Lactobacillus rhamnosus GG Promotes Recovery of the Colon Barrier in Septic Mice through Accelerating ISCs Regeneration
Abstract
1. Introduction
2. Material and Methods
2.1. Ethics Statement
2.2. Mice Treatment and Septic Model Building
2.3. Preparation of LGG Supernatant
2.4. Enteroids Establishment and Maintenance
2.5. Enteroids Treated with TNF-α and Pre-Treated with the Supernatant of LGG
2.6. Histological Analysis
2.7. LGG Detection in Fecal Samples
2.8. Quantitative PCR
2.9. RNA-Sequence
2.10. Statistical Analysis
3. Results
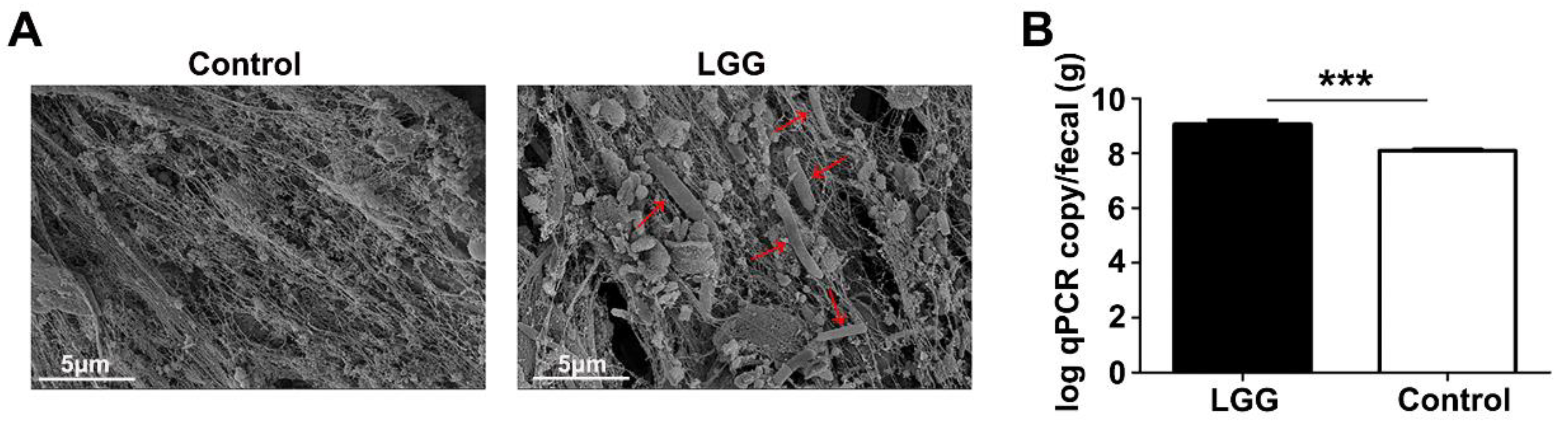
3.1. Colonization of LGG on Colon Mucosa
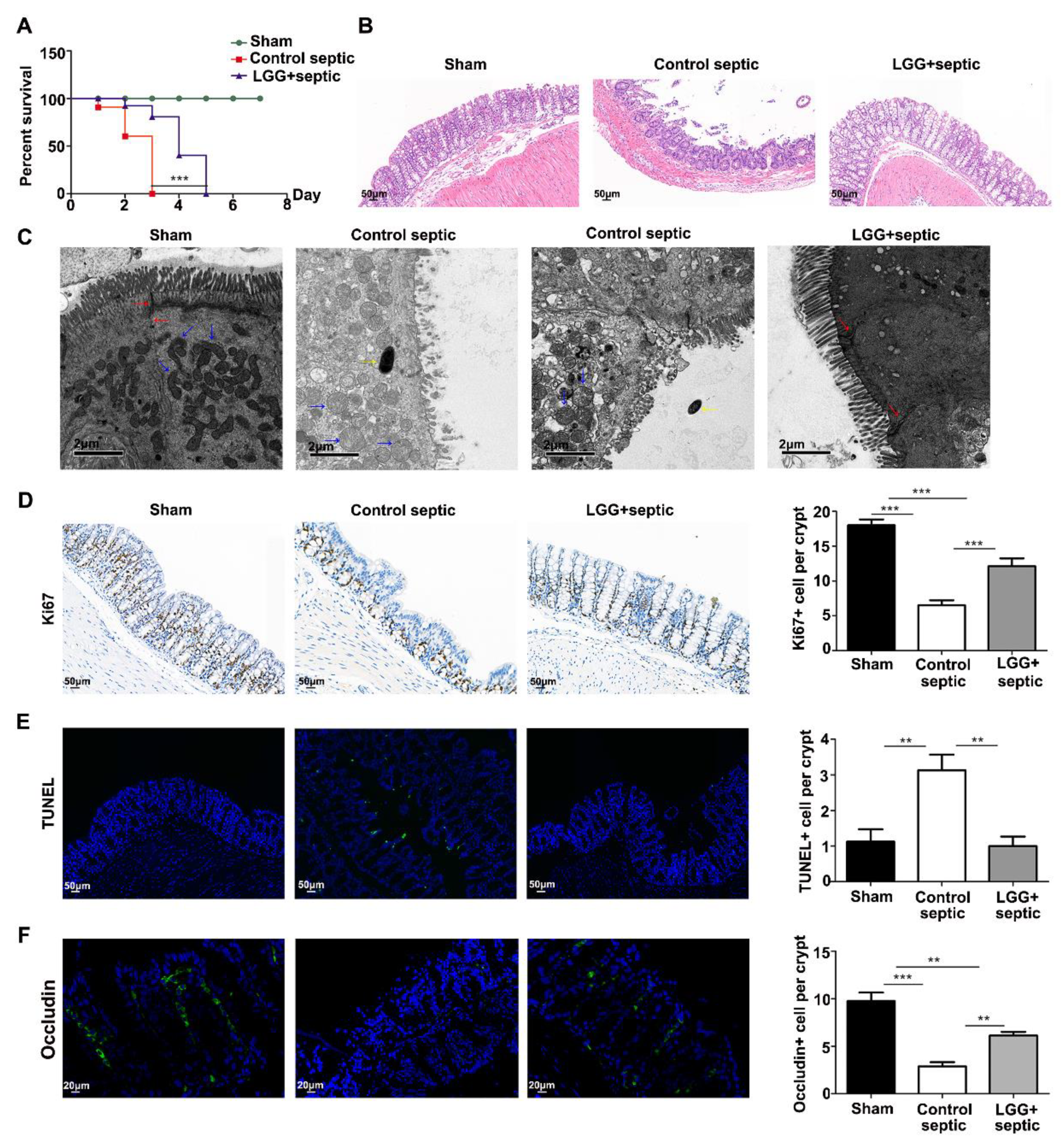
3.2. LGG Ameliorates Intestinal Barrier Injury in Septic Mice
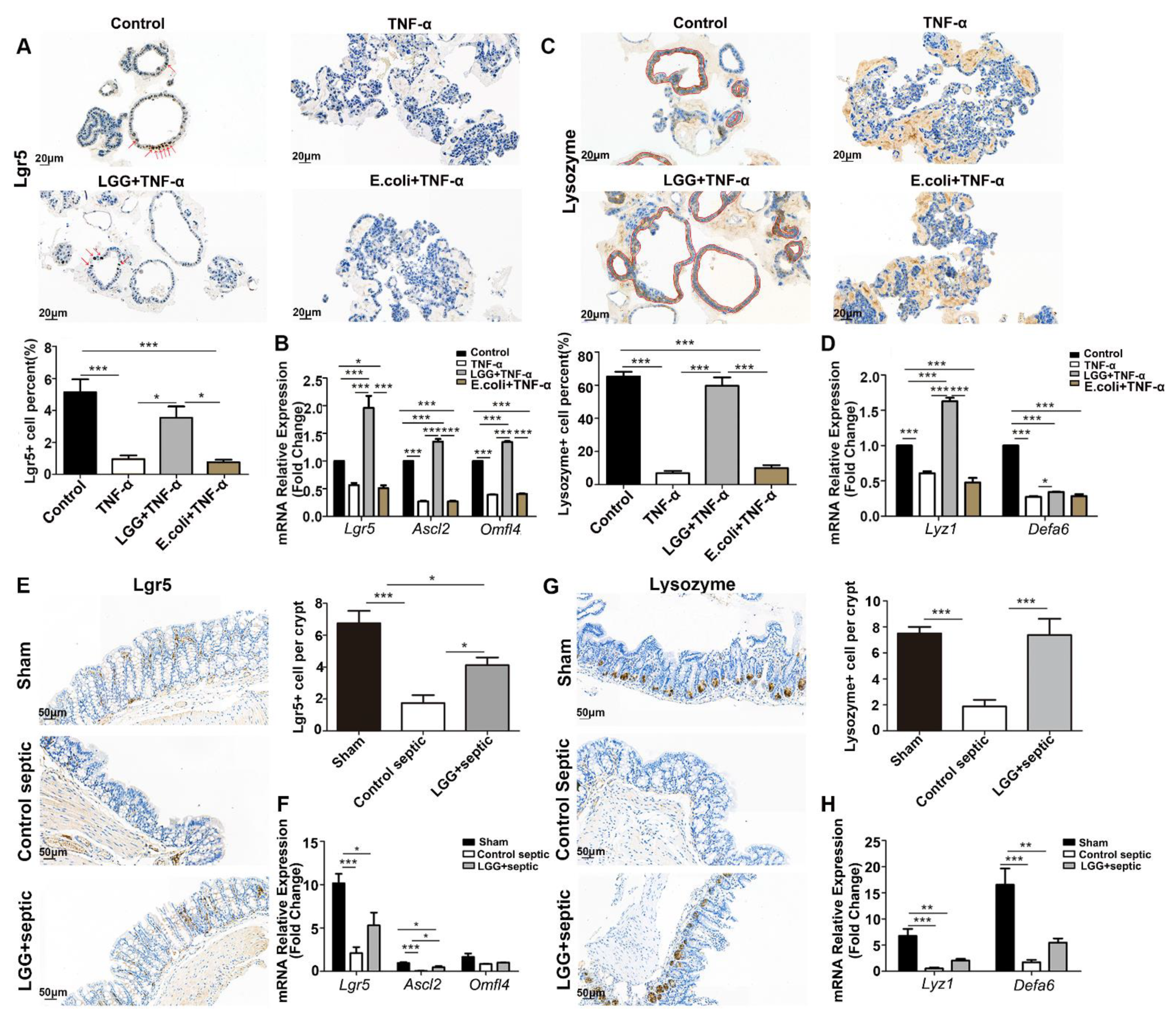
3.3. LGG Promotes Growth of Colonoids and Recovery from TNF-α Injury
3.4. LGG Promotes the Activation and Proliferation of ISCs after Damage
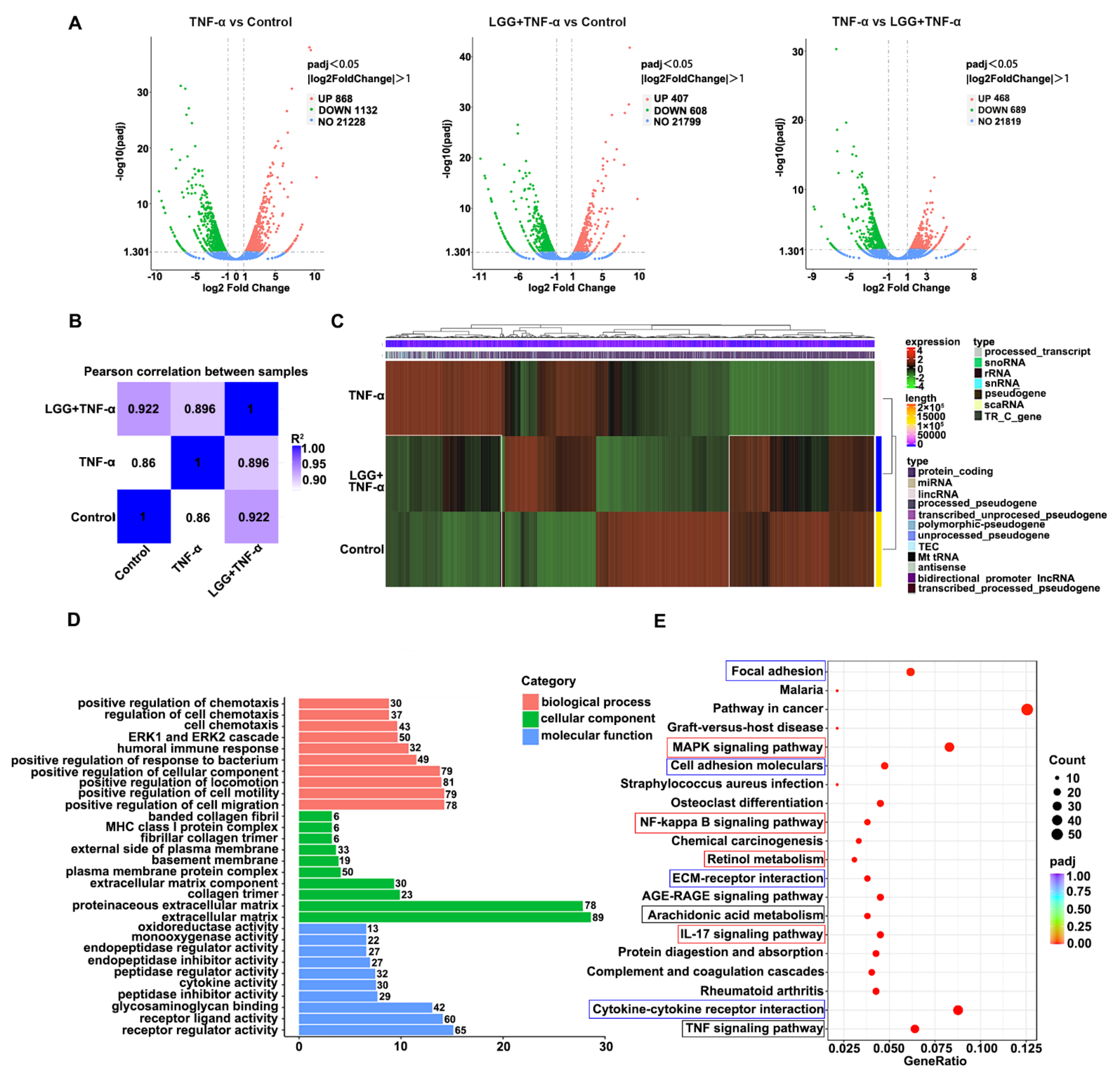
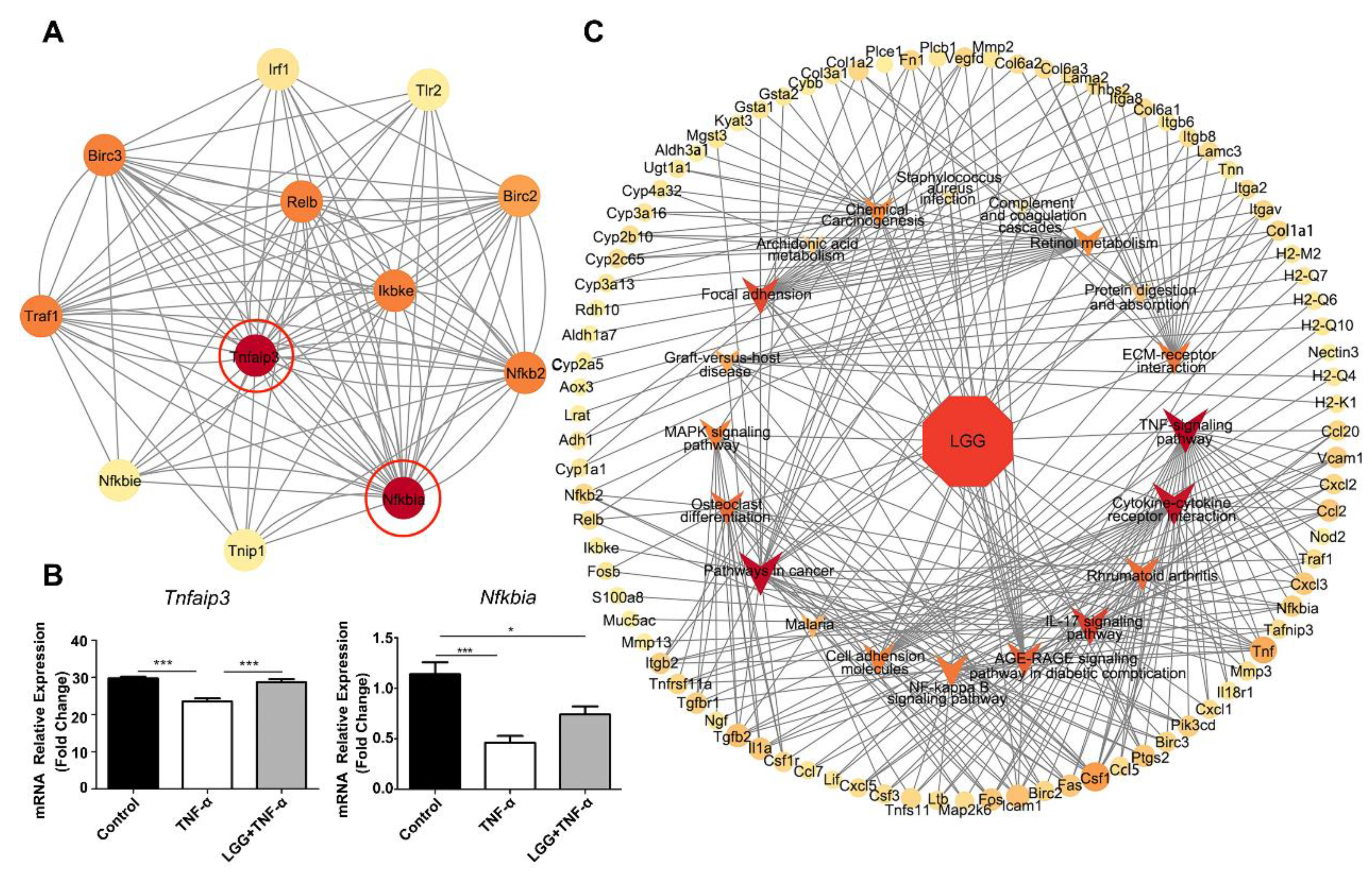
3.5. LGG Pretreatment Remolds the Transcriptional Profile in TNF-Injured Colonoids Injured by TNF-α
3.6. Construction of LGG-Pathway-Target Network and Module Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ISC | intestinal stem cells |
| LGG | Lactobacillus rhamnosus GG |
| RNA-seq | RNA-sequencing |
