Time-Restricted Feeding Modifies the Fecal Lipidome and the Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animal Experimentation
2.3. Fecal Lipid Analysis
2.4. Fecal Bile Acid Analysis
2.5. Fecal Microbiome Analysis
2.6. Statistical Analysis
3. Results
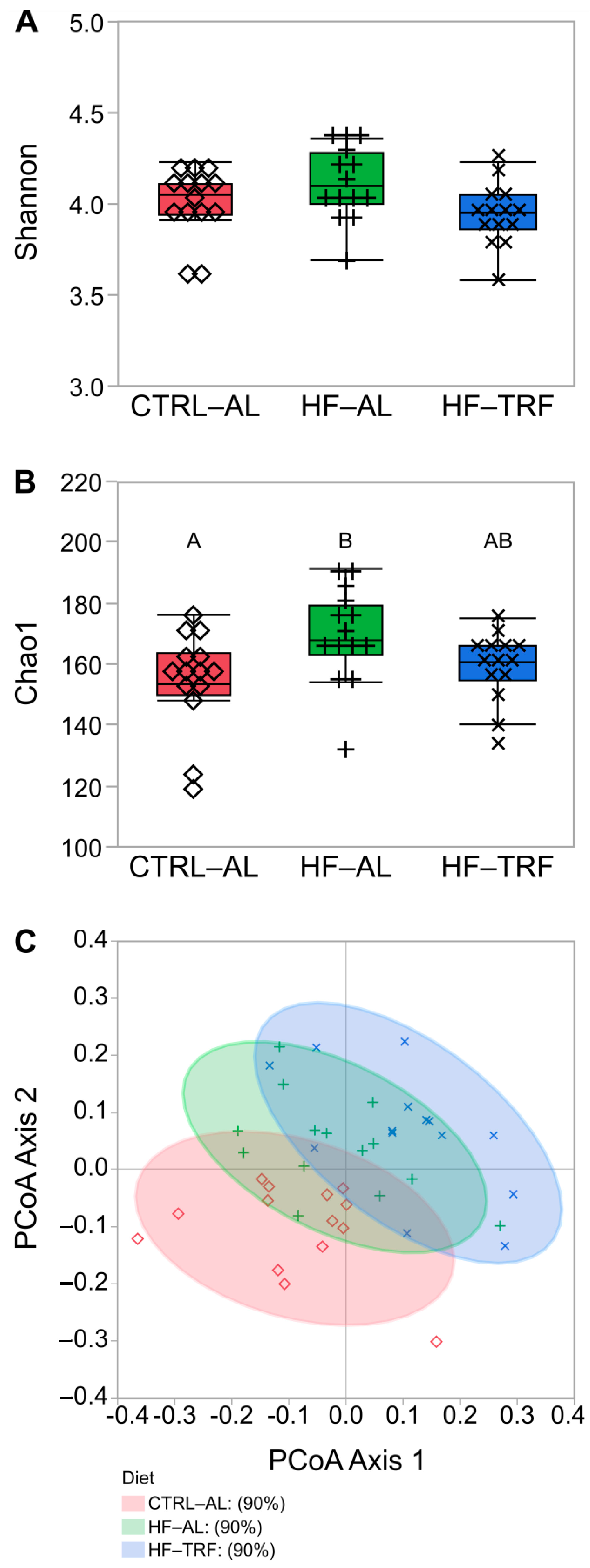
3.1. Fecal Bacterial Taxa
3.2. Fecal Lipid Content
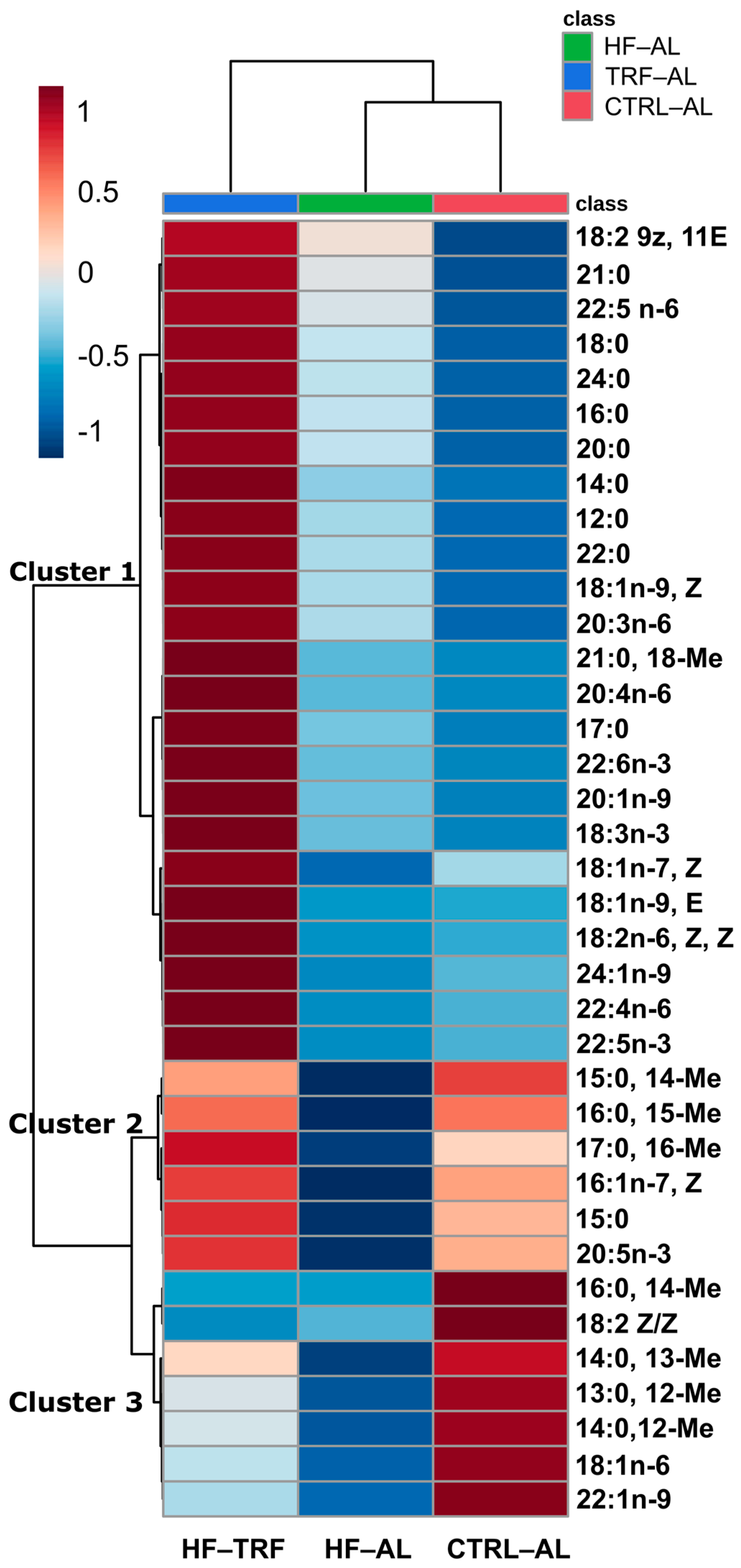
3.2.1. Fecal Fatty Acids
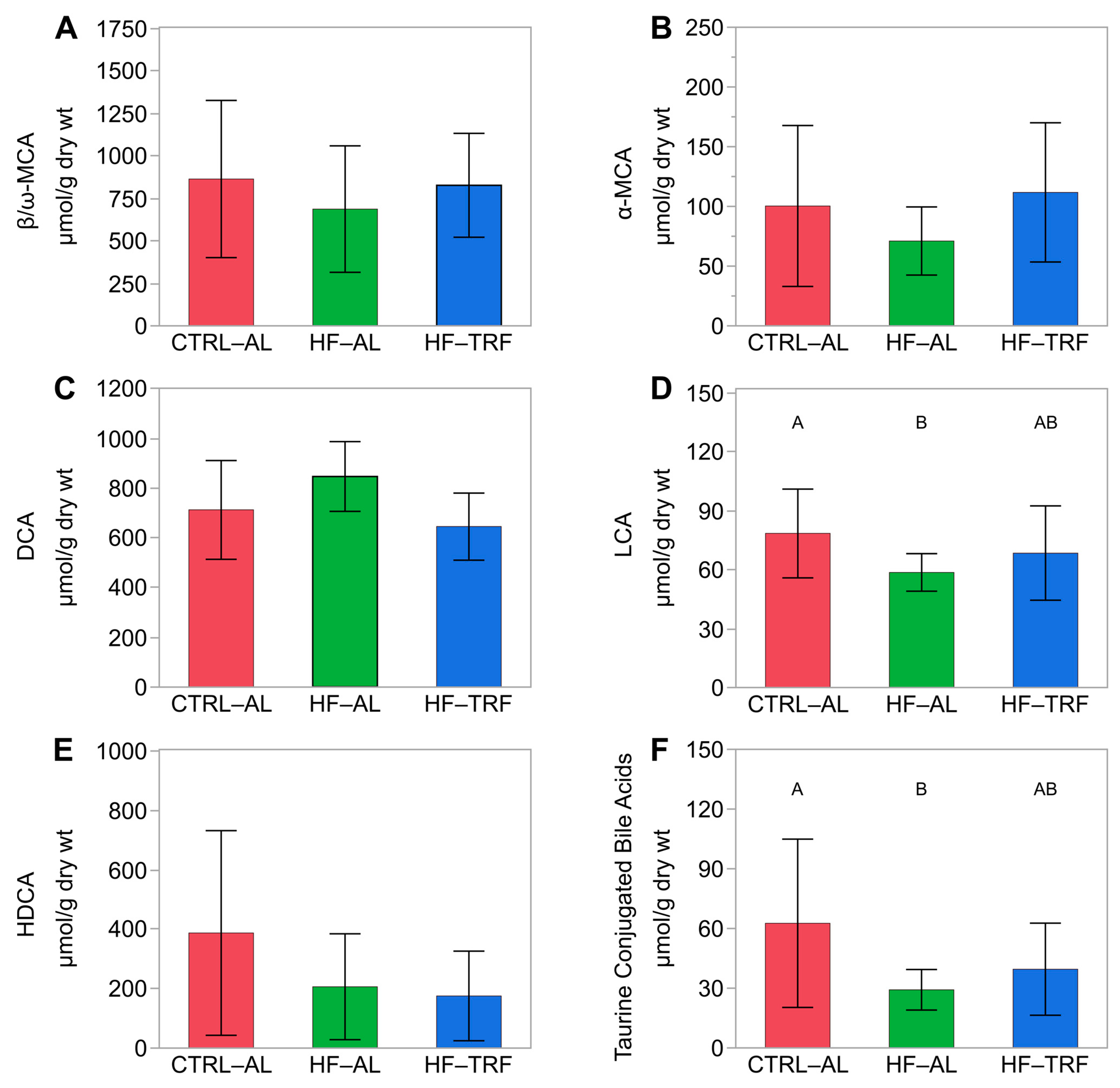
3.2.2. Fecal Bile Acids
3.2.3. Fecal Short Chain Fatty Acids
3.3. Relationship of Fecal Lipids to Fecal Microbes
3.3.1. Fatty Acids
3.3.2. Bile Acids
3.3.3. Short-Chain Fatty Acids
4. Discussion
4.1. Diet and TRF Altered the Composition of Fecal Microbiota
4.2. Diet and TRF Altered the Content and Composition of Fecal Fatty Acids
4.3. Diet and TRF Affected Bile Acids and SCFAs Associated with Microbial Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ward, Z.J.; Bleich, S.N.; Long, M.W.; Gortmaker, S.L. Association of Body Mass Index with Health Care Expenditures in the United States by Age and Sex. PLoS ONE 2021, 16, e0247307. [Google Scholar] [CrossRef] [PubMed]
- Manoogian, E.N.C.; Zadourian, A.; Lo, H.C.; Gutierrez, N.R.; Shoghi, A.; Rosander, A.; Pazargadi, A.; Ormiston, C.K.; Wang, X.; Sui, J.; et al. Feasibility of Time-Restricted Eating and Impacts on Cardiometabolic Health in 24-h Shift Workers: The Healthy Heroes Randomized Control Trial. Cell Metab. 2022, 34, 1442–1456.e7. [Google Scholar] [CrossRef] [PubMed]
- Chaix, A.; Zarrinpar, A.; Miu, P.; Panda, S. Time-Restricted Feeding Is a Preventative and Therapeutic Intervention against Diverse Nutritional Challenges. Cell Metab. 2014, 20, 991–1005. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, S.; Yan, L. Time-Restricted Feeding Reduces Adiposity in Mice Fed a High-Fat Diet. Nutr. Res. 2016, 36, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Świątkiewicz, I.; Woźniak, A.; Taub, P.R. Time-Restricted Eating and Metabolic Syndrome: Current Status and Future Perspectives. Nutrients 2021, 13, 221. [Google Scholar] [CrossRef]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of Feeding and the Intrinsic Circadian Clock Drive Rhythms in Hepatic Gene Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef]
