLD by Modulating Lipid Metabolism and Improving Mitochondrial Function. Nutrients 2019, 11, 541. [Google Scholar] [CrossRef]
- Yammine, A.; Nury, T.; Vejux, A.; Latruffe, N.; Vervandier-Fasseur, D.; Samadi, M.; Greige-Gerges, H.; Auezova, L.; Lizard, G. Prevention of 7-Ketocholesterol-Induced Overproduction of Reactive Oxygen Species, Mitochondrial Dysfunction and Cell Death with Major Nutrients (Polyphenols, ω3 and ω9 Unsaturated Fatty Acids) of the Mediterranean Diet on N2a Neuronal Cells. Molecules 2020, 25, 2296. [Google Scholar] [CrossRef]
- Chodari, L.; Aytemir, M.D.; Vahedi, P.; Alipour, M.; Vahed, S.Z.; Khatibi, S.M.H.; Ahmadian, E.; Ardalan, M.; Eftekhari, A. Targeting Mitochondrial Biogenesis with Polyphenol Compounds. Oxidative Med. Cell. Longev. 2021, 2021, 4946711. [Google Scholar] [CrossRef]
- Teixeira, J.; Chavarria, D.; Borges, F.; Wojtczak, L.; Wieckowski, M.R.; Karkucinska-Wieckowska, A.; Oliveira, P.J. Dietary Polyphenols and Mitochondrial Function: Role in Health and Disease. Curr. Med. Chem. 2019, 26, 3376–3406. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet—Dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Swanson, K.; Jin, W.; Coley, C.; Eiden, P.; Gao, H.; Guzman-Perez, A.; Hopper, T.; Kelley, B.; Mathea, M.; et al. Analyzing Learned Molecular Representations for Property Prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. [Google Scholar] [CrossRef] [PubMed]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Wu, Z.; Ramsundar, B.; Feinberg, E.N.; Gomes, J.; Geniesse, C.; Pappu, A.S.; Leswing, K.; Pande, V. MoleculeNet: A benchmark for molecular machine learning. Chem. Sci. 2017, 9, 513–530. [Google Scholar] [CrossRef]
- Crinelli, R.; Zara, C.; Galluzzi, L.; Buffi, G.; Ceccarini, C.; Smietana, M.; Mari, M.; Magnani, M.; Fraternale, A. Activation of NRF2 and ATF4 Signaling by the Pro-Glutathione Molecule I-152, A Co-Drug of N-Acetyl-Cysteine and Cysteamine. Antioxidants 2021, 10, 175. [Google Scholar] [CrossRef]
- Nifli, A.-P.; Theodoropoulos, P.A.; Munier, S.; Castagnino, C.; Roussakis, E.; Katerinopoulos, H.E.; Vercauteren, J.; Castanas, E. Quercetin Exhibits a Specific Fluorescence in Cellular Milieu: A Valuable Tool for the Study of Its Intracellular Distribution. J. Agric. Food Chem. 2007, 55, 2873–2878. [Google Scholar] [CrossRef]
- Fiorani, M.; Guidarelli, A.; Blasa, M.; Azzolini, C.; Candiracci, M.; Piatti, E.; Cantoni, O. Mitochondria accumulate large amounts of quercetin: Prevention of mitochondrial damage and release upon oxidation of the extramitochondrial fraction of the flavonoid. J. Nutr. Biochem. 2010, 21, 397–404. [Google Scholar] [CrossRef]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef]