| CLP | cecum ligation and puncture |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DEGs | Differentially-expressed genes |
| PPI | Protein–protein interaction |
References
- Haussner, F.; Chakraborty, S.; Halbgebauer, R.; Huber-Lang, M. Challenge to the Intestinal Mucosa During Sepsis. Front. Immunol. 2019, 10, 891. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.C.; Lacy, B.E.; Talley, N.J. Irritable Bowel Syndrome. N. Engl. J. Med. 2017, 376, 2566–2578. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, W.; Lan, P.; Mou, X. The microbiome in inflammatory bowel diseases: From pathogenesis to therapy. Protein Cell 2021, 12, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Haak, B.W.; Wiersinga, W.J. The role of the gut microbiota in sepsis. Lancet Gastroenterol. Hepatol. 2017, 2, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Allegretti, J.R.; Mullish, B.H.; Kelly, C.; Fischer, M. The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet 2019, 394, 420–431. [Google Scholar] [CrossRef]
- Haak, B.W.; Prescott, H.C.; Wiersinga, W.J. Therapeutic Potential of the Gut Microbiota in the Prevention and Treatment of Sepsis. Front. Immunol. 2018, 9, 2042. [Google Scholar] [CrossRef] [PubMed]
- Capurso, L. Thirty Years of Lactobacillus rhamnosus GG: A Review. J. Clin. Gastroenterol. 2019, 53, S1–S41. [Google Scholar]
