Dynamics of Serologic Change to Gluten in Celiac Disease Patients
Abstract
:1. Introduction
2. Materials and Methods
3. Results
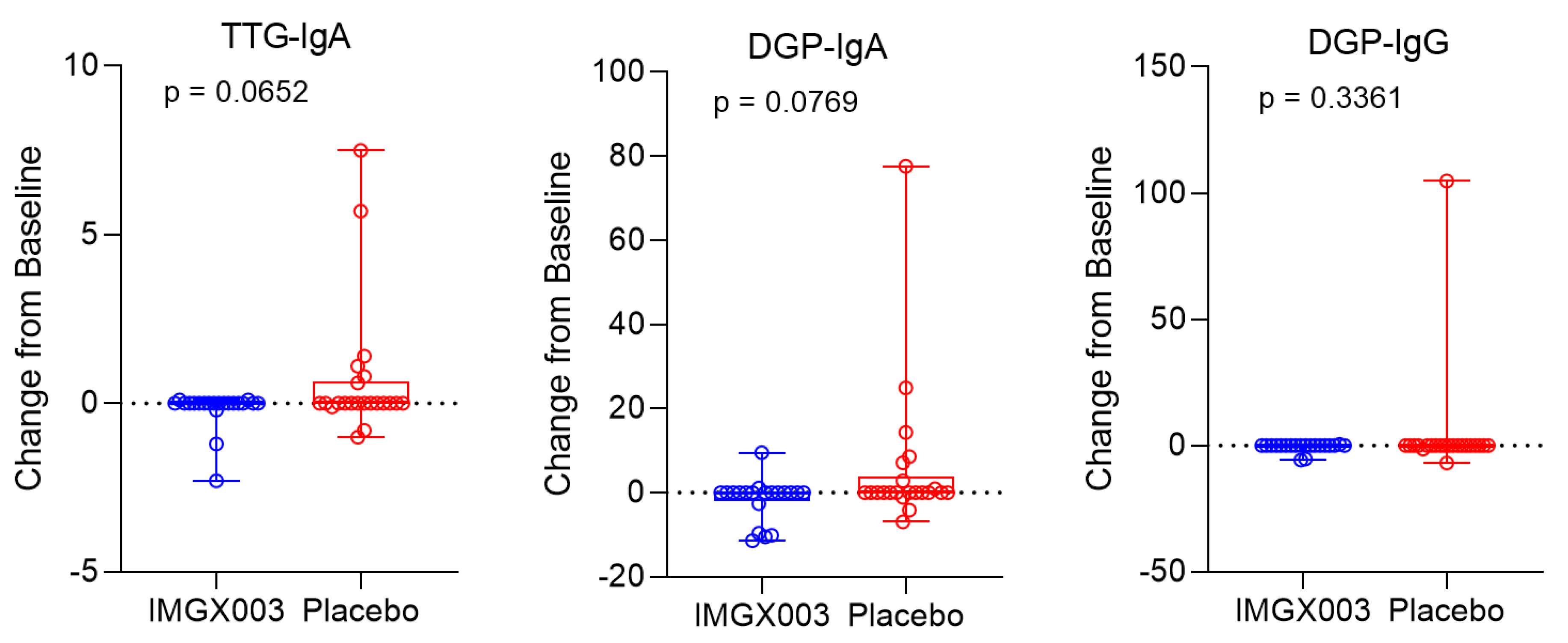
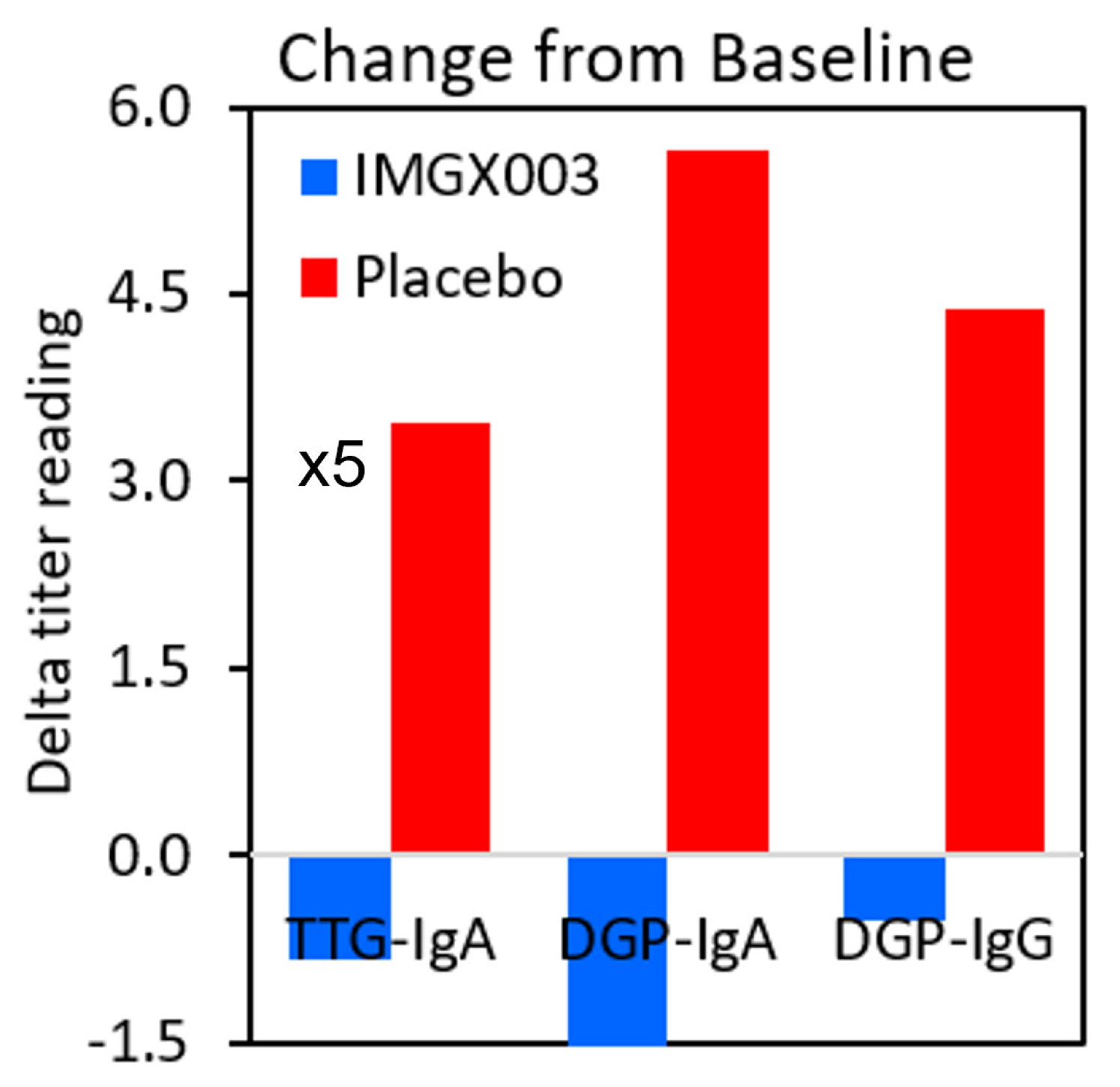
3.1. Serologic Change Dynamics
3.2. Correlation of tTG IgA vs. tTG IgA AB Assays
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ditah, I.C.; Nadeau, A.M.; Rubio-Tapia, A.; Marietta, E.V.; Brantner, T.L.; Camilleri, M.J.; Rajkumar, V.S.; Landgren, O.; Everhart, J.E.; Murray, J.A. Trends and racial/ethnic disparities in gluten- sensitive problems in the United States: Findings from the National Health and Nutrition Examination Surveys from 1988 to 2012. Am. J. Gastroenterol. 2015, 110, 455–461. [Google Scholar]
- Schuppan, D.; Junker, Y.; Barisani, D. Celiac disease: From pathogenesis to novel therapies. Gastroenterology 2009, 137, 1912–1933. [Google Scholar] [CrossRef]
- Lohi, S.; Mustalahti, K.; Kaukinen, K.; Laurila, K.; Collin, P.; Rissanen, H.; Lohi, O.; Bravi, E.; Gasparin, M.; Reunanen, A.; et al. Increasing prevalence of celiac disease over time. Aliment. Pharmacol. Ther. 2007, 26, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Lerner, B.A.; Vo LT, P.; Yates, S.; Rundle, A.G.; Green, P.H.; Lebwohl, B. Detection of gluten in gluten-free labeled restaurant food: Analysis of crowd-sourced data. Am. J. Gastroenterol. 2019, 114, 792–797. [Google Scholar] [CrossRef]
- Kelly, C.P.; Bai, J.C.; Liu, E.; Leffler, D.A. Celiac disease: Clinical spectrum and management. Gastroenterol 2015, 148, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Green, P.H.R.; Lebwohl, B.; Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 2015, 135, 1099–1106. [Google Scholar] [CrossRef]
- Guandalini, S.; Tundia, N.; Thakkar, R.; Macaulay, D.; Essenmacher, K.; Fuldcore, M. Direct cost in patients with celiac disease in the U.S.A.: A retrospective claims analysis. Dig. Dis. Sci. 2016, 10, 2823–2830. [Google Scholar] [CrossRef]
- Wolf, J.; Petroff, D.; Richter, T.; Auth, M.K.; Uhlig, H.H.; Laass, M.W.; Lauenstein, P.; Krahl, A.; Händel, N.; de Laffolie, J.; et al. Validation of Antibody-Based Strategies for Diagnosis of Pediatric Celiac Disease without Biopsy. Gastroenterol 2017, 153, 410–4219. [Google Scholar] [CrossRef]
