Dysbiosis of the Subgingival Microbiome and Relation to Periodontal Disease in Association with Obesity and Overweight
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approvals
2.2. Study Design
2.3. Inclusion Criteria
2.4. Anthropometric Measurements
2.5. Oral Examination and Periodontal Assessment
2.6. Subgingival Plaque Samples
2.7. Nanopore Sequencing of 16S rRNA Gene from the Subgingival Plaques
2.8. Statistical and Bioinformatics Analyses
3. Results
3.1. Demographic and Clinical Characteristics of Participants
3.2. Bacterial Abundance and Distribution
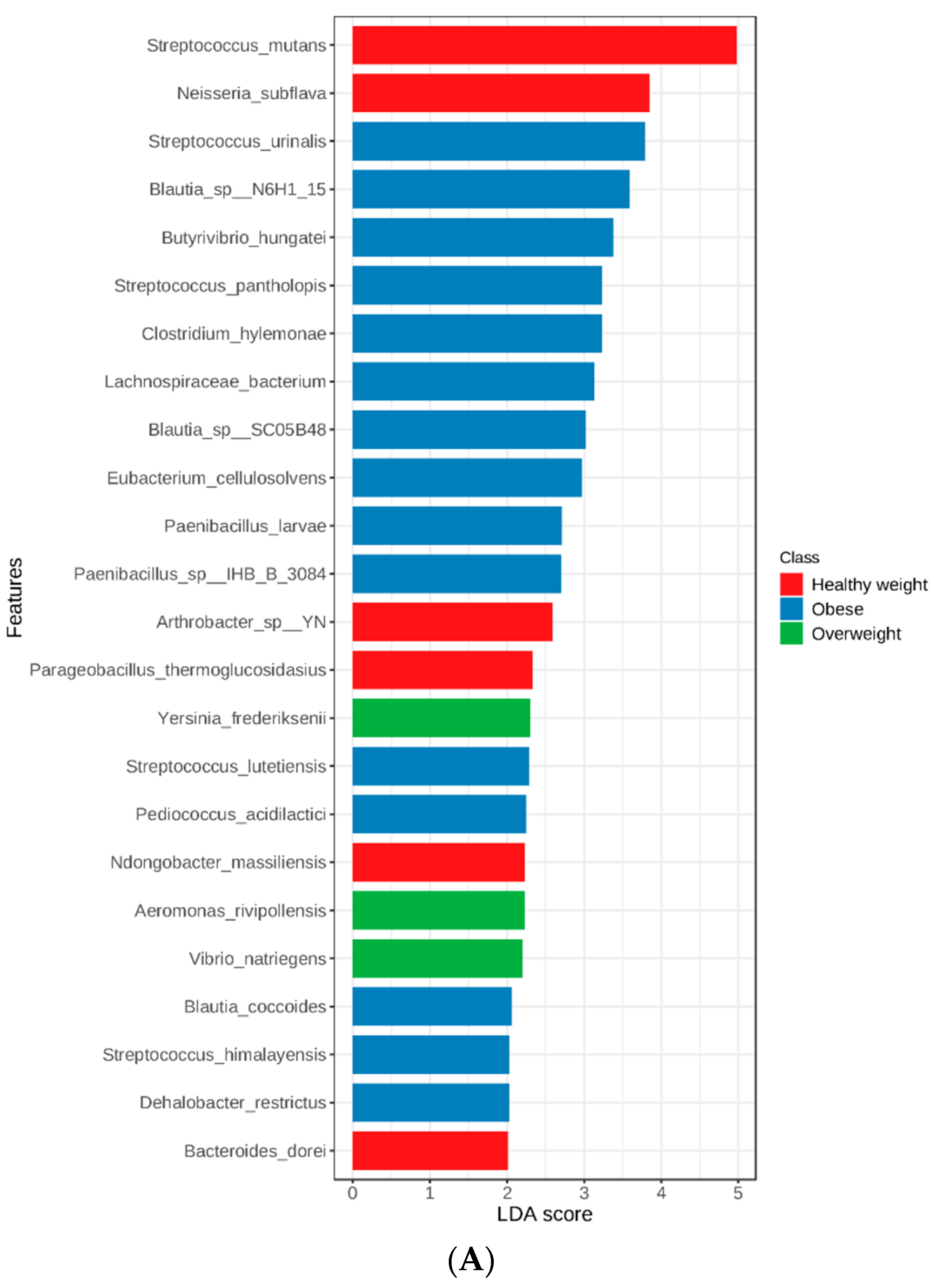
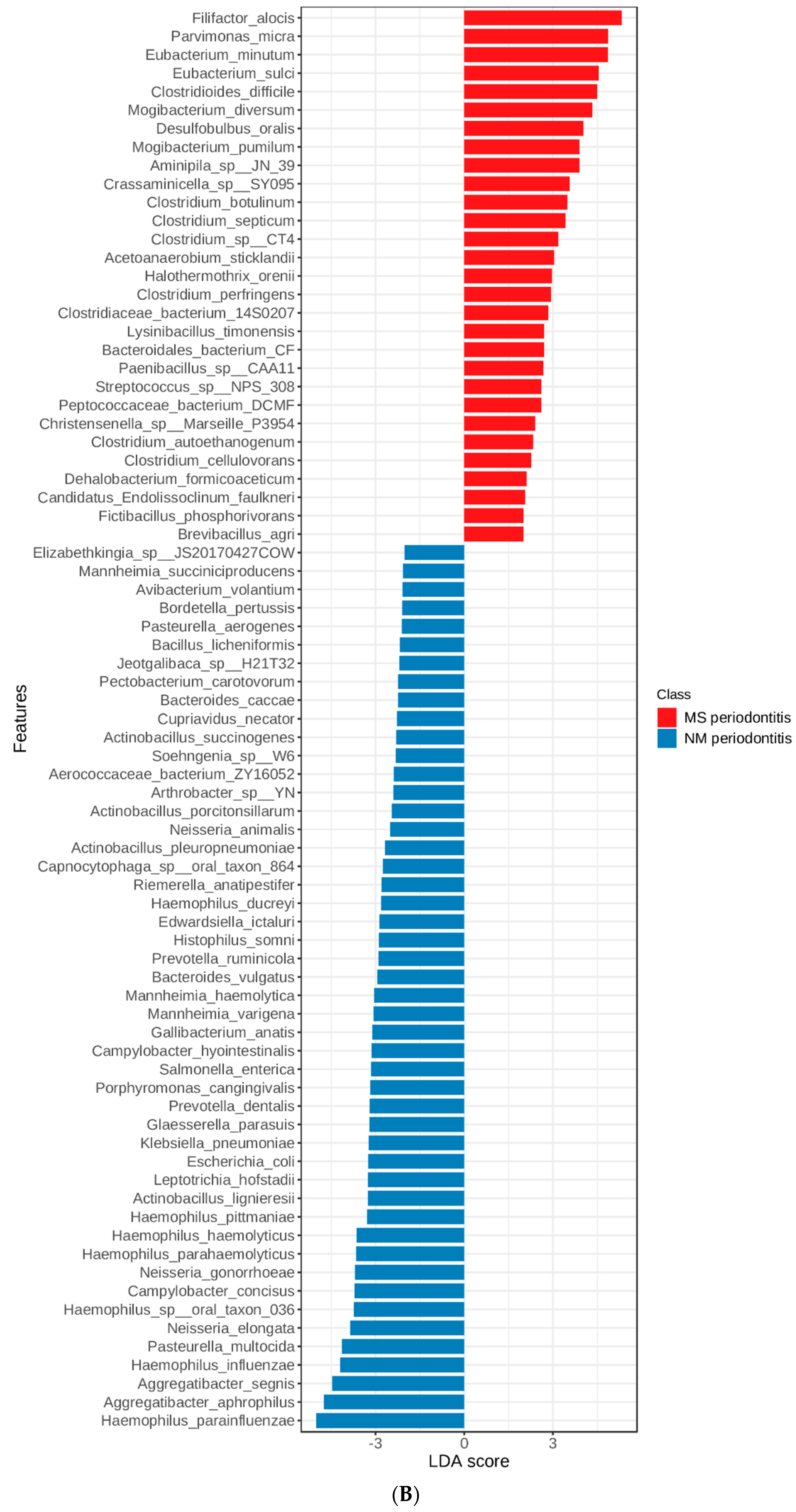
3.3. Linear Discriminant Analysis (LDA) Effect Size (LEfSe)
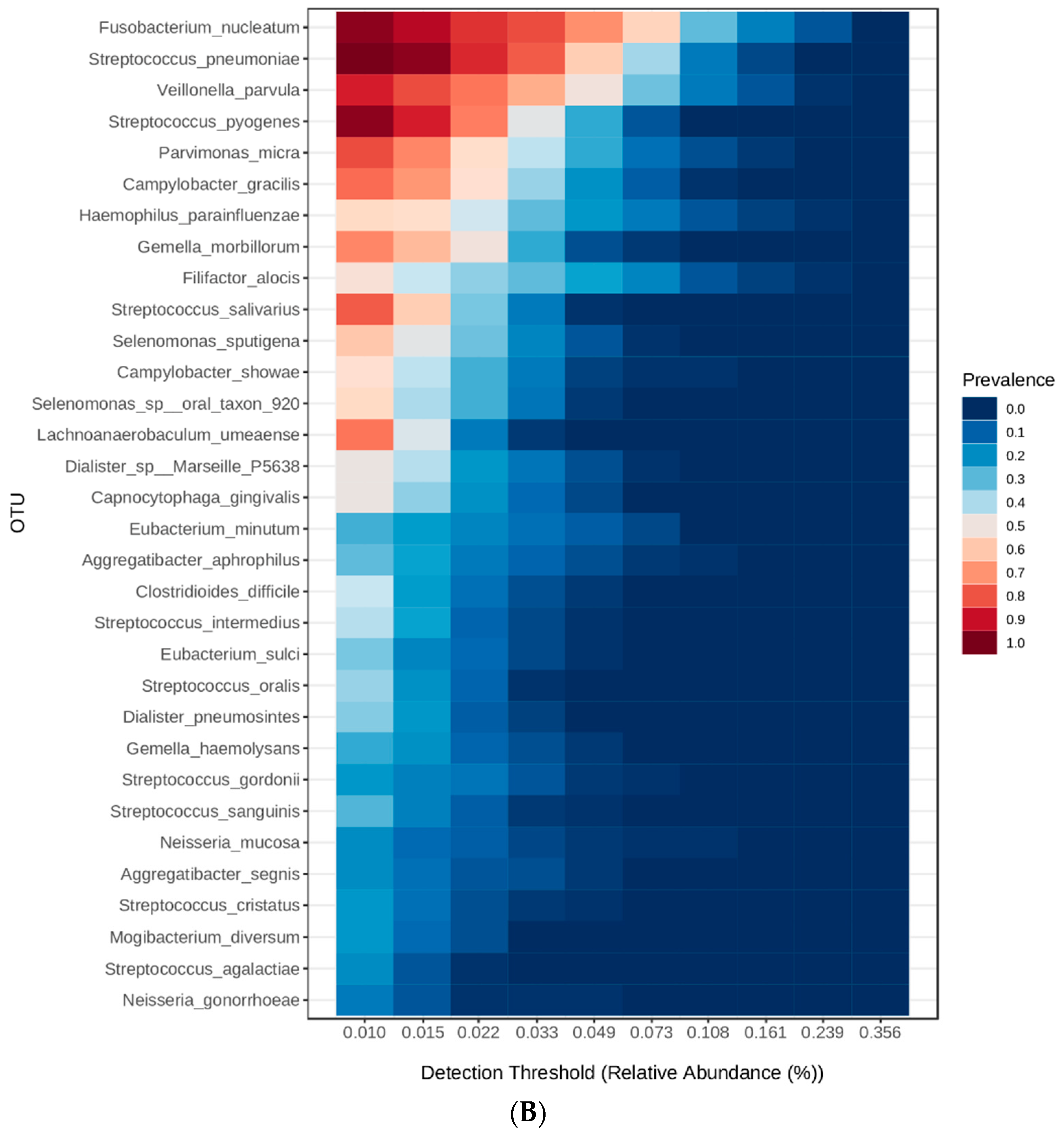
3.4. Heatmaps for Determination of Microbial Signatures
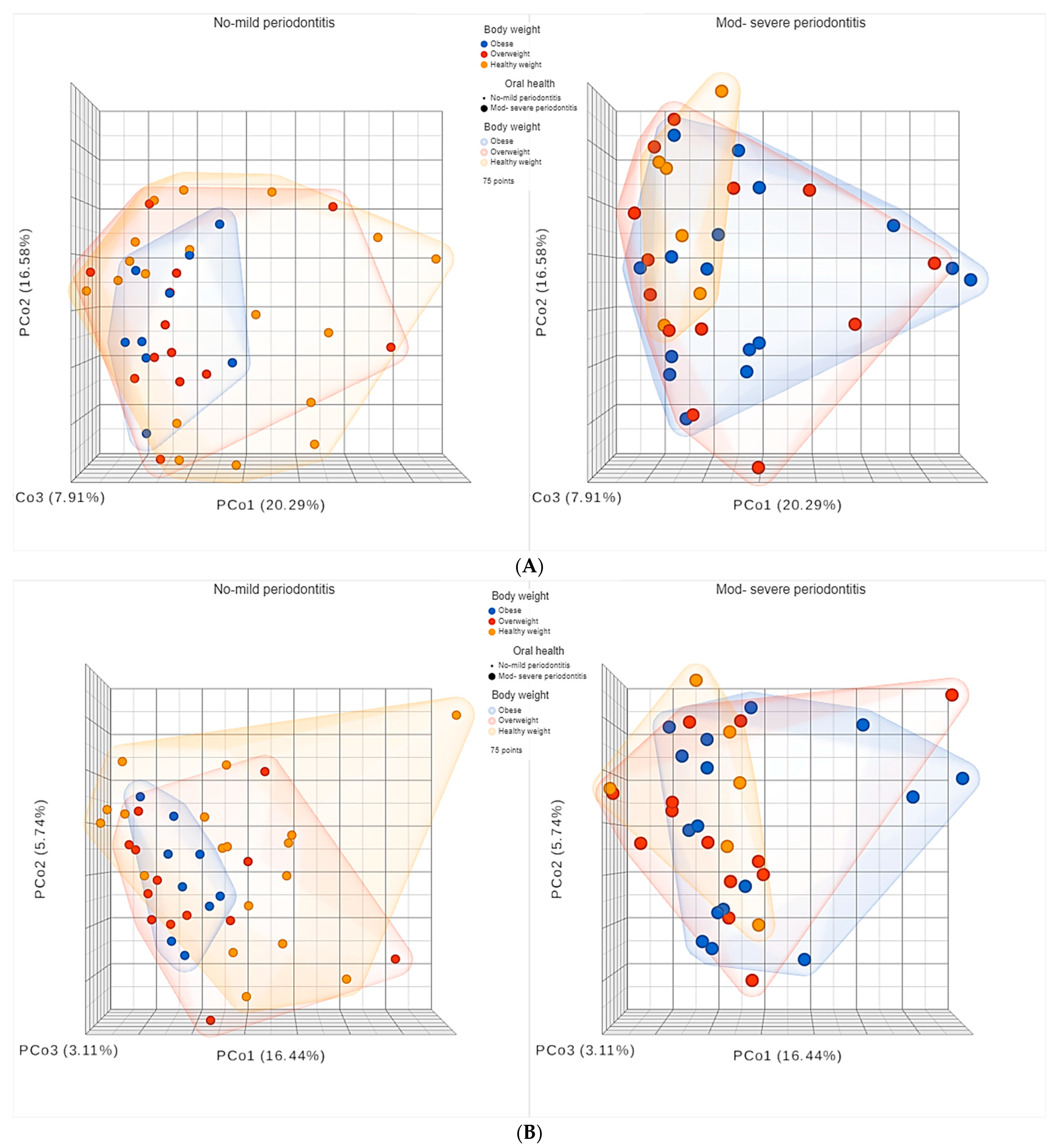
3.5. Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Okati-Aliabad, H.; Ansari-Moghaddam, A.; Kargar, S.; Jabbari, N. Prevalence of Obesity and Overweight among Adults in the Middle East Countries from 2000 to 2020: A Systematic Review and Meta-Analysis. J. Obes. 2022, 2022, 8074837. [Google Scholar] [CrossRef] [PubMed]
- Agha, M.; Agha, R. The Rising Prevalence of Obesity: Part A: Impact on Public Health. Int. J. Surg. Oncol. 2017, 2, e17. [Google Scholar] [CrossRef] [PubMed]