- Konstantinidou, V.; Covas, M.-I.; Solà, R.; Fitó, M. Up-to date knowledge on the in vivo transcriptomic effect of the Mediterranean diet in humans. Mol. Nutr. Food Res. 2013, 57, 772–783. [Google Scholar] [CrossRef]
- Konstantinidou, V.; Covas, M.; Muñoz-Aguayo, D.; Khymenets, O.; de la Torre, R.; Saez, G.; Tormos, M.D.C.; Toledo, E.; Marti, A.; Ruiz-Gutiérrez, V.; et al. In vivo nutrigenomic effects of virgin olive oil polyphenols within the frame of the Mediterranean diet: A randomized controlled trial. FASEB J. 2010, 24, 2546–2557. [Google Scholar] [CrossRef]
- Waterhouse, A.L. Wine Phenolics. Ann. N. Y. Acad. Sci. 2002, 957, 21–36. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Xiang, J.; Wang, C.; Johnson, J.B.; Beta, T. Diverse polyphenol components contribute to antioxidant activity and hypoglycemic potential of mulberry varieties. LWT 2023, 173, 114308. [Google Scholar] [CrossRef]
- Brandes, M.S.; Gray, N.E. NRF2 as a Therapeutic Target in Neurodegenerative Diseases. ASN Neuro 2020, 12, 1759091419899782. [Google Scholar] [CrossRef]
- Cuadrado, A. Brain-Protective Mechanisms of Transcription Factor NRF2: Toward a Common Strategy for Neurodegenerative Diseases. Annu. Rev. Pharmacol. Toxicol. 2022, 62, 255–277. [Google Scholar] [CrossRef]
- Magaña, A.A.; Kamimura, N.; Soumyanath, A.; Stevens, J.F.; Maier, C.S. Caffeoylquinic acids: Chemistry, biosynthesis, occurrence, analytical challenges, and bioactivity. Plant J. 2021, 107, 1299–1319. [Google Scholar] [CrossRef]
- González-Burgos, E.; Gómez-Serranillos, M.P. Effect of Phenolic Compounds on Human Health. Nutrients 2021, 13, 3922. [Google Scholar] [CrossRef]
- Chen, X.; Yang, J.H.; Cho, S.S.; Kim, J.H.; Xu, J.; Seo, K.; Ki, S.H. 5-Caffeoylquinic acid ameliorates oxidative stress-mediated cell death via Nrf2 activation in hepatocytes. Pharm. Biol. 2020, 58, 999–1005. [Google Scholar] [CrossRef]
- Shi, Y.; Liang, X.-C.; Zhang, H.; Wu, Q.-L.; Qu, L.; Sun, Q. Quercetin protects rat dorsal root ganglion neurons against high glucose-induced injury in vitro through Nrf-2/HO-1 activation and NF-κB inhibition. Acta Pharmacol. Sin. 2013, 34, 1140–1148. [Google Scholar] [CrossRef]
- Li, K.; Zhang, M.; Chen, H.; Peng, J.; Jiang, F.; Shi, X.; Bai, Y.; Jian, M.; Jia, Y. Anthocyanins from black peanut skin protect against UV-B induced keratinocyte cell and skin oxidative damage through activating Nrf 2 signaling. Food Funct. 2019, 10, 6815–6828. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Huang, L.; Yu, J. Effects of blueberry anthocyanins on retinal oxidative stress and inflammation in diabetes through Nrf2/HO-1 signaling. J. Neuroimmunol. 2016, 301, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, D.K.; Meena, A.; Dubey, V.; Masood, N.; Luqman, S. Rutin protects t-butyl hydroperoxide-induced oxidative impairment via modulating the Nrf2 and iNOS activity. Phytomedicine 2018, 55, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Ramyaa, P.; Krishnaswamy, R.; Padma, V.V. Quercetin modulates OTA-induced oxidative stress and redox signalling in HepG2 cells—Up regulation of Nrf2 expression and down regulation of NF-κB and COX-2. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 681–692. [Google Scholar] [CrossRef]