- Serra, D.; Mayr, U.; Boni, A.; Lukonin, I.; Rempfler, M.; Challet Meylan, L.; Stadler, M.B.; Strnad, P.; Papasaikas, P.; Vischi, D.; et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 2019, 569, 66–72. [Google Scholar] [CrossRef]
- Zou, W.Y.; Blutt, S.E.; Crawford, S.E.; Ettayebi, K.; Zeng, X.L.; Saxena, K.; Ramani, S.; Karandikar, U.C.; Zachos, N.C.; Estes, M.K. Human Intestinal Enteroids: New Models to Study Gastrointestinal Virus Infections. Methods Mol. Biol. 2019, 1576, 229–247. [Google Scholar] [CrossRef]
- Yoo, J.H.; Donowitz, M. Intestinal enteroids/organoids: A novel platform for drug discovery in inflammatory bowel diseases. World J. Gastroenterol. 2019, 25, 4125–4147. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [PubMed]
- Rittirsch, D.; Huber-Lang, M.S.; Flierl, M.A.; Ward, P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009, 4, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Russell, W.M.; Douglas-escobar, M.; Hauser, N.; Lopez, M.; Neu, J. Live and heat-killed Lactoba cillus rhamnosus GG: Effects on proinflammatory and anti-inflammatory cytokines/chemokines in gastrostomy-fed infant rats. Pediatr. Res. 2009, 66, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.J.; Fuchs, E. Relocation sustains intestinal stem-cell numbers. Nature 2022, 607, 451–452. [Google Scholar] [CrossRef]
- Lewin, K. The Paneth cell in disease. Gut 1969, 10, 804–811. [Google Scholar] [CrossRef]
