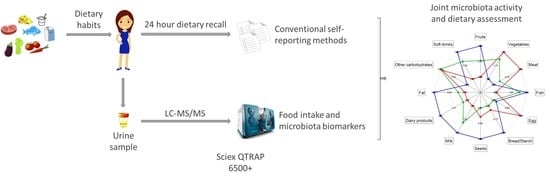
Joint Microbiota Activity and Dietary Assessment through Urinary Biomarkers by LC-MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design, Population, and Sample Collection
2.2. Dietary Assessment
2.3. Standards and Reagents
2.4. UHPLC-MS/MS Determination of Nutrition and Microbiota Biomarkers
2.5. Method Validation
2.6. Data Availability and Statistical Analysis
3. Results and Discussion
3.1. UHPLC-MS/MS Method Validation
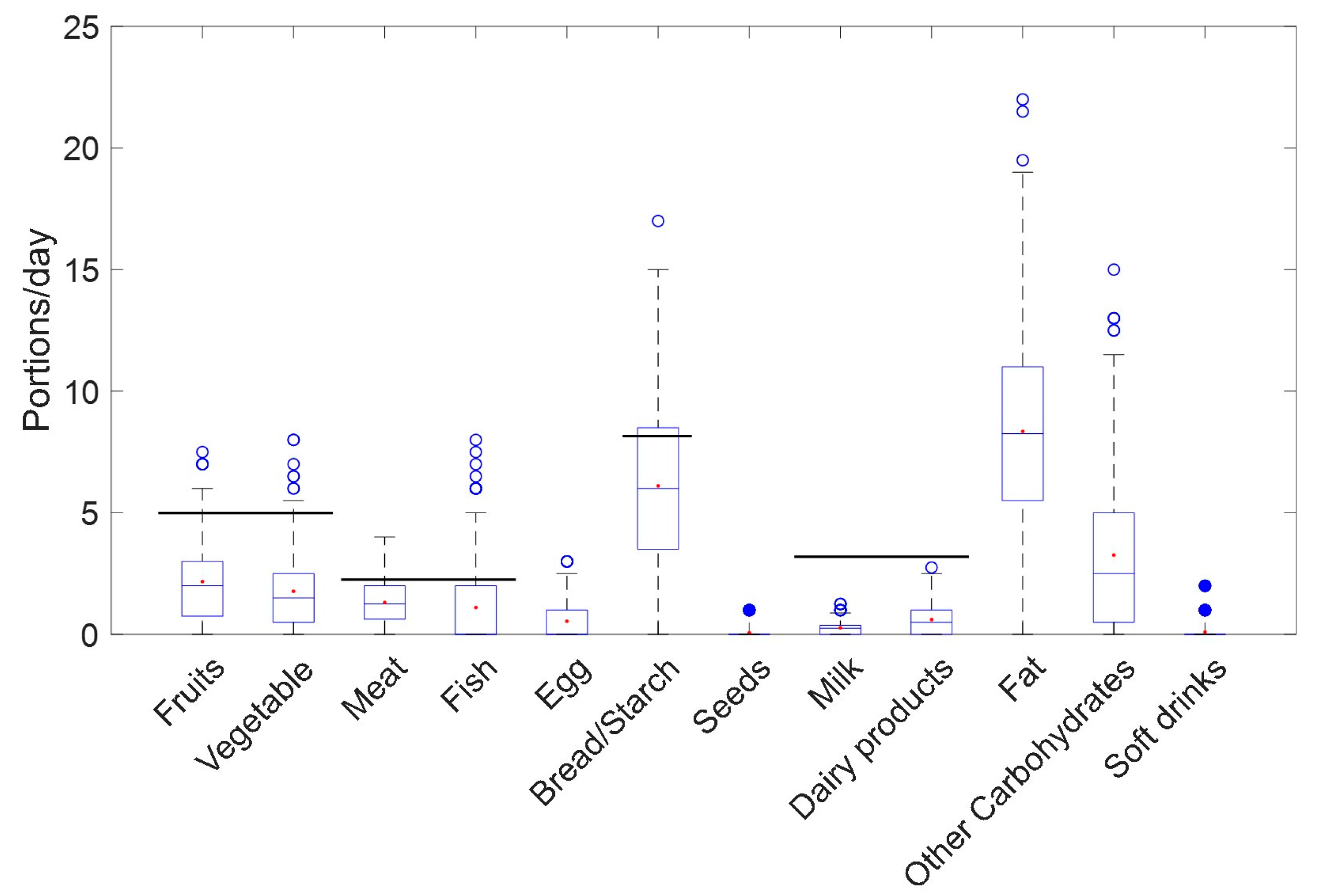
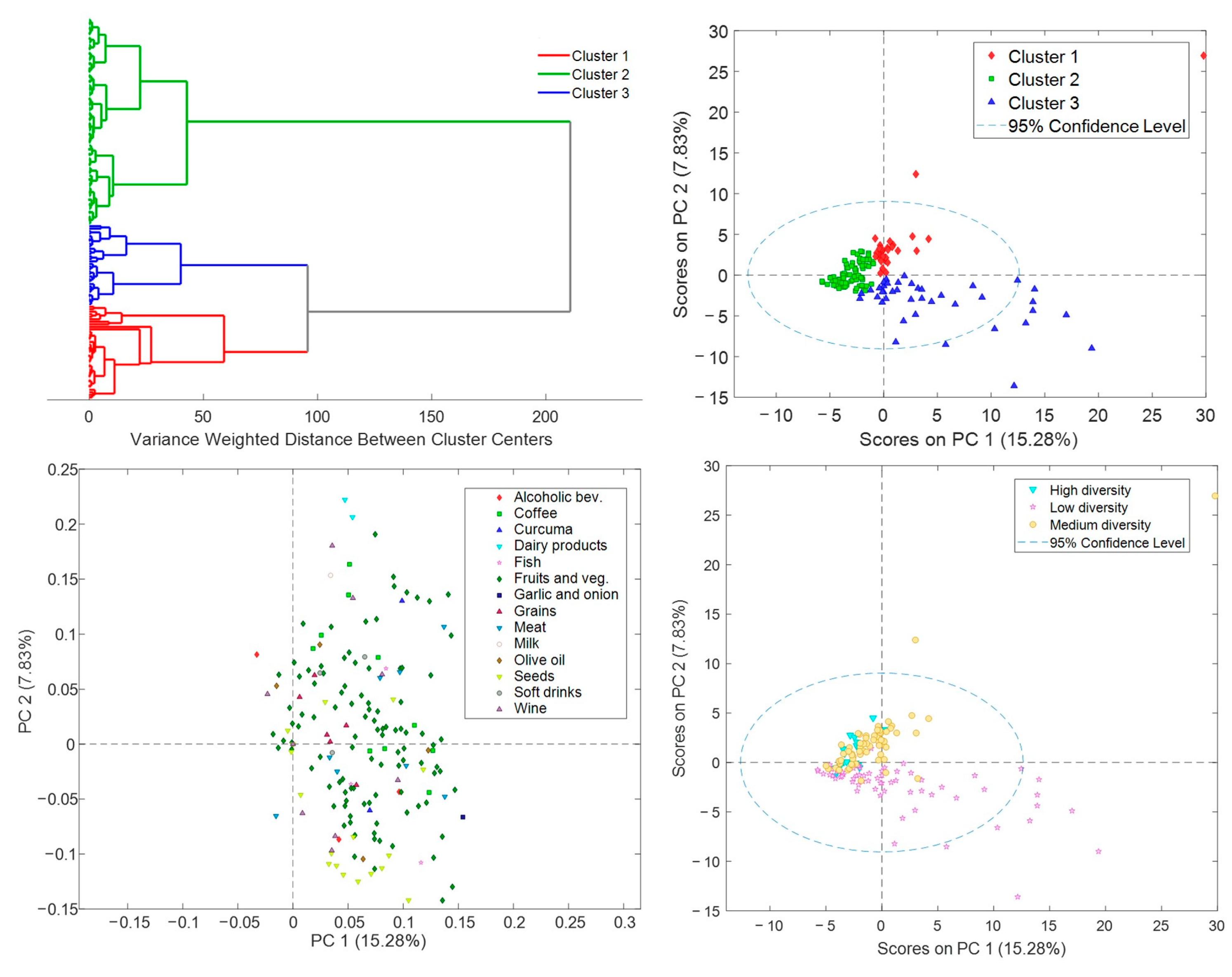
3.2. R24h Results
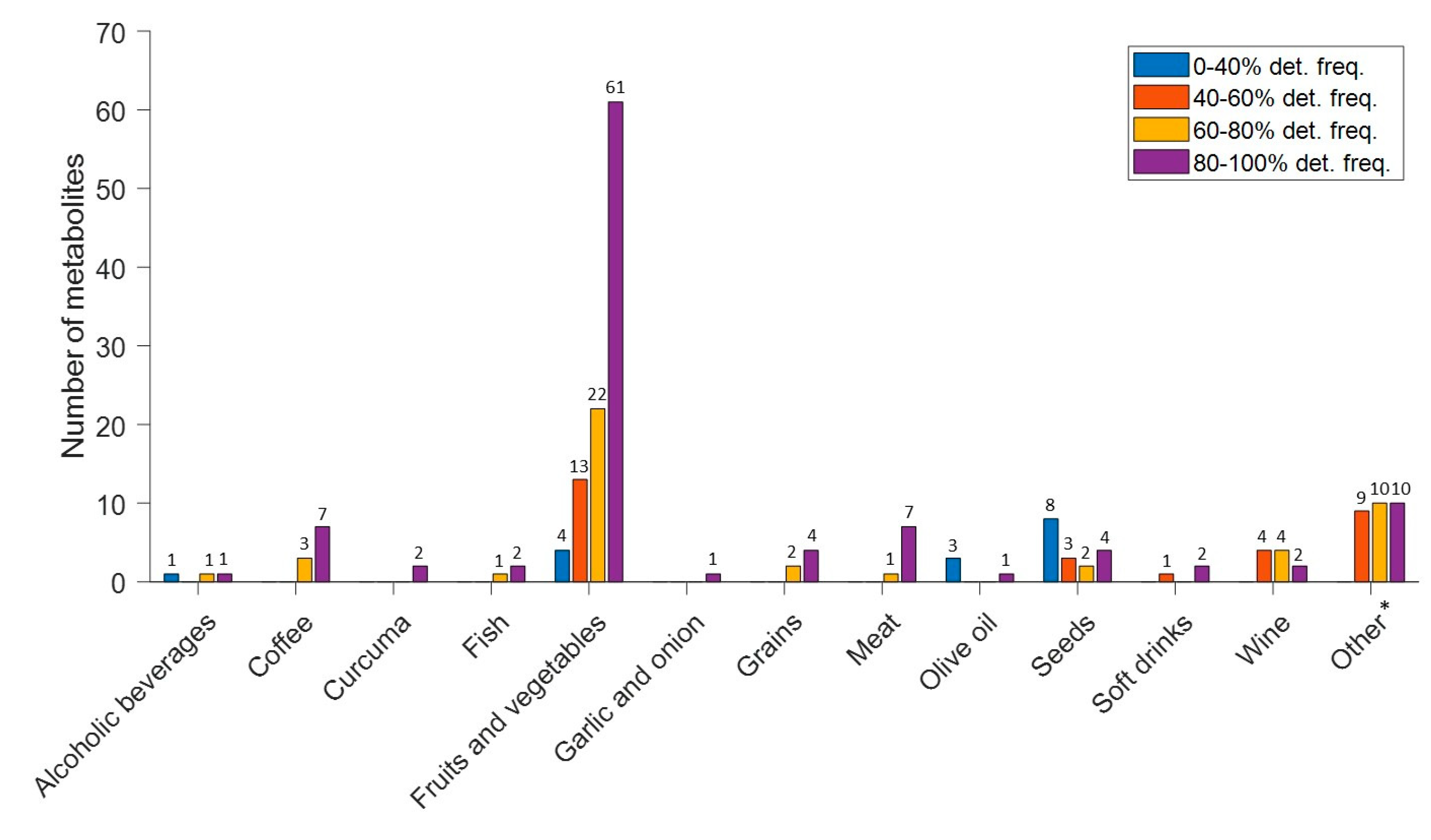
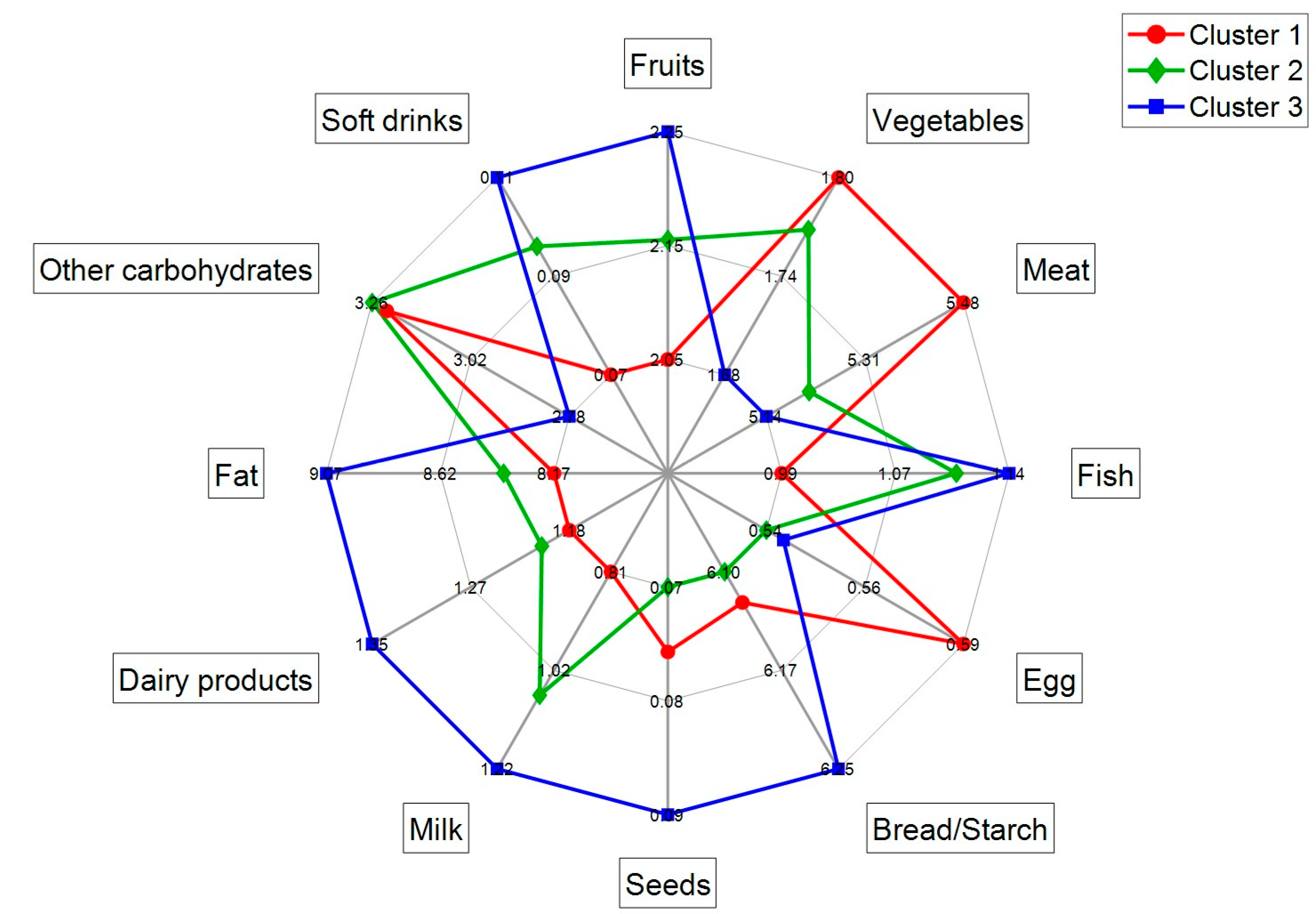
3.3. Biomarker Profiles of Lactating Mothers
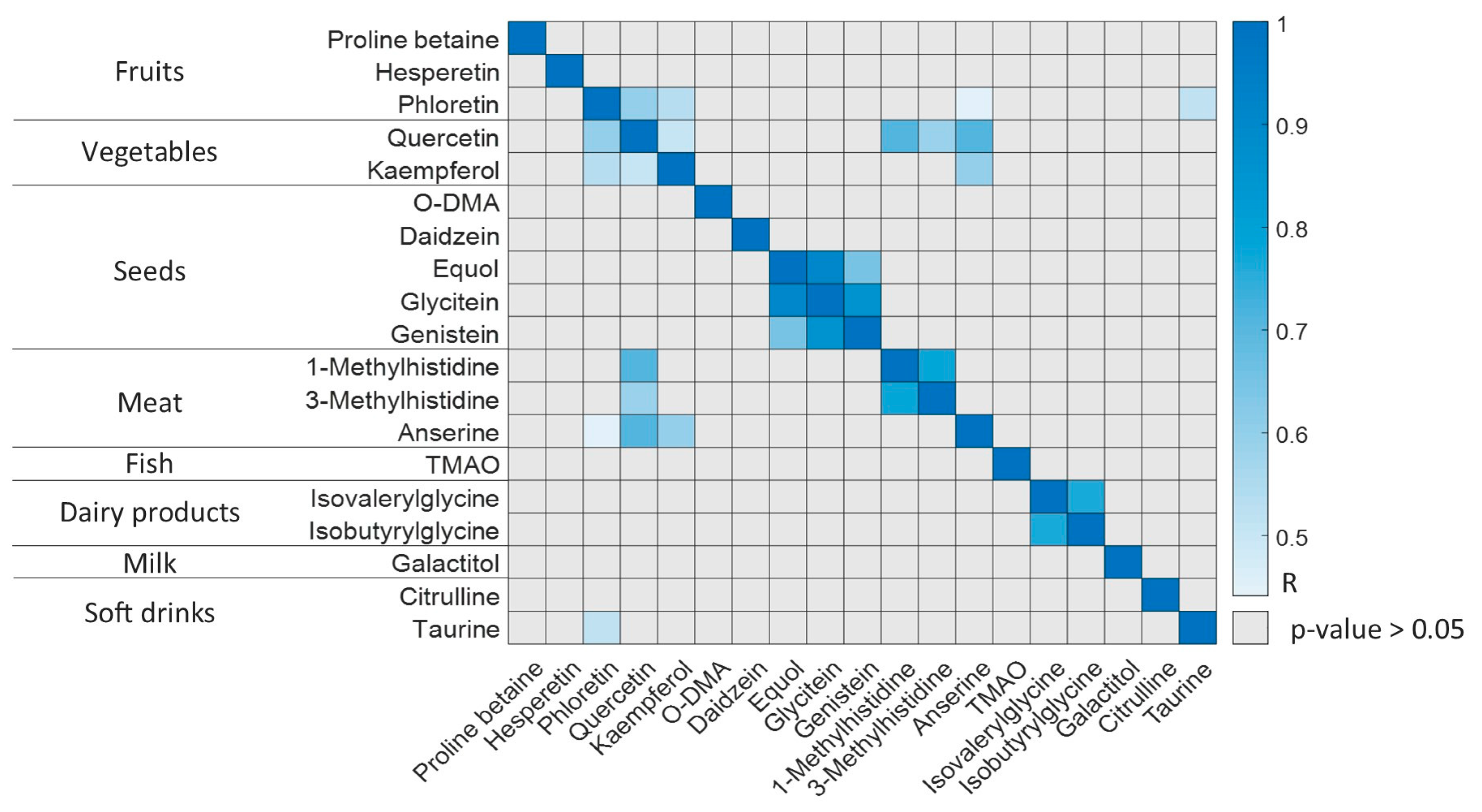
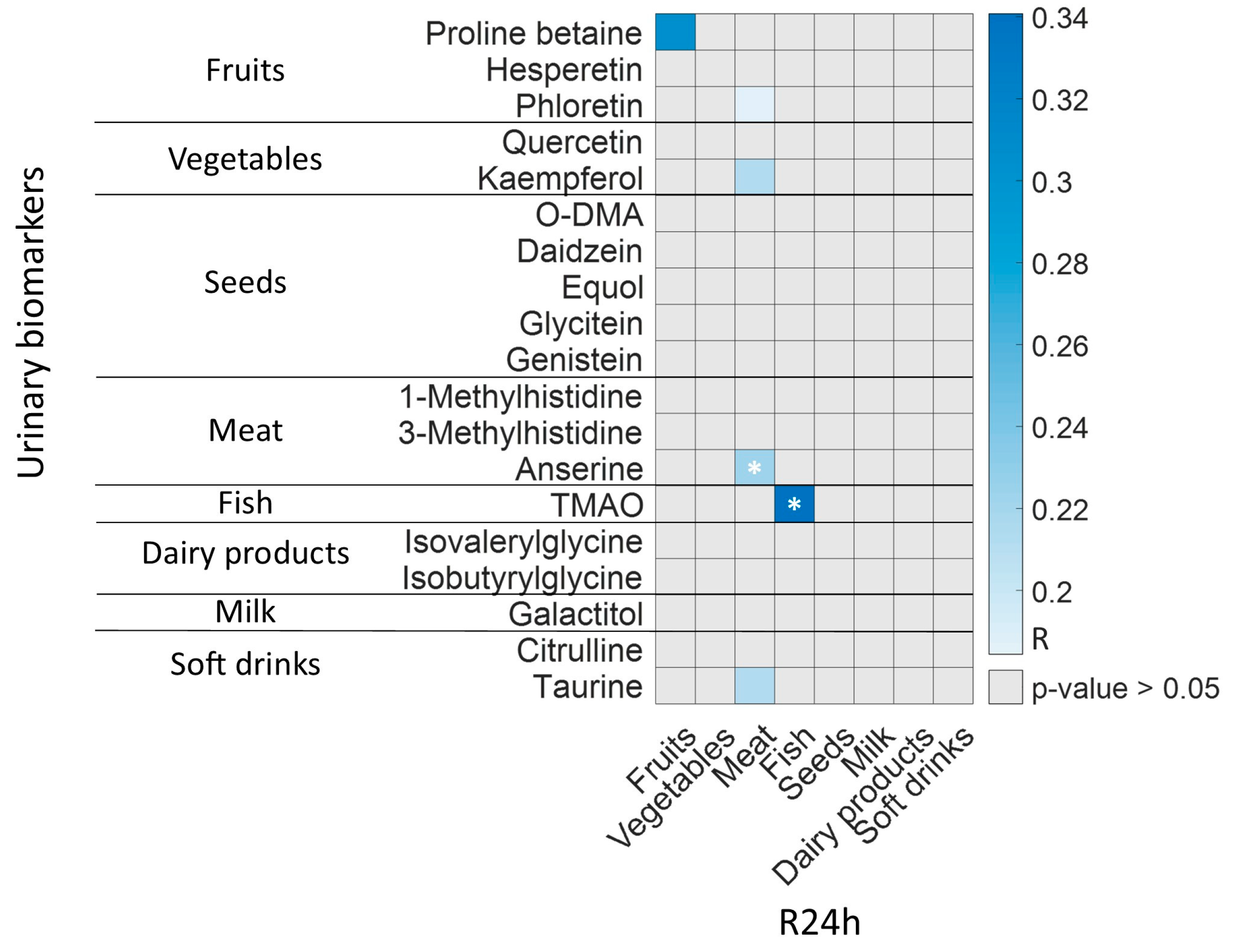
3.4. R24h and BFIs in Lactating Mothers
3.5. Microbiota Activity Biomarkers in Lactating Mothers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brennan, L. 7—Metabotyping: Moving towards personalised nutrition. In Metabolomics as a Tool in Nutrition Research; Sébédio, J.-L., Brennan, L., Eds.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Sawston, UK, 2015; pp. 137–144. ISBN 978-1-78242-084-2. [Google Scholar]
- García-Muñoz Rodrigo, F.; Díez Recinos, A.L.; García-Alix Pérez, A.; Figueras Aloy, J.; Vento Torres, M. Changes in perinatal care and outcomes in newborns at the limit of viability in Spain: The EPI-SEN Study. Neonatology 2015, 107, 120–129. [Google Scholar] [CrossRef]
- Vohr, B.R.; Poindexter, B.B.; Dusick, A.M.; McKinley, L.T.; Higgins, R.D.; Langer, J.C.; Poole, W.K.; National Institute of Child Health and Human Development National Research Network. Persistent beneficial effects of breast milk ingested in the neonatal intensive care unit on outcomes of extremely low birth weight infants at 30 months of age. Pediatrics 2007, 120, e953–e959. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human Milk Composition: Nutrients and Bioactive Factors. Pediatr. Clin. N. Am. 2013, 60, 49–74. [Google Scholar] [CrossRef] [PubMed]
