Cytidine Alleviates Dyslipidemia and Modulates the Gut Microbiota Composition in ob/ob Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Animals and Administration
2.2. Oral Glucose Tolerance Test (OGTT)
2.3. Biochemical Indicator Detection and Histopathological Analysis
2.4. Gut Microbiome Analysis
2.5. Statistical Analysis
3. Results
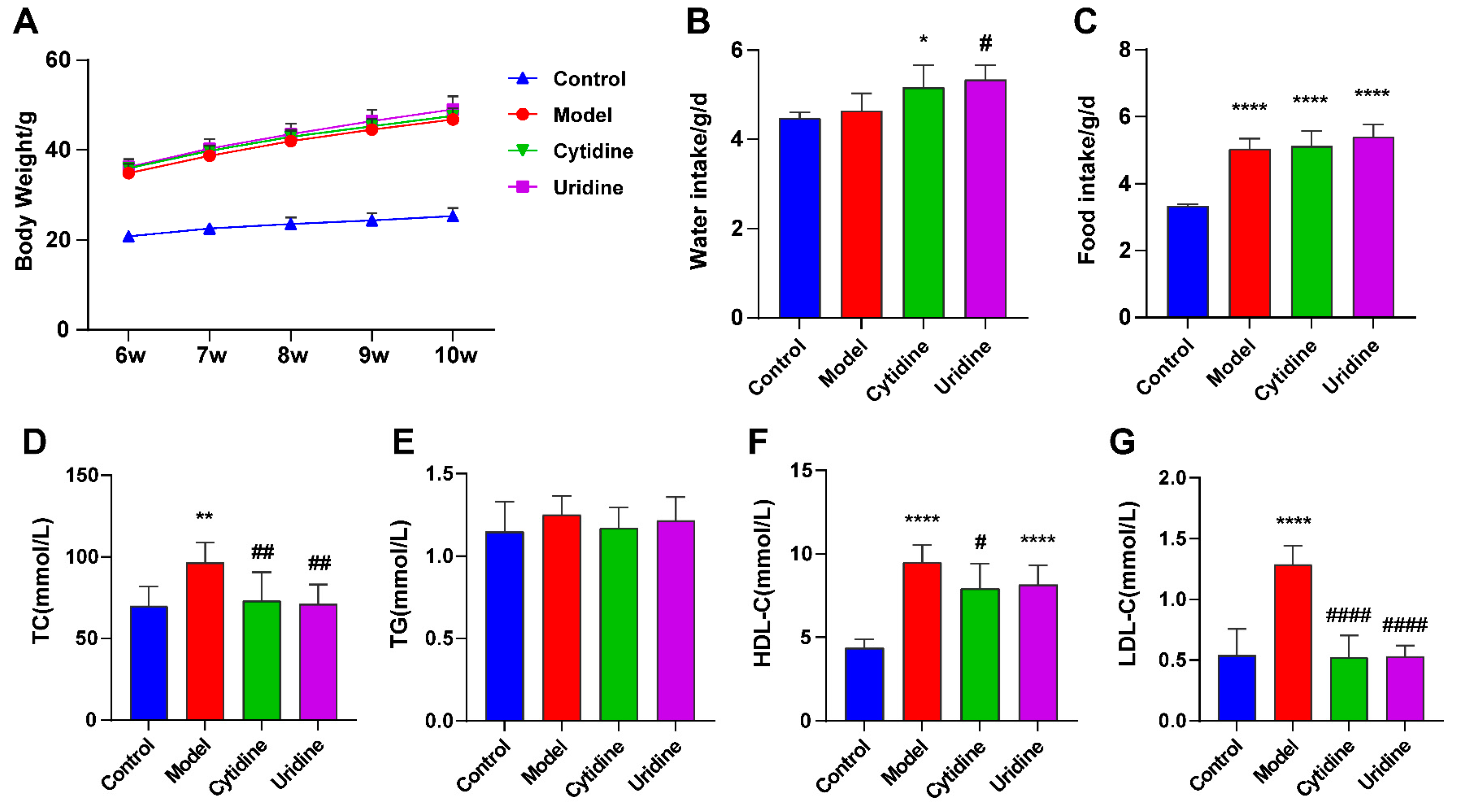
3.1. Effect of Cytidine on Body Weight, Water and Food Intake, and Serum Lipid Level
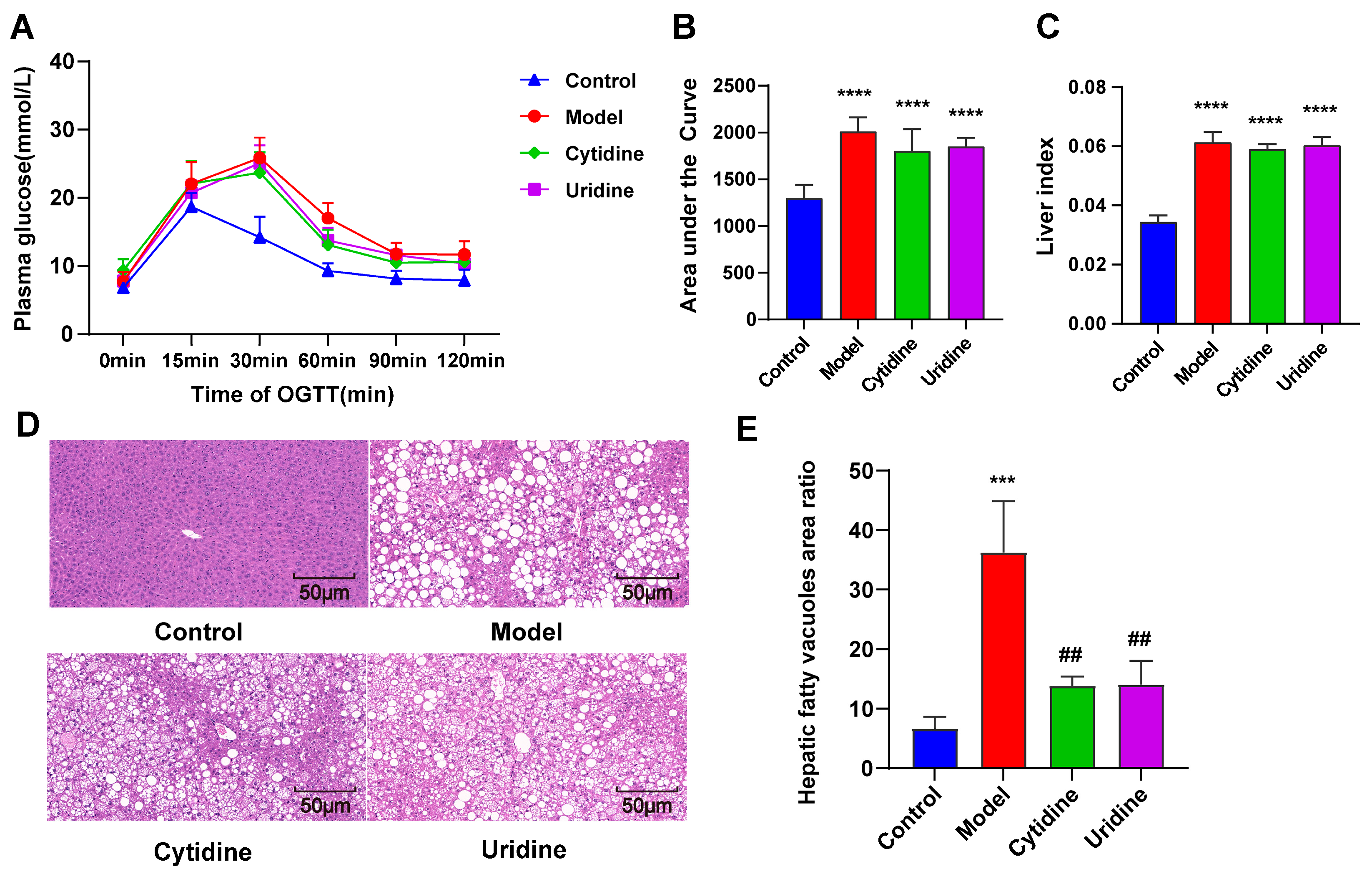
3.2. Effect of Cytidine on OGTT, Liver Index, and Hepatic Steatosis
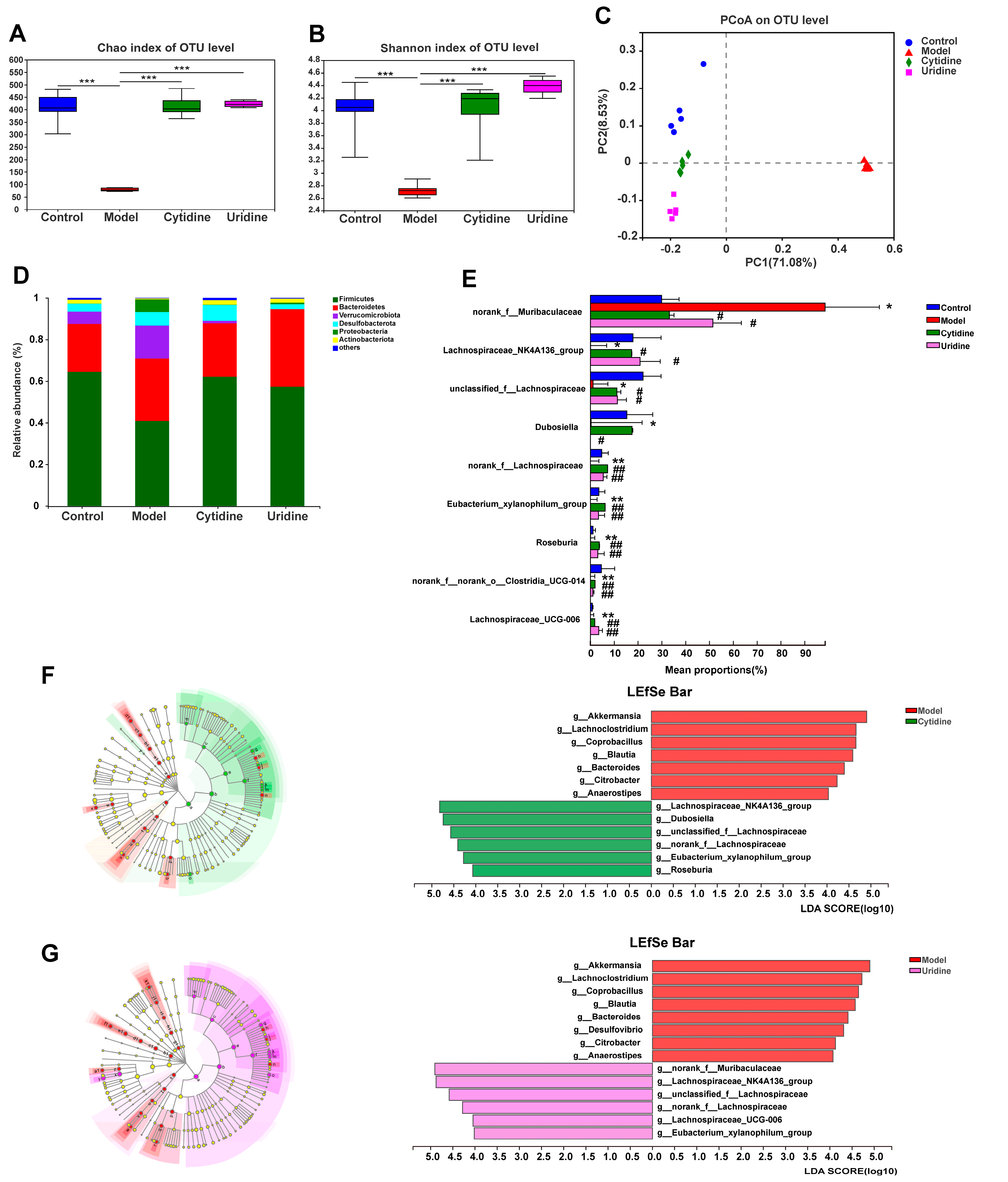
3.3. Effect of Cytidine on Gut Microbiota Composition in ob/ob Mice
3.4. Correlation Analysis of the Gut Microbiota Taxa
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adel Mehraban, M.S.; Tabatabaei-Malazy, O.; Rahimi, R.; Daniali, M.; Khashayar, P.; Larijani, B. Targeting dyslipidemia by herbal medicines: A systematic review of meta-analyses. J. Ethnopharmacol. 2021, 280, 114407. [Google Scholar] [CrossRef]
- Jia, X.; Xu, W.; Zhang, L.; Li, X.; Wang, R.; Wu, S. Impact of Gut Microbiota and Microbiota-Related Metabolites on Hyperlipidemia. Front. Cell. Infect. Microbiol. 2021, 11, 634780. [Google Scholar] [CrossRef]
- Schoeler, M.; Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef]
- Sun, S.S.; Wang, K.; Ma, K.; Bao, L.; Liu, H.W. An insoluble polysaccharide from the sclerotium of Poria cocos improves hyperglycemia, hyperlipidemia and hepatic steatosis in ob/ob mice via modulation of gut microbiota. Chin. J. Nat. Med. 2019, 17, 3–14. [Google Scholar] [CrossRef]