- Gao, J.; Li, Y.; Wan, Y.; Hu, T.; Liu, L.; Yang, S.; Gong, Z.; Zeng, Q.; Wei, Y.; Yang, W.; et al. A Novel Postbiotic from Lactobacillus rhamnosus GG with a Beneficial Effect on Intestinal Barrier Function. Front. Microbiol. 2019, 10, 477. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Lee, A.; Huang, S.; Gao, J.; Spence, J.R.; Owyang, C. Lactobacillus rhamnosus GG prevents epithelial barrier dysfunction induced by interferon-gamma and fecal supernatants from irritable bowel syndrome patients in human intestinal enteroids and colonoids. Gut Microbes 2019, 10, 59–76. [Google Scholar] [CrossRef]
- Segers, M.E.; Lebeer, S. Towards a better understanding of Lactobacillus rhamnosus GG—host interactions. Microb. Cell Factories 2014, 13 (Suppl. 1), S7. [Google Scholar] [CrossRef]
- Petrova, M.I.; Imholz, N.C.; Verhoeven, T.L.; Balzarini, J.; Van Damme, E.J.; Schols, D.; Vanderleyden, J.; Lebeer, S. Lectin-Like Molecules of Lactobacillus rhamnosus GG Inhibit Pathogenic Escherichia coli and Salmonella Biofilm Formation. PLoS ONE 2016, 11, e0161337. [Google Scholar] [CrossRef]
- Tong, L.; Zhang, X.; Hao, H.; Liu, Q.; Zhou, Z.; Liang, X.; Liu, T.; Gong, P.; Zhang, L.; Zhai, Z.; et al. Lactobacillus rhamnosus GG Derived Extracellular Vesicles Modulate Gut Microbiota and Attenuate Inflammatory in DSS-Induced Colitis Mice. Nutrients 2021, 13, 3319. [Google Scholar] [CrossRef]