- Innis, S.M. Human milk and formula fatty acids. J. Pediatr. 1992, 120, S56–S61. [Google Scholar] [CrossRef] [PubMed]
- Brenna, J.T.; Varamini, B.; Jensen, R.G.; Diersen-Schade, D.A.; Boettcher, J.A.; Arterburn, L.M. Docosahexaenoic and arachidonic acid concentrations in human breast milk worldwide. Am. J. Clin. Nutr. 2007, 85, 1457–1464. [Google Scholar] [CrossRef]
- Neuhouser, M.L.; Tinker, L.; Shaw, P.A.; Schoeller, D.; Bingham, S.A.; Horn, L.V.; Beresford, S.A.A.; Caan, B.; Thomson, C.; Satterfield, S.; et al. Use of recovery biomarkers to calibrate nutrient consumption self-reports in the Women’s Health Initiative. Am. J. Epidemiol. 2008, 167, 1247–1259. [Google Scholar] [CrossRef]
- Lafay, L.; Mennen, L.; Basdevant, A.; Charles, M.A.; Borys, J.M.; Eschwège, E.; Romon, M. Does energy intake underreporting involve all kinds of food or only specific food items? Results from the Fleurbaix Laventie Ville Santé (FLVS) study. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 1500–1506. [Google Scholar] [CrossRef]
- Jenab, M.; Slimani, N.; Bictash, M.; Ferrari, P.; Bingham, S.A. Biomarkers in nutritional epidemiology: Applications, needs and new horizons. Hum. Genet. 2009, 125, 507–525. [Google Scholar] [CrossRef]
- Scalbert, A.; Brennan, L.; Manach, C.; Andres-Lacueva, C.; Dragsted, L.O.; Draper, J.; Rappaport, S.M.; van der Hooft, J.J.J.; Wishart, D.S. The food metabolome: A window over dietary exposure. Am. J. Clin. Nutr. 2014, 99, 1286–1308. [Google Scholar] [CrossRef] [PubMed]
- Maruvada, P.; Lampe, J.W.; Wishart, D.S.; Barupal, D.; Chester, D.N.; Dodd, D.; Djoumbou-Feunang, Y.; Dorrestein, P.C.; Dragsted, L.O.; Draper, J.; et al. Perspective: Dietary Biomarkers of Intake and Exposure—Exploration with Omics Approaches. Adv. Nutr. 2019, 11, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F.; et al. Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies. Mol. Nutr. Food Res. 2019, 63, 1800384. [Google Scholar] [CrossRef]
- González-Domínguez, R. (Ed.) Mass Spectrometry for Metabolomics; Methods in Molecular Biology; Springer US: New York, NY, USA, 2023; Volume 2571, ISBN 978-1-07-162698-6. [Google Scholar]