- Liang, N.; Kitts, D.D. Amelioration of Oxidative Stress in Caco-2 Cells Treated with Pro-inflammatory Proteins by Chlorogenic Acid Isomers via Activation of the Nrf2–Keap1–ARE-Signaling Pathway. J. Agric. Food Chem. 2018, 66, 11008–11017. [Google Scholar] [CrossRef]
- Schadich, E.; Hlaváč, J.; Volná, T.; Varanasi, L.; Hajdúch, M.; Džubák, P. Effects of Ginger Phenylpropanoids and Quercetin on Nrf2-ARE Pathway in Human BJ Fibroblasts and HaCaT Keratinocytes. BioMed Res. Int. 2016, 2016, 2173275. [Google Scholar] [CrossRef]
- Tian, R.; Yang, Z.; Lu, N.; Peng, Y.-Y. Quercetin, but not rutin, attenuated hydrogen peroxide-induced cell damage via heme oxygenase-1 induction in endothelial cells. Arch. Biochem. Biophys. 2019, 676, 108157. [Google Scholar] [CrossRef]
- Egert, S.; Wolffram, S.; Bosy-Westphal, A.; Boesch-Saadatmandi, C.; Wagner, A.E.; Frank, J.; Rimbach, G.; Mueller, M.J. Daily Quercetin Supplementation Dose-Dependently Increases Plasma Quercetin Concentrations in Healthy Humans. J. Nutr. 2008, 138, 1615–1621. [Google Scholar] [CrossRef]
- Erlund, I.; Kosonen, T.; Alfthan, G.; Mäenpää, J.; Perttunen, K.; Kenraali, J.; Parantainen, J.; Aro, A. Pharmacokinetics of quercetin from quercetin aglycone and rutin in healthy volunteers. Eur. J. Clin. Pharmacol. 2000, 56, 545–553. [Google Scholar] [CrossRef]
- Sun, L.; Xu, G.; Dong, Y.; Li, M.; Yang, L.; Lu, W. Quercetin Protects against Lipopolysaccharide-Induced Intestinal Oxidative Stress in Broiler Chickens through Activation of Nrf2 Pathway. Molecules 2020, 25, 1053. [Google Scholar] [CrossRef]
- Liu, Y.-W.; Liu, X.-L.; Kong, L.; Zhang, M.-Y.; Chen, Y.-J.; Zhu, X.; Hao, Y.-C. Neuroprotection of quercetin on central neurons against chronic high glucose through enhancement of Nrf2/ARE/glyoxalase-1 pathway mediated by phosphorylation regulation. Biomed. Pharmacother. 2018, 109, 2145–2154. [Google Scholar] [CrossRef]
- Huang, C.-S.; Lii, C.-K.; Lin, A.-H.; Yeh, Y.-W.; Yao, H.-T.; Li, C.-C.; Wang, T.-S.; Chen, H.-W. Protection by chrysin, apigenin, and luteolin against oxidative stress is mediated by the Nrf2-dependent up-regulation of heme oxygenase 1 and glutamate cysteine ligase in rat primary hepatocytes. Arch. Toxicol. 2012, 87, 167–178. [Google Scholar] [CrossRef]
- Kitakaze, T.; Makiyama, A.; Yamashita, Y.; Ashida, H. Low dose of luteolin activates Nrf2 in the liver of mice at start of the active phase but not that of the inactive phase. PLoS ONE 2020, 15, e0231403. [Google Scholar] [CrossRef]