- Ibarra-Martinez, D.; Munoz-Ortega, M.H.; Quintanar-Stephano, A.; Martinez-Hernandez, S.L.; Avila-Blanco, M.E.; Ventura-Juarez, J. Antibacterial activity of supernatants of Lactoccocus lactis, Lactobacillus rhamnosus, Pediococcus pentosaceus and curcumin against Aeromonas hydrophila. In vitro study. Vet. Res. Commun. 2022, 46, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Murphey, E.D. CLP-induced impairment of innate immune function is caused by exposure to the cecal lumenal contents and not the tissue trauma or tissue ischemia/necrosis components. Microbes Infect. 2012, 14, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, H.; Chen, Y.; Yang, Y. Probiotic Lactobacillus rhamnosus GG reduces mortality of septic mice by modulating gut microbiota composition and metabolic profiles. Nutrition 2020, 78, 110863. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- de Sousa e Melo, F.; de Sauvage, F.J. Cellular Plasticity in Intestinal Homeostasis and Disease. Cell Stem Cell 2019, 24, 54–64. [Google Scholar] [CrossRef]
- Clevers, H.; Loh, K.M.; Nusse, R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014, 346, 1248012. [Google Scholar] [CrossRef]
- Srinivasan, T.; Than, E.B.; Bu, P.; Tung, K.L.; Chen, K.Y.; Augenlicht, L.; Lipkin, S.M.; Shen, X. Notch signalling regulates asymmetric division and inter-conversion between lgr5 and bmi1 expressing intestinal stem cells. Sci. Rep. 2016, 6, 26069. [Google Scholar] [CrossRef]
- Lin, X.; Gaudino, S.J.; Jang, K.K.; Bahadur, T.; Singh, A.; Banerjee, A.; Beaupre, M.; Chu, T.; Wong, H.T.; Kim, C.-K.; et al. IL-17RA-signaling in Lgr5+ intestinal stem cells induces expression of transcription factor ATOH1 to promote secretory cell lineage commitment. Immunity 2022, 55, 237–253.e238. [Google Scholar] [CrossRef]
- Gudas, L.J.; Wagner, J.A. Retinoids regulate stem cell differentiation. J. Cell. Physiol. 2011, 226, 322–330. [Google Scholar] [CrossRef]
- Zaph, C.; Troy, A.E.; Taylor, B.C.; Berman-Booty, L.D.; Guild, K.J.; Du, Y.; Yost, E.A.; Gruber, A.D.; May, M.J.; Greten, F.R.; et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature 2007, 446, 552–556. [Google Scholar] [CrossRef]
- Brischetto, C.; Krieger, K.; Klotz, C.; Krahn, I.; Kunz, S.; Kolesnichenko, M.; Mucka, P.; Heuberger, J.; Scheidereit, C.; Schmidt-Ullrich, R. NF-kappaB determines Paneth versus goblet cell fate decision in the small intestine. Development 2021, 148, dev199683. [Google Scholar] [CrossRef] [PubMed]