- Clarke, E.D.; Rollo, M.E.; Pezdirc, K.; Collins, C.E.; Haslam, R.L. Urinary biomarkers of dietary intake: A review. Nutr. Rev. 2020, 78, 364–381. [Google Scholar] [CrossRef]
- Bar, N.; Korem, T.; Weissbrod, O.; Zeevi, D.; Rothschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weinberger, A.; Le Roy, C.I.; et al. A reference map of potential determinants for the human serum metabolome. Nature 2020, 588, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhernakova, D.V.; Kurilshikov, A.; Andreu-Sánchez, S.; Wang, D.; Augustijn, H.E.; Vich Vila, A.; Lifelines Cohort Study; Weersma, R.K.; Medema, M.H.; et al. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat. Med. 2022, 28, 2333–2343. [Google Scholar] [CrossRef]
- Hedrick, V.E.; Dietrich, A.M.; Estabrooks, P.A.; Savla, J.; Serrano, E.; Davy, B.M. Dietary biomarkers: Advances, limitations and future directions. Nutr. J. 2012, 11, 109. [Google Scholar] [CrossRef]
- Playdon, M.C.; Sampson, J.N.; Cross, A.J.; Sinha, R.; Guertin, K.A.; Moy, K.A.; Rothman, N.; Irwin, M.L.; Mayne, S.T.; Stolzenberg-Solomon, R.; et al. Comparing metabolite profiles of habitual diet in serum and urine. Am. J. Clin. Nutr. 2016, 104, 776–789. [Google Scholar] [CrossRef] [PubMed]
- Sébédio, J.-L. Metabolomics, Nutrition, and Potential Biomarkers of Food Quality, Intake, and Health Status. In Advances in Food and Nutrition Research; Elsevier: Amsterdam, The Netherlands, 2017; Volume 82, pp. 83–116. ISBN 978-0-12-812633-2. [Google Scholar]
- Brouwer-Brolsma, E.M.; Brennan, L.; Drevon, C.A.; van Kranen, H.; Manach, C.; Dragsted, L.O.; Roche, H.M.; Andres-Lacueva, C.; Bakker, S.J.L.; Bouwman, J.; et al. Combining traditional dietary assessment methods with novel metabolomics techniques: Nutrition-Society Summer Meeting/Conference on New Technology in Nutrition Research and Practice / Symposium 2 on Use of Biomarkers in Dietary Assessment and Dietary Exposure. Proc. Nutr. Soc. 2017, 76, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Laly, M.; Chang, H.C.; Prior, R.L.; Fang, N.; Ronis, M.J.J.; Badger, T.M. Isoflavone conjugates are underestimated in tissues using enzymatic hydrolysis. J. Agric. Food Chem. 2005, 53, 6858–6863. [Google Scholar] [CrossRef]