- Dong, Y.; Xing, Y.; Sun, J.; Sun, W.; Xu, Y.; Quan, C. Baicalein Alleviates Liver Oxidative Stress and Apoptosis Induced by High-Level Glucose through the Activation of the PERK/Nrf2 Signaling Pathway. Molecules 2020, 25, 599. [Google Scholar] [CrossRef]
- Tan, X.; Yang, Y.; Xu, J.; Zhang, P.; Deng, R.; Mao, Y.; He, J.; Chen, Y.; Zhang, Y.; Ding, J.; et al. Luteolin Exerts Neuroprotection via Modulation of the p62/Keap1/Nrf2 Pathway in Intracerebral Hemorrhage. Front. Pharmacol. 2020, 10, 1551. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Z.; Wu, S.; Li, X.; Sang, Y.; Li, J.; Niu, Y.; Ding, H. Myricetin alleviates cuprizone-induced behavioral dysfunction and demyelination in mice by Nrf2 pathway. Food Funct. 2016, 7, 4332–4342. [Google Scholar] [CrossRef]

| No. | Targets | Description |
|---|---|---|
| 1 | NR-AR | assay to identify small molecule agonists of the androgen receptor (AR) signaling pathway using the MDA cell line |
| 2 | NR-AR-LBD | assay to identify small molecule agonists of the androgen receptor (AR) signaling pathway |
| 3 | NR-AhR | assay to identify small molecules that activate the aryl hydrocarbon receptor (AhR) signaling pathway |
| 4 | NR-Aromatase | assay to identify aromatase inhibitors |
| 5 | NR-ER | assay to identify small molecule agonists of the estrogen receptor alpha (ER-alpha) signaling pathway using the BG1 cell line |
| 6 | NR-ER-LBD | assay to identify small molecule agonists of the estrogen receptor alpha (ER-alpha) signaling pathway |
| 7 | NR-PPAR-gamma | assay to identify small molecule agonists of the peroxisome proliferator-activated receptor gamma (PPARg) signaling pathway |
| 8 | SR-ARE | assay for small molecule agonists of the antioxidant response element (ARE) signaling pathway |
| 9 | SR-ATAD5 | assay for small molecules that induce genotoxicity in human embryonic kidney cells expressing luciferase-tagged ATAD5 |
| 10 | SR-HSE | assay for small molecule activators of the heat shock response signaling pathway |
| 11 | SR-MMP | assay for small molecule disruptors of the mitochondrial membrane potential |
| 12 | SR-p53 | assay for small molecule agonists of the p53 signaling pathway |
| No | Compound | Content (μg/g dw) | Mean Content (%) |
|---|---|---|---|
| 1 | 3-O-Caffeoylquinic acid (3-CQA) | 4003.53 ± 16.17 | 48.41 |
| 2 | 3-O-p-Coumaroylquinic acid (3-p-CoQA) | 199.83 ± 2.19 | 2.42 |
| 3 | Caffeoylquinic acid dehydrodimer | 39.21 ± 0.24 | 0.47 |
| 4 | 3-O-Feruloylquinic acid (3-FQA) | 196.73 ± 2.43 | 2.38 |
| 5 | 4-O-Caffeoylquinic acid (4-CQA) | 299.96 ± 1.98 | 3.63 |
| 6 | Caffeoylquinic acid dehydrodimer isomer | 134.21 ± 2.03 | 1.62 |
| ∑ Hydroxycinnamic acid derivatives | 4873.42 ± 26.12 | 58.93 | |
| 7 | Cyanidin 3-O-glucoside | 564.77 ± 6.80 | 6.83 |