- Kabiri, Z.; Greicius, G.; Zaribafzadeh, H.; Hemmerich, A.; Counter, C.M.; Virshup, D.M. Wnt signaling suppresses MAPK-driven proliferation of intestinal stem cells. J. Clin. Investig. 2018, 128, 3806–3812. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ma, L.; Hu, T.; Liu, J.; Chen, B.; Yang, P.; Liu, Z. A20 Restores Impaired Intestinal Permeability and Inhibits Th2 Response in Mice with Colitis. Dig. Dis. Sci. 2020, 65, 1340–1347. [Google Scholar] [CrossRef]
- Vereecke, L.; Vieira-Silva, S.; Billiet, T.; van Es, J.H.; Mc Guire, C.; Slowicka, K.; Sze, M.; van den Born, M.; De Hertogh, G.; Clevers, H.; et al. A20 controls intestinal homeostasis through cell-specific activities. Nat. Commun. 2014, 5, 5103. [Google Scholar] [CrossRef] [PubMed]
- Mikuda, N.; Schmidt-Ullrich, R.; Kargel, E.; Golusda, L.; Wolf, J.; Hopken, U.E.; Scheidereit, C.; Kuhl, A.A.; Kolesnichenko, M. Deficiency in IkappaBalpha in the intestinal epithelium leads to spontaneous inflammation and mediates apoptosis in the gut. J. Pathol. 2020, 251, 160–174. [Google Scholar] [CrossRef]
- Pararajasingam, A.; Uwagwu, J. Lactobacillus: The not so friendly bacteria. BMJ Case Rep. 2017, 2017, bcr2016218423. [Google Scholar] [CrossRef]

| Target Genes | Primer Sense (5′-3′) | Primer Antisense (5′-3′) |
|---|---|---|
| Lgr5 | CCTACTCGAAGACTTACCCAGT | GCATTGGGGTGAATGATAGCA |
| Ascl2 | AAGCACACCTTGACTGGTACG | AAGTGGACGTTTGCACCTTCA |
| Olfm4 | CAGCCACTTTCCAATTTCACTG | GCTGGACATACTCCTTCACCTTA |