- Quifer-Rada, P.; Martínez-Huélamo, M.; Lamuela-Raventos, R.M. Is enzymatic hydrolysis a reliable analytical strategy to quantify glucuronidated and sulfated polyphenol metabolites in human fluids? Food Funct. 2017, 8, 2419–2424. [Google Scholar] [CrossRef]
- González-Domínguez, R.; Urpi-Sarda, M.; Jáuregui, O.; Needs, P.W.; Kroon, P.A.; Andrés-Lacueva, C. Quantitative Dietary Fingerprinting (QDF)—A Novel Tool for Comprehensive Dietary Assessment Based on Urinary Nutrimetabolomics. J. Agric. Food Chem. 2019, 68, 1851–1861. [Google Scholar] [CrossRef]
- Feliciano, R.P.; Mecha, E.; Bronze, M.R.; Rodriguez-Mateos, A. Development and validation of a high-throughput micro solid-phase extraction method coupled with ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry for rapid identification and quantification of phenolic metabolites in human plasma and urine. J. Chromatogr. A 2016, 1464, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.P.E.; Abd El Mohsen, M.M.; Minihane, A.-M.; Mathers, J.C. Biomarkers of the intake of dietary polyphenols: Strengths, limitations and application in nutrition research. Br. J. Nutr. 2008, 99, 12–22. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.-Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Redondo-Useros, N.; Nova, E.; González-Zancada, N.; Díaz, L.E.; Gómez-Martínez, S.; Marcos, A. Microbiota and Lifestyle: A Special Focus on Diet. Nutrients 2020, 12, 1776. [Google Scholar] [CrossRef] [PubMed]
- Moles, L.; Otaegui, D. The Impact of Diet on Microbiota Evolution and Human Health. Is Diet an Adequate Tool for Microbiota Modulation? Nutrients 2020, 12, 1654. [Google Scholar] [CrossRef]
- Berding, K.; Vlckova, K.; Marx, W.; Schellekens, H.; Stanton, C.; Clarke, G.; Jacka, F.; Dinan, T.G.; Cryan, J.F. Diet and the Microbiota–Gut–Brain Axis: Sowing the Seeds of Good Mental Health. Adv. Nutr. 2021, 12, 1239–1285. [Google Scholar] [CrossRef] [PubMed]