- Suvan, J.; D’Aiuto, F. Assessment and Management of Oral Health in Obesity. Curr. Obes. Rep. 2013, 2, 142–149. [Google Scholar] [CrossRef]
- Bourgeois, D.; Gonçalves, L.S.; da Costa Lima-Junior, J.; Carrouel, F. Editorial: The Oral Microbiome Is a Key Factor in Oral and Systemic Health. Front. Microbiol. 2022, 13, 855668. [Google Scholar] [CrossRef]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The Gut Microbiota Suppresses Insulin-Mediated Fat Accumulation via the Short-Chain Fatty Acid Receptor GPR43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus Report of Workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. 1), S173–S182. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and Grading of Periodontitis: Framework and Proposal of a New Classification and Case Definition. J. Periodontol. 2018, 89 (Suppl. 1), S159–S172. [Google Scholar] [CrossRef] [PubMed]
- Szkaradkiewicz, A.K.; Karpinski, T.M. Microbiology of Chronic Periodontitis. J. Biol. Earth Sci. 2013, 3, 14–20. [Google Scholar]
- Suzuki, N.; Yoneda, M.; Hirofuji, T. Mixed Red-Complex Bacterial Infection in Periodontitis. Int. J. Dent. 2013, 2013, 587279. [Google Scholar] [CrossRef]