| 8 | Cyanidin 3-O-rutinoside | 856.68 ± 9.16 | 10.36 |
| 9 | Peonidin 3-O-glucoside | 203.47 ± 1.18 | 2.46 |
| 10 | Peonidin 3-O-rutinoside | 752.20 ± 10.47 | 9.10 |
| ∑ Anthocyanins | 2377.12 ± 6.67 | 28.75 | |
| 11 | 4-(vanilloyloxy)-2,6,6-trimethylcyclohexene-1-carboxylic acid | 7.83 ± 0.15 | 0.09 |
| ∑ Hydroxybenzoic acid derivatives | 7.83 ± 0.15 | 0.09 | |
| 12 | Apigenin pentoside | 6.05 ± 0.08 | 0.07 |
| 13 | Apigenin pentoside isomer | 10.00 ± 0.19 | 0.12 |
| 14 | Quercetin-hexoside-pentoside | 75.05 ± 1.15 | 0.91 |
| 15 | Rutin | 167.49 ± 1.07 | 2.03 |
| 16 | Quercetin 3-O-galactoside | 222.91 ± 3.44 | 2.70 |
| 17 | Quercetin 3-O-xyloside | 64.54 ± 0.78 | 0.78 |
| 18 | Quercetin 3-O-arabinoside | 84.47 ± 0.56 | 1.02 |
| 19 | Quercetin pentoside | 284.72 ± 1.12 | 3.44 |
| 20 | Quercetin 3-O-rhamnoside | 95.75 ± 0.32 | 1.16 |
| ∑ Flavonoid derivatives | 1010.97 ± 8.60 | 12.23 | |
| Total phenolic compounds | 8269.33 ± 41.54 |
| No | Compound | NR-AR | NR-AR-LBD | NR-AhR | NR-Aromatase | NR-ER | NR-ER-LBD | NR-PPAR-gamma | SR-ARE | SR-ATAD5 | SR-HSE | SR-MMP | SR-p53 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3-O-Caffeoylquinic acid (3-CQA) | 0.0532 | 0.0376 | 0.0502 | 0.0132 | 0.1047 | 0.0563 | 0.1382 | 0.1536 | 0.0801 | 0.0630 | 0.0620 | 0.1135 |
| 2 | 3-O-p-Coumaroylquinic acid (3-p-CoQA) | 0.1437 | 0.1448 | 0.0215 | 0.0427 | 0.2578 | 0.0927 | 0.1310 | 0.2224 | 0.0963 | 0.0519 | 0.1232 | 0.1615 |
| 3 | Caffeoylquinic acid dehydrodimer | 0.0567 | 0.0508 | 0.0681 | 0.0344 | 0.1281 | 0.0703 | 0.2012 | 0.1889 | 0.1135 | 0.0801 | 0.1115 | 0.2238 |
| 4 | 3-O-Feruloylquinic acid (3-FQA) | 0.1070 | 0.0874 | 0.0333 | 0.0284 | 0.1441 | 0.0530 | 0.1064 | 0.1739 | 0.0833 | 0.0497 | 0.0779 | 0.1357 |
| 5 | 4-O-Caffeoylquinic acid (4-CQA) | 0.0654 | 0.0636 | 0.0546 | 0.0290 | 0.1437 | 0.0851 | 0.1920 | 0.2689 | 0.1382 | 0.0993 | 0.1301 | 0.2166 |
| 6 | Caffeoylquinic acid dehydrodimer isomer | 0.0567 | 0.0508 | 0.0681 | 0.0344 | 0.1281 | 0.0703 | 0.2012 | 0.1889 | 0.1135 | 0.0801 | 0.1115 | 0.2238 |
| 7 | Cyanidin 3-O-glucoside | 0.0705 | 0.0826 | 0.2448 | 0.1242 | 0.3776 | 0.2138 | 0.0850 | 0.4916 | 0.1196 | 0.0986 | 0.6072 | 0.5227 |
| 8 | Cyanidin 3-O-rutinoside | 0.0319 | 0.0348 | 0.1342 | 0.0545 | 0.2239 | 0.1055 | 0.0837 | 0.3123 | 0.0979 | 0.0571 | 0.1884 | 0.3870 |
| 9 | Peonidin 3-O-glucoside | 0.0707 | 0.0771 | 0.2316 | 0.1255 | 0.3334 | 0.1616 | 0.0890 | 0.4232 | 0.1151 | 0.0799 | 0.5015 | 0.4953 |
| 10 | Peonidin 3-O-rutinoside | 0.0460 | 0.0533 | 0.0892 | 0.0745 | 0.2964 | 0.1192 | 0.0764 | 0.2642 | 0.0918 | 0.0448 | 0.2428 | 0.4113 |
| 11 | 4-(vanilloyloxy)-2,6,6-trimethylcyclohexene-1-carboxylic acid | 0.0149 | 0.0061 | 0.0826 | 0.0364 | 0.0798 | 0.0504 | 0.0578 | 0.1316 | 0.0368 | 0.0775 | 0.2620 | 0.0619 |