- Dahl, W.J.; Rivero Mendoza, D.; Lambert, J.M. Diet, nutrients and the microbiome. Prog. Mol. Biol. Transl. Sci. 2020, 171, 237–263. [Google Scholar] [CrossRef]
- Bibbò, S.; Ianiro, G.; Giorgio, V.; Scaldaferri, F.; Masucci, L.; Gasbarrini, A.; Cammarota, G. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4742–4749. [Google Scholar]
- Kolodziejczyk, A.A.; Zheng, D.; Elinav, E. Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- Chassaing, B.; Gewirtz, A.T. Gut Microbiome and Metabolism. In Physiology of the Gastrointestinal Tract; Elsevier: Amsterdam, The Netherlands, 2018; pp. 775–793. ISBN 978-0-12-809954-4. [Google Scholar]
- Ramos-Garcia, V.; Ten-Doménech, I.; Moreno-Giménez, A.; Campos-Berga, L.; Parra-Llorca, A.; Ramón, A.; Vaya, M.J.; Mohareb, F.; Molitor, C.; Refinetti, P.; et al. Fact-based nutrition for infants and lactating mothers—The Nutrishield study. Front. Pediatr. 2023; Accepted for publication. [Google Scholar] [CrossRef]
- Hinojosa Nogueira, D.J.; Romero Molina, D.; Giménez Asensio, M.J.; González Alzaga, B.; López Flores, I.; Pastoriza de la Cueva, S.; Rufián Henares, J.Á.; Hernández Jérez, A.F.; Lacasaña, M. Validity and Reproducibility of a Food Frequency Questionnaire to Assess Nutrients Intake of Pregnant Women in the South-East of Spain. Nutrients 2021, 13, 3032. [Google Scholar] [CrossRef]
- Conway, J.M.; Ingwersen, L.A.; Moshfegh, A.J. Accuracy of dietary recall using the USDA five-step multiple-pass method in men: An observational validation study. J. Am. Diet. Assoc. 2004, 104, 595–603. [Google Scholar] [CrossRef]
- Panagiotakos, D.B.; Pitsavos, C.; Stefanadis, C. Dietary patterns: A Mediterranean diet score and its relation to clinical and biological markers of cardiovascular disease risk. Nutr. Metab. Cardiovasc. Dis. 2006, 16, 559–568. [Google Scholar] [CrossRef]
- FDA. Guidance for Industry: Bioanalytical Method Validation; Food and Drug Administration, Center for Drug Evaluation and Research, Center for Veterinary Medicine: Rockville, MD, USA, 2018; Volume 44. [Google Scholar]