- Lerner, A.; Ramesh, A.; Matthias, T. Serologic Diagnosis of Celiac Disease: New Biomarkers. Gastroenterol. Clin. N. Am. 2019, 48, 207–317. [Google Scholar] [CrossRef]
- Tonutti, E.; Visentini, D.; Picierno, A.; Bizzaro, N.; Villalta, D.; Tozzoli, R.; Kodermaz, G.; Carroccio, A.; Iacono, G.; Teresi, S.; et al. Diagnostic efficacy of the ELISA test for the detection of deamidated anti-gliadin peptide antibodies in the diagnosis and monitoring of celiac disease. J. Clin. Lab. Anal. 2009, 23, 165–171. [Google Scholar] [CrossRef]
- Hujoel, I.A.; Reilly, N.R.; Rubio-Tapia, A. Celiac disease: Clinical features and diagnosis. Gastroenterol. Clin. N. Am. 2019, 48, 19–37. [Google Scholar] [CrossRef]
- Husby, S.; Murray, J.A.; Katzka, D.A. AGA Clinical Practice Update on Diagnosis and Monitoring of Celiac Disease—Changing Utility of Serology and Histologic Measures: Expert Review. Gastroenterology 2019, 156, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Valitutti, F.; Leonard, M.M.; Kenyon, V.; Montuori, M.; Piemontese, P.; Francavilla, R.; Malamisura, B.; Norsa, L.; Calvi, A.; Lionetti, M.E.; et al. Early Antibody Dynamics in a Prospective Cohort of Children at Risk of Celiac Disease. Am. J. Gastroenterol. 2023, 118, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Catassi, G.N.; Pulvirenti, A.; Monachesi, C.; Catassi, C.; Lionetti, E. Diagnostic Accuracy of IgA Anti-Transglutaminase and IgG Anti-Deamidated Gliadin for Diagnosis of Celiac Disease in Children under Two Years of Age: A Systematic Review and Meta-Analysis. Nutrients 2021, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Syage, J.A.; Murray, J.A.; Green, P.H.R.; Khosla, C. Latiglutenase Improves Symptoms in Seropositive Celiac Disease Patients While on a Gluten-Free Diet. Dig. Dis. Sci. 2017, 62, 2428–2432. [Google Scholar] [CrossRef]
- Syage, J.A.; Green, P.H.; Khosla, C.; Adelman, D.C.; Sealey-Voyksner, J.A.; Murray, J.A. Latiglutenase treatment for celiac disease: Symptom and quality of life improvement for seropositive patients on a gluten-free diet. GastroHep 2019, 1, 293–301. [Google Scholar] [CrossRef]
- Lahdeaho, M.-L.; Kaukinen, K.; Laurila, K.; Adelman, D.C.; Maki, M. Glutenase ALV003 Attenuates Gluten-Induced Mucosal Injury in Patients With Celiac Disease. Gastroenterology 2014, 146, 1649–1658. [Google Scholar] [CrossRef]