- Fu, Y.-W.; Li, X.-X.; Xu, H.-Z.; Gong, Y.-Q.; Yang, Y. Effects of Periodontal Therapy on Serum Lipid Profile and Proinflammatory Cytokines in Patients with Hyperlipidemia: A Randomized Controlled Trial. Clin. Oral. Investig. 2016, 20, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Hotamisligil, G.S. Obesity-Induced Inflammatory Changes in Adipose Tissue. J. Clin. Investig. 2003, 112, 1785–1788. [Google Scholar] [CrossRef]
- Gregor, M.F.; Hotamisligil, G.S. Inflammatory Mechanisms in Obesity. Annu. Rev. Immunol. 2011, 29, 415–445. [Google Scholar] [CrossRef]
- Haffajee, A.D.; Socransky, S.S. Relation of Body Mass Index, Periodontitis and Tannerella Forsythia. J. Clin. Periodontol. 2009, 36, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Goodson, J.M.; Groppo, D.; Halem, S.; Carpino, E. Is Obesity an Oral Bacterial Disease? J. Dent. Res. 2009, 88, 519–523. [Google Scholar] [CrossRef]
- Feng, Z.; Weinberg, A. Role of Bacteria in Health and Disease of Periodontal Tissues. Periodontol. 2000 2006, 40, 50–76. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J.; Grossman, N.S. Periodontal Disease as a Specific, Albeit Chronic, Infection: Diagnosis and Treatment. Clin. Microbiol. Rev. 2001, 14, 727–752. [Google Scholar] [CrossRef]
- Burt, B. Research, Science and Therapy Committee of the American Academy of Periodontology Position Paper: Epidemiology of Periodontal Diseases. J. Periodontol. 2005, 76, 1406–1419. [Google Scholar] [CrossRef]