- Healthy Eating during Pregnancy and Breastfeeding—Booklet for Mothers; World Health Organization: Geneva, Switzerland, 2021.
- Lloyd, A.J.; Beckmann, M.; Favé, G.; Mathers, J.C.; Draper, J. Proline betaine and its biotransformation products in fasting urine samples are potential biomarkers of habitual citrus fruit consumption. Br. J. Nutr. 2011, 106, 812–824. [Google Scholar] [CrossRef]
- Cheung, W.; Keski-Rahkonen, P.; Assi, N.; Ferrari, P.; Freisling, H.; Rinaldi, S.; Slimani, N.; Zamora-Ros, R.; Rundle, M.; Frost, G.; et al. A metabolomic study of biomarkers of meat and fish intake. Am. J. Clin. Nutr. 2017, 105, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Krogholm, K.S.; Bredsdorff, L.; Alinia, S.; Christensen, T.; Rasmussen, S.E.; Dragsted, L.O. Free fruit at workplace intervention increases total fruit intake: A validation study using 24 h dietary recall and urinary flavonoid excretion. Eur. J. Clin. Nutr. 2010, 64, 1222–1228. [Google Scholar] [CrossRef]
- Gibbons, H.; McNulty, B.A.; Nugent, A.P.; Walton, J.; Flynn, A.; Gibney, M.J.; Brennan, L. A metabolomics approach to the identification of biomarkers of sugar-sweetened beverage intake. Am. J. Clin. Nutr. 2015, 101, 471–477. [Google Scholar] [CrossRef]
- Cross, A.J.; Major, J.M.; Sinha, R. Urinary biomarkers of meat consumption. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, L.G.; Winning, H.; Savorani, F.; Toft, H.; Larsen, T.M.; Dragsted, L.O.; Astrup, A.; Engelsen, S.B. Assessment of the Effect of High or Low Protein Diet on the Human Urine Metabolome as Measured by NMR. Nutrients 2012, 4, 112–131. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.G.; Smith, L.H. Biochemistry and physiology of taurine and taurine derivatives. Physiol. Rev. 1968, 48, 424–511. [Google Scholar] [CrossRef]
- Garcia-Aloy, M.; Rabassa, M.; Casas-Agustench, P.; Hidalgo-Liberona, N.; Llorach, R.; Andres-Lacueva, C. Novel strategies for improving dietary exposure assessment: Multiple-data fusion is a more accurate measure than the traditional single-biomarker approach. Trends Food Sci. Technol. 2017, 69, 220–229. [Google Scholar] [CrossRef]