- Cândido, F.G.; Valente, F.X.; Grześkowiak, Ł.M.; Moreira, A.P.B.; Rocha, D.M.U.P.; Alfenas, R.d.C.G. Impact of Dietary Fat on Gut Microbiota and Low-Grade Systemic Inflammation: Mechanisms and Clinical Implications on Obesity. Int. J. Food Sci. Nutr. 2018, 69, 125–143. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab. 2014, 20, 1006–1017. [Google Scholar] [CrossRef]
- Albrecht, U. Timing to Perfection: The Biology of Central and Peripheral Circadian Clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef]
- Longo, V.D.; Panda, S. Fasting, Circadian Rhythms, and Time Restricted Feeding in Healthy Lifespan. Cell Metab. 2016, 23, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; An, Y.; Tang, H.; Wang, Y. Alterations of Bile Acids and Gut Microbiota in Obesity Induced by High Fat Diet in Rat Model. J. Agric. Food Chem. 2019, 67, 3624–3632. [Google Scholar] [CrossRef] [PubMed]
- Lam, Y.Y.; Ha, C.W.Y.; Hoffmann, J.M.A.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of Dietary Fat Profile on Gut Permeability and Microbiota and Their Relationships with Metabolic Changes in Mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.P.B.; Texeira, T.F.S.; Ferreira, A.B.; Peluzio, M.D.C.G.; Alfenas, R.D.C.G. Influence of a High-Fat Diet on Gut Microbiota, Intestinal Permeability and Metabolic Endotoxaemia. Br. J. Nutr. 2012, 108, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary Lipids, Gut Microbiota and Lipid Metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef]
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective Increases of Bifidobacteria in Gut Microflora Improve High-Fat-Diet-Induced Diabetes in Mice through a Mechanism Associated with Endotoxaemia. Diabetologia 2007, 50, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-A.; Gu, W.; Lee, I.-A.; Joh, E.-H.; Kim, D.-H. High Fat Diet-Induced Gut Microbiota Exacerbates Inflammation and Obesity in Mice via the TLR4 Signaling Pathway. PLoS ONE 2012, 7, e47713. [Google Scholar] [CrossRef]