- Yu, Y.; Lu, J.; Sun, L.; Lyu, X.; Chang, X.Y.; Mi, X.; Hu, M.G.; Wu, C.; Chen, X. Akkermansia muciniphila: A potential novel mechanism of nuciferine to improve hyperlipidemia. Biomed. Pharmacother. 2021, 133, 111014. [Google Scholar] [CrossRef]
- Sanchez-Pozo, A.; Gil, A. Nucleotides as semiessential nutritional components. Br. J. Nutr. 2002, 87 (Suppl. S1), S135–S137. [Google Scholar] [CrossRef]
- Green Corkins, K.; Shurley, T. What’s in the Bottle? A Review of Infant Formulas. Nutr. Clin. Pract. 2016, 31, 723–729. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, Z.V.; Gordillo, R.; An, Y.; Zhang, C.; Liang, Q.; Yoshino, J.; Cautivo, K.M.; De Brabander, J.; Elmquist, J.K.; et al. An adipo-biliary-uridine axis that regulates energy homeostasis. Science 2017, 355, eaaf5375. [Google Scholar] [CrossRef]
- Le, T.T.; Ziemba, A.; Urasaki, Y.; Hayes, E.; Brotman, S.; Pizzorno, G. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. J. Lipid Res. 2013, 54, 1044–1057. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, C.; Zhai, Z.; Deng, Z.Y.; De Jonge, H.R.; Wu, X.; Ruan, Z. Uridine attenuates obesity, ameliorates hepatic lipid accumulation and modifies the gut microbiota composition in mice fed with a high-fat diet. Food Funct. 2021, 12, 1829–1840. [Google Scholar] [CrossRef]
- Urasaki, Y.; Pizzorno, G.; Le, T.T. Uridine affects liver protein glycosylation, insulin signaling, and heme biosynthesis. PLoS ONE 2014, 9, e99728. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, Z.V.; Gordillo, R.; Zhu, Y.; Ali, A.; Zhang, C.; Wang, X.; Shao, M.; Zhang, Z.; Iyengar, P.; et al. Adipocyte Xbp1s overexpression drives uridine production and reduces obesity. Mol. Metab. 2018, 11, 1–17. [Google Scholar] [CrossRef]