- Brennan, L. Metabolomics: A tool to aid dietary assessment in nutrition. Curr. Opin. Food Sci. 2017, 16, 96–99. [Google Scholar] [CrossRef]
- Praticò, G.; Gao, Q.; Scalbert, A.; Vergères, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Pedapati, S.H.; Afman, L.A.; Wishart, D.S.; et al. Guidelines for Biomarker of Food Intake Reviews (BFIRev): How to conduct an extensive literature search for biomarker of food intake discovery. Genes Nutr. 2018, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.C.; Matthews, C.E.; Sampson, J.N.; Stolzenberg-Solomon, R.Z.; Zheng, W.; Cai, Q.; Tan, Y.T.; Chow, W.-H.; Ji, B.-T.; Liu, D.K.; et al. Human metabolic correlates of body mass index. Metabolomics 2014, 10, 259–269. [Google Scholar] [CrossRef]
- Schutte, B.A.M.; van den Akker, E.B.; Deelen, J.; van de Rest, O.; van Heemst, D.; Feskens, E.J.M.; Beekman, M.; Slagboom, P.E. The effect of standardized food intake on the association between BMI and 1H-NMR metabolites. Sci. Rep. 2016, 6, 38980. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef]
| Parameters | Mothers (N = 59) |
|---|---|
| Age (years), mean (SD) | 36 (5) |
| Weight (kg), mean (SD) | 62 (14) |
| BMI (kg/m2), mean (SD) | 24 (5) |
| C-Section delivery, N (%) | 28 (48) |
| Antibiotic therapy, N (%) | 3 (5) |
| Dietary supplements, N (%) | 52 (88) |
| MedDietScore *, mean (SD) | 30 (5) |
| Category | Metabolite | Range (µmol/g Creat) | Median (µmol/g Creat) | IQR (25–75) | Detection Frecuency (%) |
|---|---|---|---|---|---|
| Microbiota | Phenylpropionylglycine | 0.007–58 | 0.03 | 0.4 | 72 |
| 3-IPA | 0.012–8 | 2 | 3 | 3 | |
| L-Kynurenine | 0.009–21 | 0.5 | 2 | 80 | |
| 3-IAA | 0.012–176 | 1.3 | 7 | 99 | |
| L-Tyrosine | 0.014–2829 | 16 | 43 | 98 | |
| Hippuric Acid | 11–8496 | 255 | 598 | 100 | |
| Gallic Acid | 0.013–10 | 0.4 | 0.4 | 62 | |
| Ferullic Acid Sulphate | 0.02–126 | 0.4 | 3 | 74 | |
| Fruits | Proline betaine | 1.4–2568 | 88 | 315 | 100 |
| Hesperetin | 0.02–11 | 0.05 | 0.13 | 64 | |
| Phloretin | 0.004–2 | 0.02 | 0.10 | 29 | |
| Vegetables | Quercetin | 0.03–76 | 0.4 | 0.2 | 48 |