- Machate, D.J.; Figueiredo, P.S.; Marcelino, G.; Guimarães, R.D.C.A.; Hiane, P.A.; Bogo, D.; Pinheiro, V.A.Z.; Oliveira, L.C.S.D.; Pott, A. Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis. Int. J. Mol. Sci. 2020, 21, 4093. [Google Scholar] [CrossRef]
- Raatz, S.K.; Conrad, Z.; Johnson, L.K.; Picklo, M.J.; Jahns, L. Relationship of the Reported Intakes of Fat and Fatty Acids to Body Weight in US Adults. Nutrients 2017, 9, 438. [Google Scholar] [CrossRef]
- Žáček, P.; Bukowski, M.; Mehus, A.; Johnson, L.; Zeng, H.; Raatz, S.; Idso, J.P.; Picklo, M. Dietary Saturated Fatty Acid Type Impacts Obesity-Induced Metabolic Dysfunction and Plasma Lipidomic Signatures in Mice. J. Nutr. Biochem. 2019, 64, 32–44. [Google Scholar] [CrossRef]
- Jackman, J.A.; Yoon, B.K.; Li, D.; Cho, N.-J. Nanotechnology Formulations for Antibacterial Free Fatty Acids and Monoglycerides. Molecules 2016, 21, 305. [Google Scholar] [CrossRef] [PubMed]
- Shilling, M.; Matt, L.; Rubin, E.; Visitacion, M.P.; Haller, N.A.; Grey, S.F.; Woolverton, C.J. Antimicrobial Effects of Virgin Coconut Oil and Its Medium-Chain Fatty Acids on Clostridium Difficile. J. Med. Food 2013, 16, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjære, E.; Midtbø, L.K.; et al. High-Fat Feeding Rather than Obesity Drives Taxonomical and Functional Changes in the Gut Microbiota in Mice. Microbiome 2017, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Ikushiro, H.; Seo, H.S.; Shin, K.-O.; Kim, Y.I.; Kim, J.Y.; Lee, Y.-M.; Yano, T.; Holleran, W.M.; Elias, P.; et al. ER Stress Stimulates Production of the Key Antimicrobial Peptide, Cathelicidin, by Forming a Previously Unidentified Intracellular S1P Signaling Complex. Proc. Natl. Acad. Sci. USA 2016, 113, E1334–E1342. [Google Scholar] [CrossRef] [PubMed]