- Niu, K.; Bai, P.; Yang, B.; Feng, X.; Qiu, F. Asiatic acid alleviates metabolism disorders in ob/ob mice: Mechanistic insights. Food Funct. 2022, 13, 6934–6946. [Google Scholar] [CrossRef]
- Xia, J.F.; Hu, P.; Liang, Q.L.; Zou, T.T.; Wang, Y.M.; Luo, G.A. Correlations of creatine and six related pyrimidine metabolites and diabetic nephropathy in Chinese type 2 diabetic patients. Clin. Biochem. 2010, 43, 957–962. [Google Scholar] [CrossRef]
- Hamann, A.; Matthaei, S. Regulation of energy balance by leptin. Exp. Clin. Endocrinol. Diabetes 1996, 104, 293–300. [Google Scholar] [CrossRef]
- Urasaki, Y.; Pizzorno, G.; Le, T.T. Chronic Uridine Administration Induces Fatty Liver and Pre-Diabetic Conditions in Mice. PLoS ONE 2016, 11, e0146994. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Yang, M.; Liu, Y.; Xie, H.; Wen, Z.; Zhang, Y.; Wu, C.; Huang, L.; Wu, J.; Xie, C.; Wang, T.; et al. Gut Microbiota Composition and Structure of the Ob/Ob and Db/Db Mice. Int. J. Endocrinol. 2019, 2019, 1394097. [Google Scholar] [CrossRef]
- Gargari, G.; Deon, V.; Taverniti, V.; Gardana, C.; Denina, M.; Riso, P.; Guardamagna, O.; Guglielmetti, S. Evidence of dysbiosis in the intestinal microbial ecosystem of children and adolescents with primary hyperlipidemia and the potential role of regular hazelnut intake. FEMS Microbiol. Ecol. 2018, 94, fiy045. [Google Scholar] [CrossRef]
- Huang, F.; Zheng, X.; Ma, X.; Jiang, R.; Zhou, W.; Zhou, S.; Zhang, Y.; Lei, S.; Wang, S.; Kuang, J.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota and bile acid metabolism. Nat. Commun. 2019, 10, 4971. [Google Scholar] [CrossRef]