| Kaempferol | 0.02–2 | 0.2 | 0.6 | 93 | |
| Seeds | O-DMA | 0.007–5 | 0.02 | 0.3 | 61 |
| Daidzein | 0.005–3 | 0.4 | 0.9 | 96 | |
| Equol | 0.012–10 | 0.3 | 0.05 | 34 | |
| Glycitein | 0.006–1 | 0.05 | 0.2 | 25 | |
| Genistein | 0.4–2 | 0.03 | 0.02 | 2 | |
| Meat | 1-Methylhistidine | 4–1784 | 48 | 95 | 100 |
| 3-Methylhistidine | 0.2–4555 | 31 | 87 | 100 | |
| Anserine | 0.6–304 | 2 | 10 | 88 | |
| Fish | TMAO | 3–3368 | 142 | 473 | 100 |
| Dairy products | Isovalerylglycine | 0.005–91 | 3 | 13 | 87 |
| Isobutyrylglycine | 0.7–125 | 3 | 9 | 93 | |
| Milk | Galactitol | 1.2–2424 | 22 | 67 | 92 |
| Soft drinks | Citrulline | 0.09–60 | 7 | 11 | 95 |
| Taurine | 0.2–2341 | 55 | 207 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Garcia, V.; Ten-Doménech, I.; Moreno-Giménez, A.; Campos-Berga, L.; Parra-Llorca, A.; Gormaz, M.; Vento, M.; Karipidou, M.; Poulimeneas, D.; Mamalaki, E.; et al. Joint Microbiota Activity and Dietary Assessment through Urinary Biomarkers by LC-MS/MS. Nutrients 2023, 15, 1894. https://doi.org/10.3390/nu15081894
Ramos-Garcia V, Ten-Doménech I, Moreno-Giménez A, Campos-Berga L, Parra-Llorca A, Gormaz M, Vento M, Karipidou M, Poulimeneas D, Mamalaki E, et al. Joint Microbiota Activity and Dietary Assessment through Urinary Biomarkers by LC-MS/MS. Nutrients. 2023; 15(8):1894. https://doi.org/10.3390/nu15081894
Chicago/Turabian StyleRamos-Garcia, Victoria, Isabel Ten-Doménech, Alba Moreno-Giménez, Laura Campos-Berga, Anna Parra-Llorca, María Gormaz, Máximo Vento, Melina Karipidou, Dimitrios Poulimeneas, Eirini Mamalaki, and et al. 2023. "Joint Microbiota Activity and Dietary Assessment through Urinary Biomarkers by LC-MS/MS" Nutrients 15, no. 8: 1894. https://doi.org/10.3390/nu15081894
APA StyleRamos-Garcia, V., Ten-Doménech, I., Moreno-Giménez, A., Campos-Berga, L., Parra-Llorca, A., Gormaz, M., Vento, M., Karipidou, M., Poulimeneas, D., Mamalaki, E., Bathrellou, E., & Kuligowski, J. (2023). Joint Microbiota Activity and Dietary Assessment through Urinary Biomarkers by LC-MS/MS. Nutrients, 15(8), 1894. https://doi.org/10.3390/nu15081894