- Wolf, C.; Siegel, J.B.; Tinberg, C.; Camarca, A.; Gianfrani, C.; Paski, S.; Guan, R.; Montelione, G.; Baker, D.; Pultz, I.S. Engineering of kuma030: A gliadin peptidase that rapidly degrades immunogenic gliadin peptides in gastric conditions. J. Am. Chem. Soc. 2015, 137, 13106–13113. [Google Scholar] [CrossRef]
- Leffler, D.A.; Kelly, C.P.; Green, P.H.; Fedorak, R.N.; DiMarino, A.; Perrow, W.; Rasmussen, H.; Wang, C.; Bercik, P.; Bachir, N.M.; et al. Larazotide acetate for persistent symptoms of celiac disease despite a gluten-free diet: A randomized controlled trial. Gastroenterology 2015, 148, 1311–1319. [Google Scholar] [CrossRef]
- Daveson AJ, M.; Ee, H.C.; Andrews, J.M.; King, T.; Goldstein, K.E.; Dzuris, J.L.; MacDougall, J.A.; Williams, L.J.; Treohan, A.; Cooreman, M.P.; et al. Epitope-Specific Immunotherapy Targeting CD4-Positive T Cells in CeliacDisease: Safety, Pharmacokinetics, and Effects on Intestinal Histology and Plasma Cytokines with Escalating Dose Regimens of Nexvax2 in a Randomized, Double-Blind, Placebo-Controlled Phase 1 Study. EBioMedicine 2017, 26, 78–90. [Google Scholar]
- Murray, J.A.; Syage, J.A.; Wu, T.-T.; Dickason, M.A.; Ramos, A.G.; Van Dyke, C.; Horwath, I.; Lavin, P.T.; Mäki, M.; Hujoel, I.; et al. Latiglutenase Protects the Mucosa and Attenuates Symptom Severity in Patients with Celiac Disease Exposed to a Gluten Challenge. Gastroenterology 2022, 163, 1510–1521.e6. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.; Kurppa, K.; Mearin, M.L.; Ribes-Koninckx, C.; Shamir, R.; Troncone, R.; Auricchio, R.; Castillejo, G.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition Guidelines for Diagnosing Coeliac Disease. J. Pediatr. Gastroenterol. Nutr. 2020, 70, 141–156. [Google Scholar] [CrossRef]
- Kurppa, K.; Taavela, J.; Saavalainen, P.; Kaukinen, K.; Kindfors, K. Novel diagnostic techniques for celiac disease. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Lähdeaho, M.-L.; Mäki, M.; Laurila, K.; Huhtala, H.; Kaukinen, K. Small-bowel mucosal changes and antibody responses after low- and moderate-dose gluten challenge in celiac disease. BMC Gastroenterol. 