- Song, J.J.; Tian, W.J.; Kwok, L.Y.; Wang, Y.L.; Shang, Y.N.; Menghe, B.; Wang, J.G. Effects of microencapsulated Lactobacillus plantarum LIP-1 on the gut microbiota of hyperlipidaemic rats. Br. J. Nutr. 2017, 118, 481–492. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Xing, Y.; Xing, R.; Liu, Y.; Xu, Y. Changes of gut microbiota during silybin-mediated treatment of high-fat diet-induced non-alcoholic fatty liver disease in mice. Hepatol. Res. 2020, 50, 5–14. [Google Scholar] [CrossRef]
- Li, Y.; Yan, H.; Zhang, Y.; Li, Q.; Yu, L.; Li, Q.; Liu, C.; Xie, Y.; Chen, K.; Ye, F.; et al. Alterations of the Gut Microbiome Composition and Lipid Metabolic Profile in Radiation Enteritis. Front. Cell. Infect. Microbiol. 2020, 10, 541178. [Google Scholar] [CrossRef]
- Wei, J.; Zhao, Y.; Zhou, C.; Zhao, Q.; Zhong, H.; Zhu, X.; Fu, T.; Pan, L.; Shang, Q.; Yu, G. Dietary Polysaccharide from Enteromorpha clathrata Attenuates Obesity and Increases the Intestinal Abundance of Butyrate-Producing Bacterium, Eubacterium xylanophilum, in Mice Fed a High-Fat Diet. Polymers 2021, 13, 3286. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Yang, C.; Huang, S.; Lin, Z.; Chen, H.; Xu, C.; Lin, Y.; Sun, H.; Huang, F.; Lin, D.; Guo, F. Polysaccharides from Enteromorpha prolifera alleviate hypercholesterolemia via modulating the gut microbiota and bile acid metabolism. Food Funct. 2022, 13, 12194–12207. [Google Scholar] [CrossRef]
- Bachmanov, A.A.; Tordoff, M.G.; Beauchamp, G.K. Intake of umami-tasting solutions by mice: A genetic analysis. J. Nutr. 2000, 130 (Suppl. S4), 935S–941S. [Google Scholar] [CrossRef]