- Humphrey, L.L.; Fu, R.; Buckley, D.I.; Freeman, M.; Helfand, M. Periodontal Disease and Coronary Heart Disease Incidence: A Systematic Review and Meta-Analysis. J. Gen. Intern. Med. 2008, 23, 2079–2086. [Google Scholar] [CrossRef]
- Albandar, J.M. Underestimation of Periodontitis in NHANES Surveys. J. Periodontol. 2011, 82, 337–341. [Google Scholar] [CrossRef]
- Khan, S.; Bettiol, S.; Kent, K.; Barnett, T.; Peres, M.; Crocombe, L.A. Obesity and Periodontitis in Australian Adults: A Population-Based Cross-Sectional Study. Int. Dent. J. 2020, 70, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Xu, A.; Leung, W.K. Obesity, Bone Loss, and Periodontitis: The Interlink. Biomolecules 2022, 12, 865. [Google Scholar] [CrossRef] [PubMed]
- Probst, P.; Grummich, K.; Heger, P.; Zaschke, S.; Knebel, P.; Ulrich, A.; Büchler, M.W.; Diener, M.K. Blinding in Randomized Controlled Trials in General and Abdominal Surgery: Protocol for a Systematic Review and Empirical Study. Syst. Rev. 2016, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Simoni, L.; Shirka, E.; Hasimi, E.; Kabili, S.; Goda, A. Differences Among Body Mass Index (BMI) Groups in Patients Undergoing First Elective Percutaneous Coronary Intervention. Med. Arch. 2015, 69, 396–399. [Google Scholar] [CrossRef]
- Ainamo, J.; Bay, I. Problems and Proposals for Recording Gingivitis and Plaque. Int. Dent. J. 1975, 25, 229–235. [Google Scholar]