- Mehus, A.A.; Rust, B.; Idso, J.P.; Hanson, B.; Zeng, H.; Yan, L.; Bukowski, M.R.; Picklo, M.J. Time-Restricted Feeding Mice a High-Fat Diet Induces a Unique Lipidomic Profile. J. Nutr. Biochem. 2020, 88, 108531. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile Acids and the Gut Microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar] [CrossRef]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int. J. Mol. Sci. 2019, 20, 1214. [Google Scholar] [CrossRef]
- Reeves, P.G. Components of the AIN-93 Diets as Improvements in the AIN-76A Diet. J. Nutr. 1997, 127, 838S–841S. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Direct Transesterification of All Classes of Lipids in a One-Step Reaction. J. Lipid Res. 1986, 27, 114–120. [Google Scholar] [CrossRef]
- Zeng, H.; Larson, K.J.; Cheng, W.-H.; Bukowski, M.R.; Safratowich, B.D.; Liu, Z.; Hakkak, R. Advanced Liver Steatosis Accompanies an Increase in Hepatic Inflammation, Colonic, Secondary Bile Acids and Lactobacillaceae/Lachnospiraceae Bacteria in C57BL/6 Mice Fed a High-Fat Diet. J. Nutr. Biochem. 2020, 78, 108336. [Google Scholar] [CrossRef]
- Hagio, M.; Matsumoto, M.; Fukushima, M.; Hara, H.; Ishizuka, S. Improved Analysis of Bile Acids in Tissues and Intestinal Contents of Rats Using LC/ESI-MS. J. Lipid Res. 2009, 50, 173–180. [Google Scholar] [CrossRef]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic Improvement of Amplicon Marker Gene Methods for Increased Accuracy in Microbiome Studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Galachyants, Y.P.; Morozov, I.V.; Bukin, S.V.; Zakharenko, A.S.; Zemskaya, T.I. The Effect of 16S RRNA Region Choice on Bacterial Community Metabarcoding Results. Sci. Data 2019, 6, 1–14. [Google Scholar] [CrossRef]
- Strube, M.L. RibDif: Can Individual Species Be Differentiated by 16S Sequencing? Bioinform. Adv. 2021, 1, vbab020. [Google Scholar] [CrossRef]
- Daniel, H.; Gholami, A.M.; Berry, D.; Desmarchelier, C.; Hahne, H.; Loh, G.; Mondot, S.; Lepage, P.; Rothballer, M.; Walker, A.; et al. High-Fat Diet Alters Gut Microbiota Physiology in Mice. ISME J. 2014, 8, 295–308. [Google Scholar] [CrossRef]