| Lyz1 | GAGACCGAAGCACCGACTATG | CGGTTTTGACATTGTGTTCGC |
| Defa6 | CCTTCCAGGTCCAGGCTGAT | TGAGAAGTGGTCATCAGGCAC |
| Tnfaip3 | ATGCACCGATACACACTGGA | GCGTGTGTCTGTTTCCTTGA |
| Nfkbia | TGAAGGACGAGGAGTACGAGC | TTCGTGGATGATTGCCAAGTG |
| GAPDH | ATGGTGAAGGTCGGTGTGAA | TGGAAGATGGTGATGGGCTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Li, S.; Peng, C.; Gui, Q.; Li, J.; Xu, Z.; Yang, Y. Lactobacillus rhamnosus GG Promotes Recovery of the Colon Barrier in Septic Mice through Accelerating ISCs Regeneration. Nutrients 2023, 15, 672. https://doi.org/10.3390/nu15030672
Chen L, Li S, Peng C, Gui Q, Li J, Xu Z, Yang Y. Lactobacillus rhamnosus GG Promotes Recovery of the Colon Barrier in Septic Mice through Accelerating ISCs Regeneration. Nutrients. 2023; 15(3):672. https://doi.org/10.3390/nu15030672
Chicago/Turabian StyleChen, Lufang, Shumin Li, Chunting Peng, Qifeng Gui, Jinyou Li, Zherong Xu, and Yunmei Yang. 2023. "Lactobacillus rhamnosus GG Promotes Recovery of the Colon Barrier in Septic Mice through Accelerating ISCs Regeneration" Nutrients 15, no. 3: 672. https://doi.org/10.3390/nu15030672
APA StyleChen, L., Li, S., Peng, C., Gui, Q., Li, J., Xu, Z., & Yang, Y. (2023). Lactobacillus rhamnosus GG Promotes Recovery of the Colon Barrier in Septic Mice through Accelerating ISCs Regeneration. Nutrients, 15(3), 672. https://doi.org/10.3390/nu15030672