- Vanhaecke, T.; Bretin, O.; Poirel, M.; Tap, J. Drinking Water Source and Intake Are Associated with Distinct Gut Microbiota Signatures in US and UK Populations. J. Nutr. 2022, 152, 171–182. [Google Scholar] [CrossRef]
- Bowyer, R.C.E.; Schillereff, D.N.; Jackson, M.A.; Le Roy, C.; Wells, P.M.; Spector, T.D.; Steves, C.J. Associations between UK tap water and gut microbiota composition suggest the gut microbiome as a potential mediator of health differences linked to water quality. Sci. Total Environ. 2020, 739, 139697. [Google Scholar] [CrossRef]
- Hansen, T.H.; Thomassen, M.T.; Madsen, M.L.; Kern, T.; Bak, E.G.; Kashani, A.; Allin, K.H.; Hansen, T.; Pedersen, O. The effect of drinking water pH on the human gut microbiota and glucose regulation: Results of a randomized controlled cross-over intervention. Sci. Rep. 2018, 8, 16626. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, K.; Bai, P.; Zhang, J.; Feng, X.; Qiu, F. Cytidine Alleviates Dyslipidemia and Modulates the Gut Microbiota Composition in ob/ob Mice. Nutrients 2023, 15, 1147. https://doi.org/10.3390/nu15051147
Niu K, Bai P, Zhang J, Feng X, Qiu F. Cytidine Alleviates Dyslipidemia and Modulates the Gut Microbiota Composition in ob/ob Mice. Nutrients. 2023; 15(5):1147. https://doi.org/10.3390/nu15051147
Chicago/Turabian StyleNiu, Kaixia, Pengpeng Bai, Junyang Zhang, Xinchi Feng, and Feng Qiu. 2023. "Cytidine Alleviates Dyslipidemia and Modulates the Gut Microbiota Composition in ob/ob Mice" Nutrients 15, no. 5: 1147. https://doi.org/10.3390/nu15051147
APA StyleNiu, K., Bai, P., Zhang, J., Feng, X., & Qiu, F. (2023). Cytidine Alleviates Dyslipidemia and Modulates the Gut Microbiota Composition in ob/ob Mice. Nutrients, 15(5), 1147. https://doi.org/10.3390/nu15051147