- Zeigler, C.C.; Persson, G.R.; Wondimu, B.; Marcus, C.; Sobko, T.; Modéer, T. Microbiota in the Oral Subgingival Biofilm Is Associated with Obesity in Adolescence. Obesity 2012, 20, 157–164. [Google Scholar] [CrossRef]
- Simón-Soro, A.; Tomás, I.; Cabrera-Rubio, R.; Catalan, M.D.; Nyvad, B.; Mira, A. Microbial Geography of the Oral Cavity. J. Dent. Res. 2013, 92, 616–621. [Google Scholar] [CrossRef]
- Al Kawas, S.; Al-Marzooq, F.; Rahman, B.; Shearston, J.A.; Saad, H.; Benzina, D.; Weitzman, M. The Impact of Smoking Different Tobacco Types on the Subgingival Microbiome and Periodontal Health: A Pilot Study. Sci. Rep. 2021, 11, 1113. [Google Scholar] [CrossRef]
- Al-Marzooq, F.; Al Kawas, S.; Rahman, B.; Shearston, J.A.; Saad, H.; Benzina, D.; Weitzman, M. Supragingival Microbiome Alternations as a Consequence of Smoking Different Tobacco Types and Its Relation to Dental Caries. Sci. Rep. 2022, 12, 2861. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Brinker, E.; Graff, E.C.; Cao, W.; Gross, A.L.; Johnson, A.K.; Zhang, C.; Martin, D.R.; Wang, X. Whole-Genome Shotgun Metagenomic Sequencing Reveals Distinct Gut Microbiome Signatures of Obese Cats. Microbiol. Spectr. 2022, 10, e00837-22. [Google Scholar] [CrossRef] [PubMed]
- WHO. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 1 January 2023).