- Patterson, E.; Doherty, R.M.O.; Murphy, E.F.; Wall, R.; Sullivan, O.O.; Nilaweera, K.; Fitzgerald, G.F.; Cotter, P.D.; Ross, R.P.; Stanton, C. Impact of Dietary Fatty Acids on Metabolic Activity and Host Intestinal Microbiota Composition in C57BL/6J Mice. Br. J. Nutr. 2014, 111, 1905–1917. [Google Scholar] [CrossRef]
- Shen, W.; Wolf, P.G.; Carbonero, F.; Zhong, W.; Reid, T.; Gaskins, H.R.; McIntosh, M.K. Intestinal and Systemic Inflammatory Responses Are Positively Associated with Sulfidogenic Bacteria Abundance in High-Fat–Fed Male C57BL/6J Mice. J. Nutr. 2014, 144, 1181–1187. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The Human Gut Bacteria Christensenellaceae Are Widespread, Heritable, and Associated with Health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, M.; Couëdelo, L.; Meugnier, E.; Plaisancié, P.; Létisse, M.; Benoit, B.; Gabert, L.; Penhoat, A.; Durand, A.; Pineau, G.; et al. Dietary Emulsifiers from Milk and Soybean Differently Impact Adiposity and Inflammation in Association with Modulation of Colonic Goblet Cells in High-Fat Fed Mice. Mol. Nutr. Food Res. 2016, 60, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Murase, T.; Mizuno, T.; Omachi, T.; Onizawa, K.; Komine, Y.; Kondo, H.; Hase, T.; Tokimitsu, I. Dietary Diacylglycerol Suppresses High Fat and High Sucrose Diet-Induced Body Fat Accumulation in C57BL/6J Mice. J. Lipid Res. 2001, 42, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Leone, V.; Devkota, S.; Wang, Y.; Brady, M.; Chang, E. Composition of Dietary Fat Source Shapes Gut Microbiota Architecture and Alters Host Inflammatory Mediators in Mouse Adipose Tissue. J. Parenter. Enter. Nutr. 2013, 37, 746–754. [Google Scholar] [CrossRef]
- Feldman, E.B.; Russell, B.S.; Hawkins, C.B.; Forte, T. Intestinal Lymph Lipoproteins in Rats Fed Diets Enriched in Specific Fatty Acids. J. Nutr. 1983, 113, 2323–2334. [Google Scholar] [CrossRef]
- Feldman, E.B.; Russell, B.S.; Schnare, F.H.; Miles, B.C.; Doyle, E.A.; Moretti-Rojas, I. Effects of Tristearin, Triolein and Safflower Oil Diets on Cholesterol Balance in Rats. J. Nutr. 1979, 109, 2226–2236. [Google Scholar] [CrossRef]