2011, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Sarna, V.K.; Skodje, G.I.; Reims, H.M.; Risnes, L.F.; Dahal-Koirala, S.; Sollid, L.M.; Lundin, K.E. HLA-DQ:gluten tetramer test in blood gives better detection of coeliac patients than biopsy after 14-day gluten challenge. Gut 2018, 67, 1606–1613. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.P.; Goel, G.; Hardy, M.Y.; Russell, A.K.; Wang, S.; Szymczak, E.; Zhang, R.; E Goldstein, K.; Neff, K.M.; E Truitt, K.; et al. Whole blood interleukin-2 release test to detect and characterize rare circulating gluten-specific T cell responses in coeliac disease. Clin. Exp. Immunol. 2021, 204, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Tye-Din, J.A.; on behalf of the RESET CeD Study Group; Daveson, A.J.M.; Goldstein, K.E.; Hand, H.L.; Neff, K.M.; Goel, G.; Williams, L.J.; Truitt, K.E.; Anderson, R.P. Patient factors influencing acute gluten reactions and cytokine release in treated coeliac disease. BMC Med. 2020, 18, 362. [Google Scholar] [CrossRef] [PubMed]
- Choung, R.S.; Rostamkolaei, S.K.; Ju, J.M.; Marietta, E.V.; Van Dyke, C.T.; Rajasekaran, J.; Jayaraman, V.; Wang, T.; Bei, K.; Rajasekaran, K.E.; et al. Synthetic Neoepitopes of the Transglutaminase–Deamidated Gliadin Complex as Biomarkers for Diagnosing and Monitoring Celiac Disease. Gastroenterology 2019, 156, 582–591.e1. [Google Scholar] [CrossRef]
- Cartee, A.K.; Choung, R.S.; King, K.S.; Wang, S.; Dzuris, J.L.; Anderson, R.P.; Van Dyke, C.T.; Hinson, C.A.; Marietta, E.; Katzka, D.A.; et al. Plasma IL-2 and Symptoms Response after Acute Gluten Exposure in Subjects With Celiac Disease or Nonceliac Gluten Sensitivity. Am. J. Gastroenterol. 2022, 117, 319–326. [Google Scholar] [CrossRef]
- Naiyer, A.J.; Hernandez, L.; Ciaccio, E.J.; Papadakis, K.; Manavalan, J.S.; Bhagat, G.; Green, P.H.R. Comparison of Commercially Available Serologic Kits for the Detection of Celiac Disease. J. Clin. Gastroenterol. 2009, 43, 225–232. [Google Scholar] [CrossRef]
- Rashtak, S.; Ettore, M.W.; Homburger, H.A.; Murray, J.A. Combination testing for antibodies in the diagnosis of coeliac disease: Comparison of multiplex immunoassay and ELISA methods. Aliment. Pharmacol. Ther. 2008, 28, 805–813. [Google Scholar] [CrossRef]
- Castelijn, D.A.; Mulder, A.L.; van der Pol, P.; Hollander, J.C.; Kuiper, T.; Bijnens, C.; Damoiseaux, J.; Bontkes, H.J. Multicenter study to compare the diagnostic performance of CLIA vs. FEIA transglutaminase IgA assays for the diagnosis of celiac disease. Clin. Chem. Lab. Med. 2023, 61, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Taavela, J.; Kurppa, K.; Jääskeläinen, T.; Kaartinen, N.E.; Rissanen, H.; Huhtala, H.; Mäki, M.; Kaukinen, K. Trends in the prevalence rates and predictive factors of coeliac disease: A long-term nationwide follow-up study. Aliment. Pharmacol. Ther. 2023. [Google Scholar] [CrossRef] [PubMed]
- Silvester, J.A.; Kurada, S.; Szwajcer, A.; Kelly, C.P.; Leffler, D.A.; Duerksen, D.R. Tests for Serum Transglutaminase and Endomysial Antibodies Do Not Detect Most Patients With Celiac Disease and Persistent Villous Atrophy on Gluten-free Diets: A Meta-analysis. Gastroenterology 2017, 153, 689–701.e1. [Google Scholar] [CrossRef] [PubMed]

| ULN tTG-IgA | N,N | P,P | N,P | P,N | Sens | 1-Spec |
|---|---|---|---|---|---|---|
| ≥4 | 192 | 105 | 48 | 2 | 86% | 14% |
| ≥3 | 178 | 139 | 14 | 16 | 91% | 9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syage, J.; Ramos, A.; Loskutov, V.; Norum, A.; Bledsoe, A.; Choung, R.S.; Dickason, M.; Sealey-Voyksner, J.; Murray, J. Dynamics of Serologic Change to Gluten in Celiac Disease Patients. Nutrients 2023, 15, 5083. https://doi.org/10.3390/nu15245083
Syage J, Ramos A, Loskutov V, Norum A, Bledsoe A, Choung RS, Dickason M, Sealey-Voyksner J, Murray J. Dynamics of Serologic Change to Gluten in Celiac Disease Patients. Nutrients. 2023; 15(24):5083. https://doi.org/10.3390/nu15245083
Chicago/Turabian StyleSyage, Jack, Ana Ramos, Vasiliy Loskutov, Anna Norum, Adam Bledsoe, Rok Seon Choung, Matthew Dickason, Jennifer Sealey-Voyksner, and Joseph Murray. 2023. "Dynamics of Serologic Change to Gluten in Celiac Disease Patients" Nutrients 15, no. 24: 5083. https://doi.org/10.3390/nu15245083
APA StyleSyage, J., Ramos, A., Loskutov, V., Norum, A., Bledsoe, A., Choung, R. S., Dickason, M., Sealey-Voyksner, J., & Murray, J. (2023). Dynamics of Serologic Change to Gluten in Celiac Disease Patients. Nutrients, 15(24), 5083. https://doi.org/10.3390/nu15245083