- Sulaiman, N.; Elbadawi, S.; Hussein, A.; Abusnana, S.; Madani, A.; Mairghani, M.; Alawadi, F.; Sulaiman, A.; Zimmet, P.; Huse, O.; et al. Prevalence of Overweight and Obesity in United Arab Emirates Expatriates: The UAE National Diabetes and Lifestyle Study. Diabetol. Metab. Syndr. 2017, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Gorman, A.; Kaye, E.K.; Apovian, C.; Fung, T.T.; Nunn, M.; Garcia, R.I. Overweight and Obesity Predict Time to Periodontal Disease Progression in Men. J. Clin. Periodontol. 2012, 39, 107–114. [Google Scholar] [CrossRef]
- Jimenez, M.; Hu, F.B.; Marino, M.; Li, Y.; Joshipura, K.J. Prospective Associations between Measures of Adiposity and Periodontal Disease. Obesity 2012, 20, 1718–1725. [Google Scholar] [CrossRef] [PubMed]
- Chaffee, B.W.; Weston, S.J. Association between Chronic Periodontal Disease and Obesity: A Systematic Review and Meta-Analysis. J. Periodontol. 2010, 81, 1708–1724. [Google Scholar] [CrossRef]
- Saito, T.; Shimazaki, Y.; Sakamoto, M. Obesity and Periodontitis. N. Engl. J. Med. 1998, 339, 482–483. [Google Scholar] [CrossRef]
- Al-Zahrani, M.S.; Bissada, N.F.; Borawskit, E.A. Obesity and Periodontal Disease in Young, Middle-Aged, and Older Adults. J. Periodontol. 2003, 74, 610–615. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel Disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Q.; Zheng, W.; Steinwandel, M.; Blot, W.J.; Shu, X.-O.; Long, J. Oral Microbiome and Obesity in a Large Study of Low-Income and African-American Populations. J. Oral. Microbiol. 2019, 11, 1650597. [Google Scholar] [CrossRef]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’Sullivan, O.; Fouhy, F.; Clarke, S.F.; O’Toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and Energy Harvesting Capacity of the Gut Microbiota: Relationship to Diet, Obesity and Time in Mouse Models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef]
- Stephens, R.W.; Arhire, L.; Covasa, M. Gut Microbiota: From Microorganisms to Metabolic Organ Influencing Obesity. Obesity 2018, 26, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.; Hoffmann, T.; Fischer, S.; Bornstein, S.; Gräßler, J.; Noack, B. Obesity Alters Composition and Diversity of the Oral Microbiota in Patients with Type 2 Diabetes Mellitus Independently of Glycemic Control. PLoS ONE 2018, 13, e0204724. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.J. Chapter 7—Gamma Proteobacteria. In Taxonomic Guide to Infectious Diseases; Berman, J.J., Ed.; Academic Press: Boston, MA, USA, 2012; pp. 37–47. ISBN 978-0-12-415895-5. [Google Scholar]
- Kajihara, T.; Yahara, K.; Yoshikawa, M.; Haruta, A.; Kawada-Matsuo, M.; Le, M.N.-T.; Arai, C.; Takeuchi, M.; Kitamura, N.; Sugawara, Y.; et al. Oral and Rectal Colonization by Antimicrobial-Resistant Gram-Negative Bacteria and Their Association with Death among Residents of Long-Term Care Facilities: A Prospective, Multicenter, Observational, Cohort Study. GER 2022, 1–12. [Google Scholar] [CrossRef]
- Tseng, C.-H.; Wu, C.-Y. The Gut Microbiome in Obesity. J. Formos. Med. Assoc. 2019, 118, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Al-Rawi, N.; Al-Marzooq, F. The Relation between Periodontopathogenic Bacterial Levels and Resistin in the Saliva of Obese Type 2 Diabetic Patients. J. Diabetes Res. 2017, 2017, 2643079. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S.; Mahendra, J.; Kumar, A.R.P.; Singh, G.; Jayaraman, S.; Paul, R. Comparative Analysis of Subgingival Red Complex Bacteria in Obese and Normal Weight Subjects with and without Chronic Periodontitis. J. Indian Soc. Periodontol. 2017, 21, 186–191. [Google Scholar] [CrossRef]