- Haeusler, R.A.; Astiarraga, B.; Camastra, S.; Accili, D.; Ferrannini, E. Human Insulin Resistance Is Associated With Increased Plasma Levels of 12α-Hydroxylated Bile Acids. Diabetes 2013, 62, 4184–4191. [Google Scholar] [CrossRef]
- Li, H.; Limenitakis, J.P.; Fuhrer, T.; Geuking, M.B.; Lawson, M.A.; Wyss, M.; Brugiroux, S.; Keller, I.; Macpherson, J.A.; Rupp, S.; et al. The Outer Mucus Layer Hosts a Distinct Intestinal Microbial Niche. Nat. Commun. 2015, 6, 8292. [Google Scholar] [CrossRef]
- Arora, T.; Tremaroli, V. Therapeutic Potential of Butyrate for Treatment of Type 2 Diabetes. Front. Endocrinol. 2021, 12, 761834. [Google Scholar] [CrossRef]
 each animal within LF-AL;
each animal within LF-AL;  indicates individual animals within HF-AL;
indicates individual animals within HF-AL;  indicates each animal in HF-TRF (N = 14 for each diet). (A) * Bacteroidaceae were elevated in HF-TRF compared to CTRL-AL and HF-AL diets (p = 0.02). (B) ** Bifidobacteriaceae were not detected in mice undergoing HF-TRF feeding, and relative abundance was lower than CTRL-AL (p < 0.01). Two animals in the HF-AL had detectable OTUs attributable to Bifidobacteriaceae, but no differences were seen between the HF-AL and CTRL-AL or HF-TRF animals. (C) ** Christensenellaneae were elevated by HF-AL feeding compared to HF-TRF (p = 0.03), but animals with CTRL-AL feeding did not differ from HF-AL or HF-TRF. (D) ** Clostridiaceae were increased by HF-TRF compared to both CTRL-AL and HF-AL diets (p < 0.01 for both). (E) ** Enterococcaceae were increased by HF-AL feeding compared to CTRL-AL and HF-TRF (p < 0.01 and p = 0.04, respectively). (F) * Muribaculaceae were increased by CTRL-AL feeding compared to HF-AL and HF-TRF (p = 0.03 and p < 0.01, respectively). (G) * Rikenellaceae were higher in HF-AL and HF-TRF than in CTRL-AL (p = 0.04 and p < 0.01, respectively). (H) Saccharimonadaceae was reduced by HF-TRF compared to CTRL-AL (p = 0.02), but HF-AL did not differ from either CTRL-AL or HF-TRF. * One-way ANOVA with Tukey’s post hoc test. ** Steel–Dwass nonparametric test. CTRL-AL, control ad libitum feeding; HF-AL, high-fat ad libitum feeding; HF-TRF, high-fat time-restricted feeding. The asterisks are intended to denote which statistical tests were applied to each taxonomic family: non-parametric for non-Gaussian residual error and parametric for families with normally distributed residual error.
indicates each animal in HF-TRF (N = 14 for each diet). (A) * Bacteroidaceae were elevated in HF-TRF compared to CTRL-AL and HF-AL diets (p = 0.02). (B) ** Bifidobacteriaceae were not detected in mice undergoing HF-TRF feeding, and relative abundance was lower than CTRL-AL (p < 0.01). Two animals in the HF-AL had detectable OTUs attributable to Bifidobacteriaceae, but no differences were seen between the HF-AL and CTRL-AL or HF-TRF animals. (C) ** Christensenellaneae were elevated by HF-AL feeding compared to HF-TRF (p = 0.03), but animals with CTRL-AL feeding did not differ from HF-AL or HF-TRF. (D) ** Clostridiaceae were increased by HF-TRF compared to both CTRL-AL and HF-AL diets (p < 0.01 for both). (E) ** Enterococcaceae were increased by HF-AL feeding compared to CTRL-AL and HF-TRF (p < 0.01 and p = 0.04, respectively). (F) * Muribaculaceae were increased by CTRL-AL feeding compared to HF-AL and HF-TRF (p = 0.03 and p < 0.01, respectively). (G) * Rikenellaceae were higher in HF-AL and HF-TRF than in CTRL-AL (p = 0.04 and p < 0.01, respectively). (H) Saccharimonadaceae was reduced by HF-TRF compared to CTRL-AL (p = 0.02), but HF-AL did not differ from either CTRL-AL or HF-TRF. * One-way ANOVA with Tukey’s post hoc test. ** Steel–Dwass nonparametric test. CTRL-AL, control ad libitum feeding; HF-AL, high-fat ad libitum feeding; HF-TRF, high-fat time-restricted feeding. The asterisks are intended to denote which statistical tests were applied to each taxonomic family: non-parametric for non-Gaussian residual error and parametric for families with normally distributed residual error.