| 12 | Apigenin pentoside | 0.0595 | 0.0764 | 0.2636 | 0.1116 | 0.3539 | 0.1957 | 0.2096 | 0.4552 | 0.2228 | 0.1288 | 0.4121 | 0.5178 |
| 13 | Apigenin pentoside isomer | 0.0595 | 0.0764 | 0.2636 | 0.1116 | 0.3539 | 0.1957 | 0.2096 | 0.4552 | 0.2228 | 0.1288 | 0.4121 | 0.5178 |
| 14 | Quercetin hexoside-pentoside | 0.0325 | 0.0316 | 0.1042 | 0.0398 | 0.1810 | 0.0859 | 0.0843 | 0.2824 | 0.0914 | 0.0495 | 0.1301 | 0.3472 |
| 15 | Rutin | 0.0790 | 0.0885 | 0.0538 | 0.0705 | 0.4809 | 0.2301 | 0.0569 | 0.2308 | 0.0768 | 0.0366 | 0.3834 | 0.4153 |
| 16 | Quercetin 3-O-galactoside | 0.0771 | 0.1047 | 0.2179 | 0.1346 | 0.4228 | 0.2718 | 0.0977 | 0.4850 | 0.1404 | 0.1121 | 0.6302 | 0.5602 |
| 17 | Quercetin 3-O-xyloside | 0.0785 | 0.1257 | 0.3414 | 0.1941 | 0.4162 | 0.2803 | 0.1449 | 0.6347 | 0.2174 | 0.1848 | 0.7389 | 0.6537 |
| 18 | Quercetin 3-O-arabinoside | 0.0705 | 0.1062 | 0.2916 | 0.1449 | 0.4227 | 0.3137 | 0.1225 | 0.5764 | 0.1832 | 0.1581 | 0.6973 | 0.6104 |
| 19 | Quercetin pentoside | 0.0494 | 0.0731 | 0.4738 | 0.1447 | 0.2836 | 0.1910 | 0.1928 | 0.6568 | 0.2507 | 0.2220 | 0.6121 | 0.6019 |
| 20 | Quercetin 3-O-rhamnoside | 0.0744 | 0.0999 | 0.2808 | 0.1601 | 0.4283 | 0.2713 | 0.1035 | 0.5510 | 0.1595 | 0.1420 | 0.7367 | 0.5956 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colomba, M.; Benedetti, S.; Fraternale, D.; Guidarelli, A.; Coppari, S.; Freschi, V.; Crinelli, R.; Kass, G.E.N.; Gorassini, A.; Verardo, G.; et al. Nrf2-Mediated Pathway Activated by Prunus spinosa L. (Rosaceae) Fruit Extract: Bioinformatics Analyses and Experimental Validation. Nutrients 2023, 15, 2132. https://doi.org/10.3390/nu15092132
Colomba M, Benedetti S, Fraternale D, Guidarelli A, Coppari S, Freschi V, Crinelli R, Kass GEN, Gorassini A, Verardo G, et al. Nrf2-Mediated Pathway Activated by Prunus spinosa L. (Rosaceae) Fruit Extract: Bioinformatics Analyses and Experimental Validation. Nutrients. 2023; 15(9):2132. https://doi.org/10.3390/nu15092132
Chicago/Turabian StyleColomba, Mariastella, Serena Benedetti, Daniele Fraternale, Andrea Guidarelli, Sofia Coppari, Valerio Freschi, Rita Crinelli, George E. N. Kass, Andrea Gorassini, Giancarlo Verardo, and et al. 2023. "Nrf2-Mediated Pathway Activated by Prunus spinosa L. (Rosaceae) Fruit Extract: Bioinformatics Analyses and Experimental Validation" Nutrients 15, no. 9: 2132. https://doi.org/10.3390/nu15092132
APA StyleColomba, M., Benedetti, S., Fraternale, D., Guidarelli, A., Coppari, S., Freschi, V., Crinelli, R., Kass, G. E. N., Gorassini, A., Verardo, G., Roselli, C., Meli, M. A., Di Giacomo, B., & Albertini, M. C. (2023). Nrf2-Mediated Pathway Activated by Prunus spinosa L. (Rosaceae) Fruit Extract: Bioinformatics Analyses and Experimental Validation. Nutrients, 15(9), 2132. https://doi.org/10.3390/nu15092132