- Maciel, S.S.; Feres, M.; Gonçalves, T.E.D.; Zimmermann, G.S.; da Silva, H.D.P.; Figueiredo, L.C.; Duarte, P.M. Does Obesity Influence the Subgingival Microbiota Composition in Periodontal Health and Disease? J. Clin. Periodontol. 2016, 43, 1003–1012. [Google Scholar] [CrossRef]
- Silva-Boghossian, C.M.; Cesário, P.C.; Leão, A.T.T.; Colombo, A.P.V. Subgingival Microbial Profile of Obese Women with Periodontal Disease. J. Periodontol. 2018, 89, 186–194. [Google Scholar] [CrossRef]
- Vedam, V.; Ganapathy, S. Association of Oral Microbiota with Obesity in Children: Insight from Dental Physicians. Hong Kong Med. J. 2020, 26, 354. [Google Scholar] [CrossRef]
- Thomas, C.; Minty, M.; Canceill, T.; Loubières, P.; Azalbert, V.; Tercé, F.; Champion, C.; Burcelin, R.; Barthet, P.; Laurencin-Dalicieux, S.; et al. Obesity Drives an Oral Microbiota Signature of Female Patients with Periodontitis: A Pilot Study. Diagnostics 2021, 11, 745. [Google Scholar] [CrossRef]
- Genco, R.J.; LaMonte, M.J.; McSkimming, D.I.; Buck, M.J.; Li, L.; Hovey, K.M.; Andrews, C.A.; Sun, Y.; Tsompana, M.; Zheng, W.; et al. The Subgingival Microbiome Relationship to Periodontal Disease in Older Women. J. Dent. Res. 2019, 98, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Bu, S.; Sugino, K.; Garneau, N.; Nuessle, T.; Tucker, R.; Comstock, S. A Citizen Science Project: Associations of the Oral Microbiota with Body Mass Index and Measures of Sweet Taste Liking (P20-030-19). Curr. Dev. Nutr. 2019, 3, nzz040-P20. [Google Scholar] [CrossRef]
- Bombin, A.; Yan, S.; Bombin, S.; Mosley, J.D.; Ferguson, J.F. Obesity Influences Composition of Salivary and Fecal Microbiota and Impacts the Interactions between Bacterial Taxa. Physiol. Rep. 2022, 10, e15254. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Li, L.; Zhang, Y.; Wang, M.; Chen, F.; Ge, S.; Chen, B.; Yan, F. Periodontitis May Induce Gut Microbiota Dysbiosis via Salivary Microbiota. Int. J. Oral. Sci. 2022, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.S.; Hayward, M.R.; Coelho, L.P.; Li, S.S.; Costea, P.I.; Voigt, A.Y.; Wirbel, J.; Maistrenko, O.M.; Alves, R.J.; Bergsten, E.; et al. Extensive Transmission of Microbes along the Gastrointestinal Tract. eLife 2019, 8, e42693. [Google Scholar] [CrossRef]
- Kleinstein, S.E.; Nelson, K.E.; Freire, M. Inflammatory Networks Linking Oral Microbiome with Systemic Health and Disease. J. Dent. Res. 2020, 99, 1131–1139. [Google Scholar] [CrossRef]
- Konkel, J.E.; O’Boyle, C.; Krishnan, S. Distal Consequences of Oral Inflammation. Front. Immunol. 2019, 10, 1403. [Google Scholar] [CrossRef]
- Blasco-Baque, V.; Garidou, L.; Pomié, C.; Escoula, Q.; Loubieres, P.; Le Gall-David, S.; Lemaitre, M.; Nicolas, S.; Klopp, P.; Waget, A.; et al. Periodontitis Induced by Porphyromonas Gingivalis Drives Periodontal Microbiota Dysbiosis and Insulin Resistance via an Impaired Adaptive Immune Response. Gut 2017, 66, 872–885. [Google Scholar] [CrossRef]
- Allaker, R.P.; Stephen, A.S. Use of Probiotics and Oral Health. Curr. Oral. Health Rep. 2017, 4, 309–318. [Google Scholar] [CrossRef]
- Gedam, K.Y.; Katre, A.N. Efficacy of Probiotic, Chlorhexidine, and Sodium Fluoride Mouthrinses on Mutans Streptococci in 8- to 12-Year-Old Children: A Crossover Randomized Trial. LFG 2022, 15, 35–44. [Google Scholar] [CrossRef]
- Henrique Soares, K.; Firoozi, P.; Maria de Souza, G.; Beatriz Lopes Martins, O.; Gabriel Moreira Falci, S.; Rocha dos Santos, C.R. Efficacy of Probiotics Compared to Chlorhexidine Mouthwash in Improving Periodontal Status: A Systematic Review and Meta-Analysis. Int. J. Dent. 2023, 2023, e4013004. [Google Scholar] [CrossRef]