 each animal within LF-AL;
each animal within LF-AL;  indicates individual animals within HF-AL;
indicates individual animals within HF-AL;  indicates each animal in HF-TRF (N = 14 for each diet). (A) * Bacteroidaceae were elevated in HF-TRF compared to CTRL-AL and HF-AL diets (p = 0.02). (B) ** Bifidobacteriaceae were not detected in mice undergoing HF-TRF feeding, and relative abundance was lower than CTRL-AL (p < 0.01). Two animals in the HF-AL had detectable OTUs attributable to Bifidobacteriaceae, but no differences were seen between the HF-AL and CTRL-AL or HF-TRF animals. (C) ** Christensenellaneae were elevated by HF-AL feeding compared to HF-TRF (p = 0.03), but animals with CTRL-AL feeding did not differ from HF-AL or HF-TRF. (D) ** Clostridiaceae were increased by HF-TRF compared to both CTRL-AL and HF-AL diets (p < 0.01 for both). (E) ** Enterococcaceae were increased by HF-AL feeding compared to CTRL-AL and HF-TRF (p < 0.01 and p = 0.04, respectively). (F) * Muribaculaceae were increased by CTRL-AL feeding compared to HF-AL and HF-TRF (p = 0.03 and p < 0.01, respectively). (G) * Rikenellaceae were higher in HF-AL and HF-TRF than in CTRL-AL (p = 0.04 and p < 0.01, respectively). (H) Saccharimonadaceae was reduced by HF-TRF compared to CTRL-AL (p = 0.02), but HF-AL did not differ from either CTRL-AL or HF-TRF. * One-way ANOVA with Tukey’s post hoc test. ** Steel–Dwass nonparametric test. CTRL-AL, control ad libitum feeding; HF-AL, high-fat ad libitum feeding; HF-TRF, high-fat time-restricted feeding. The asterisks are intended to denote which statistical tests were applied to each taxonomic family: non-parametric for non-Gaussian residual error and parametric for families with normally distributed residual error.
indicates each animal in HF-TRF (N = 14 for each diet). (A) * Bacteroidaceae were elevated in HF-TRF compared to CTRL-AL and HF-AL diets (p = 0.02). (B) ** Bifidobacteriaceae were not detected in mice undergoing HF-TRF feeding, and relative abundance was lower than CTRL-AL (p < 0.01). Two animals in the HF-AL had detectable OTUs attributable to Bifidobacteriaceae, but no differences were seen between the HF-AL and CTRL-AL or HF-TRF animals. (C) ** Christensenellaneae were elevated by HF-AL feeding compared to HF-TRF (p = 0.03), but animals with CTRL-AL feeding did not differ from HF-AL or HF-TRF. (D) ** Clostridiaceae were increased by HF-TRF compared to both CTRL-AL and HF-AL diets (p < 0.01 for both). (E) ** Enterococcaceae were increased by HF-AL feeding compared to CTRL-AL and HF-TRF (p < 0.01 and p = 0.04, respectively). (F) * Muribaculaceae were increased by CTRL-AL feeding compared to HF-AL and HF-TRF (p = 0.03 and p < 0.01, respectively). (G) * Rikenellaceae were higher in HF-AL and HF-TRF than in CTRL-AL (p = 0.04 and p < 0.01, respectively). (H) Saccharimonadaceae was reduced by HF-TRF compared to CTRL-AL (p = 0.02), but HF-AL did not differ from either CTRL-AL or HF-TRF. * One-way ANOVA with Tukey’s post hoc test. ** Steel–Dwass nonparametric test. CTRL-AL, control ad libitum feeding; HF-AL, high-fat ad libitum feeding; HF-TRF, high-fat time-restricted feeding. The asterisks are intended to denote which statistical tests were applied to each taxonomic family: non-parametric for non-Gaussian residual error and parametric for families with normally distributed residual error.



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rust, B.M.; Picklo, M.J.; Yan, L.; Mehus, A.A.; Zeng, H. Time-Restricted Feeding Modifies the Fecal Lipidome and the Gut Microbiota. Nutrients 2023, 15, 1562. https://doi.org/10.3390/nu15071562
Rust BM, Picklo MJ, Yan L, Mehus AA, Zeng H. Time-Restricted Feeding Modifies the Fecal Lipidome and the Gut Microbiota. Nutrients. 2023; 15(7):1562. https://doi.org/10.3390/nu15071562
Chicago/Turabian StyleRust, Bret M., Matthew J. Picklo, Lin Yan, Aaron A. Mehus, and Huawei Zeng. 2023. "Time-Restricted Feeding Modifies the Fecal Lipidome and the Gut Microbiota" Nutrients 15, no. 7: 1562. https://doi.org/10.3390/nu15071562
APA StyleRust, B. M., Picklo, M. J., Yan, L., Mehus, A. A., & Zeng, H. (2023). Time-Restricted Feeding Modifies the Fecal Lipidome and the Gut Microbiota. Nutrients, 15(7), 1562. https://doi.org/10.3390/nu15071562