- Butera, A.; Gallo, S.; Maiorani, C.; Molino, D.; Chiesa, A.; Preda, C.; Esposito, F.; Scribante, A. Probiotic Alternative to Chlorhexidine in Periodontal Therapy: Evaluation of Clinical and Microbiological Parameters. Microorganisms 2020, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Syromyatnikov, M.; Nesterova, E.; Gladkikh, M.; Smirnova, Y.; Gryaznova, M.; Popov, V. Characteristics of the Gut Bacterial Composition in People of Different Nationalities and Religions. Microorganisms 2022, 10, 1866. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, B.; Selvaraju, V.; Chen, J.; Ayine, P.; Yang, L.; Ramesh Babu, J.; Geetha, T.; Taneja, V. Ethnic Variability Associating Gut and Oral Microbiome with Obesity in Children. Gut Microbes 2021, 13, 1882926. [Google Scholar] [CrossRef]
- Ang, Q.Y.; Alba, D.L.; Upadhyay, V.; Bisanz, J.E.; Cai, J.; Lee, H.L.; Barajas, E.; Wei, G.; Noecker, C.; Patterson, A.D.; et al. The East Asian Gut Microbiome Is Distinct from Colocalized White Subjects and Connected to Metabolic Health. eLife 2021, 10, e70349. [Google Scholar] [CrossRef]
- Rosselli, R.; Romoli, O.; Vitulo, N.; Vezzi, A.; Campanaro, S.; de Pascale, F.; Schiavon, R.; Tiarca, M.; Poletto, F.; Concheri, G.; et al. Direct 16S RRNA-Seq from Bacterial Communities: A PCR-Independent Approach to Simultaneously Assess Microbial Diversity and Functional Activity Potential of Each Taxon. Sci. Rep. 2016, 6, 32165. [Google Scholar] [CrossRef] [PubMed]










| Characteristics | ||
|---|---|---|
| Continuous variables | Mean | Range |
| Age (years) | 31.1 ± 10.4 | 18–55 |
| Body weight (kg) | 84.4 ± 22.9 | 53–154.1 |
| Hight (cm) | 171.5 ± 9.3 | 153–190 |
| BMI | 28.4 ± 6.7 | 18.9–56.1 |
| Waist circumference (cm) | 45.3 ± 16.2 | 20.4–108 |
| Hip circumference (cm) | 49.6 ± 19.6 | 24.4–114 |
| WHR | 0.9 ± 0.09 | 0.7–1.2 |
| Categorical variables | N | % |
| Gender | ||
| - Female | 16 | 21.3 |
| - Male | 59 | 78.7 |
| Periodontal health | ||
| - No-mild periodontitis | 40 | 53.3 |
| - Moderate-severe periodontitis | 35 | 46.7 |
| Criteria | Moderate-Severe Periodontitis | No-Mild Periodontitis | p Value | ||
|---|---|---|---|---|---|
| Mean ± SD | Range | Mean ± SD | Range | ||
| Anthropometric parameters | |||||
| Waist circumference | 44.10 ± 12.66 | 30.00–108.00 | 46.23 ± 18.83 | 20.40–102.00 | 0.414 |
| Hip circumference | 45.45 ± 12.37 | 36.60–110.00 | 53.04 ± 23.76 | 24.40–114.00 | 0.908 |
| WHR * | 0.97 ± 0.07 | 0.80–1.20 | 0.90 ± 0.09 | 0.7–1.09 | 0.007 |
| Body weight | 88.32 ± 21.45 | 60.00–154.10 | 81.06 ± 23.80 | 53.00–151.00 | 0.086 |
| BMI * | 30.08 ± 5.95 | 18.90–44.60 | 26.97 ± 7.05 | 19.15–56.10 | 0.008 |
| Periodontal health parameters # | |||||
| PD * | 2.96± 0.64 | 2.04–5.54 | 2.29 ± 0.57 | 1.10–3.96 | <0.001 |
| CAL * | 1.76 ± 1.40 | 0.43–7.29 | 0.7 ± 0.42 | 0.00–2.77 | <0.001 |
| DMFT | 7.47 ± 5.48 | 0–22 | 5.23 ± 4.8 | 0–17 | 0.067 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, B.; Al-Marzooq, F.; Saad, H.; Benzina, D.; Al Kawas, S. Dysbiosis of the Subgingival Microbiome and Relation to Periodontal Disease in Association with Obesity and Overweight. Nutrients 2023, 15, 826. https://doi.org/10.3390/nu15040826
Rahman B, Al-Marzooq F, Saad H, Benzina D, Al Kawas S. Dysbiosis of the Subgingival Microbiome and Relation to Periodontal Disease in Association with Obesity and Overweight. Nutrients. 2023; 15(4):826. https://doi.org/10.3390/nu15040826
Chicago/Turabian StyleRahman, Betul, Farah Al-Marzooq, Hiba Saad, Dalenda Benzina, and Sausan Al Kawas. 2023. "Dysbiosis of the Subgingival Microbiome and Relation to Periodontal Disease in Association with Obesity and Overweight" Nutrients 15, no. 4: 826. https://doi.org/10.3390/nu15040826
APA StyleRahman, B., Al-Marzooq, F., Saad, H., Benzina, D., & Al Kawas, S. (2023). Dysbiosis of the Subgingival Microbiome and Relation to Periodontal Disease in Association with Obesity and Overweight. Nutrients, 15(4), 826. https://doi.org/10.3390/nu15040826






