Gut–Joint Axis: Impact of Bifidobacterial Cell Wall Lipoproteins on Arthritis Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Arthritis Induction
2.3. Bacterial Enumeration in Organs
2.4. Enumeration of Spleen CD11C+ DCs and CD4+ T Cells and Culture of Splenocytes in CIA Model
2.5. In Vivo and In Vitro Lpps Uptake
2.6. Statistical Analysis
3. Results
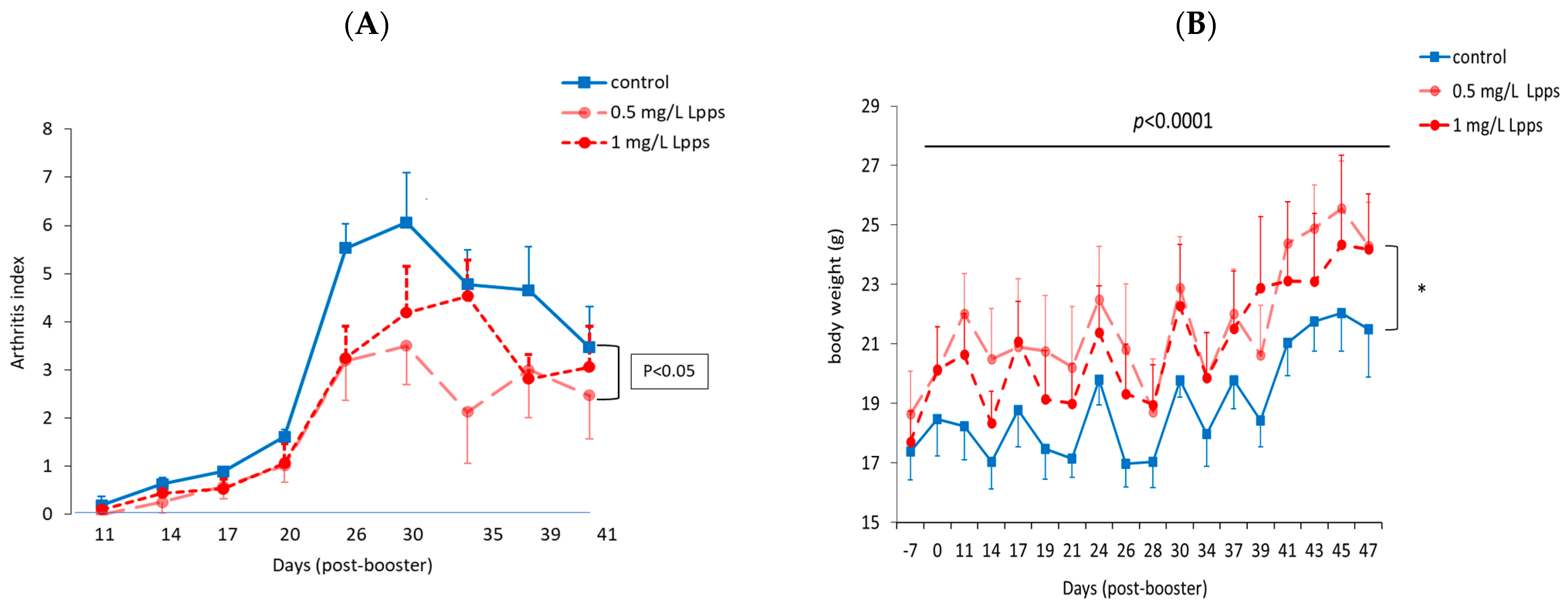
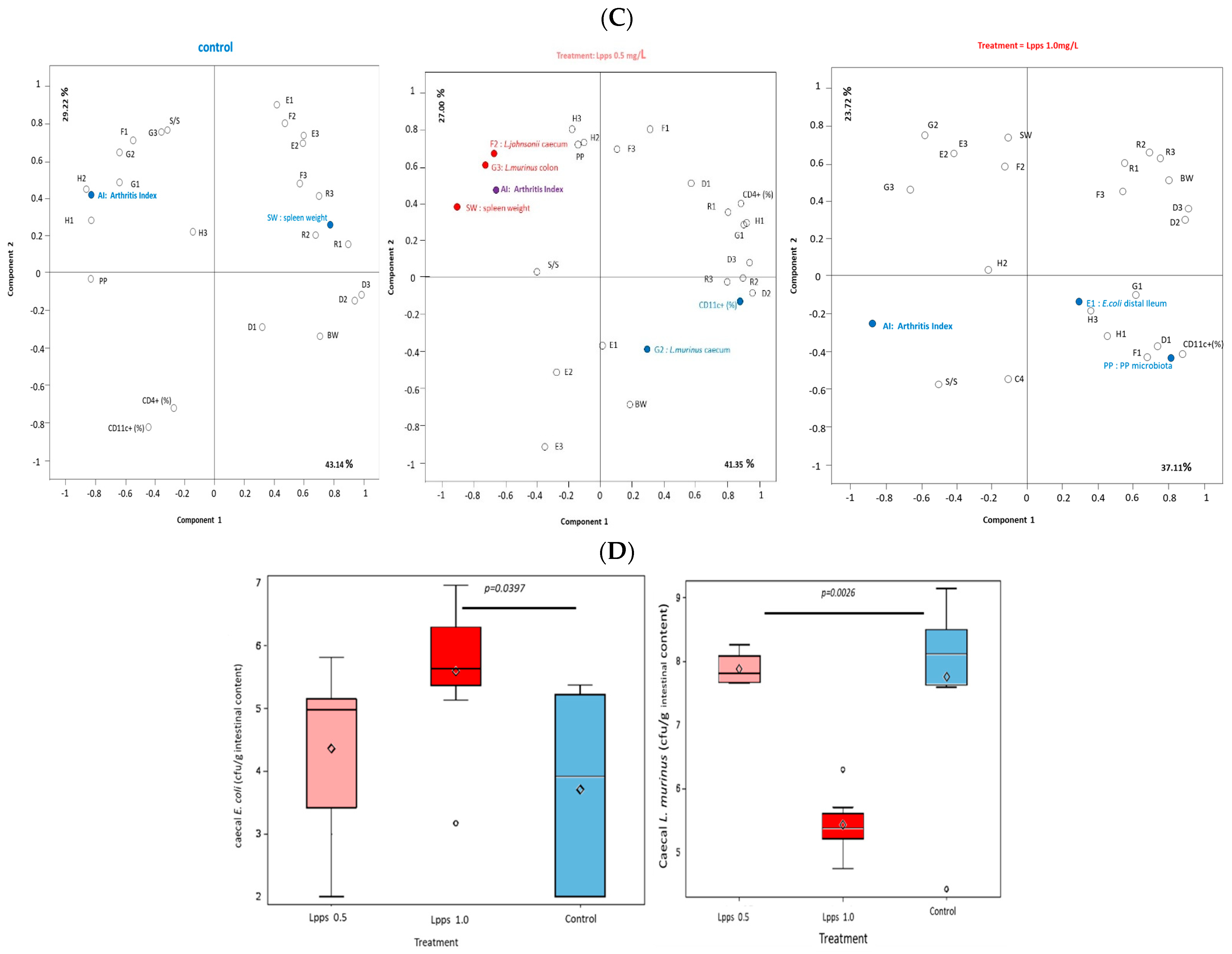
3.1. Protection against Arthritis Progression Depends on the Lpps Dose and Is Primarily Related to Ligilactobacillus murinus Intestinal Colonization
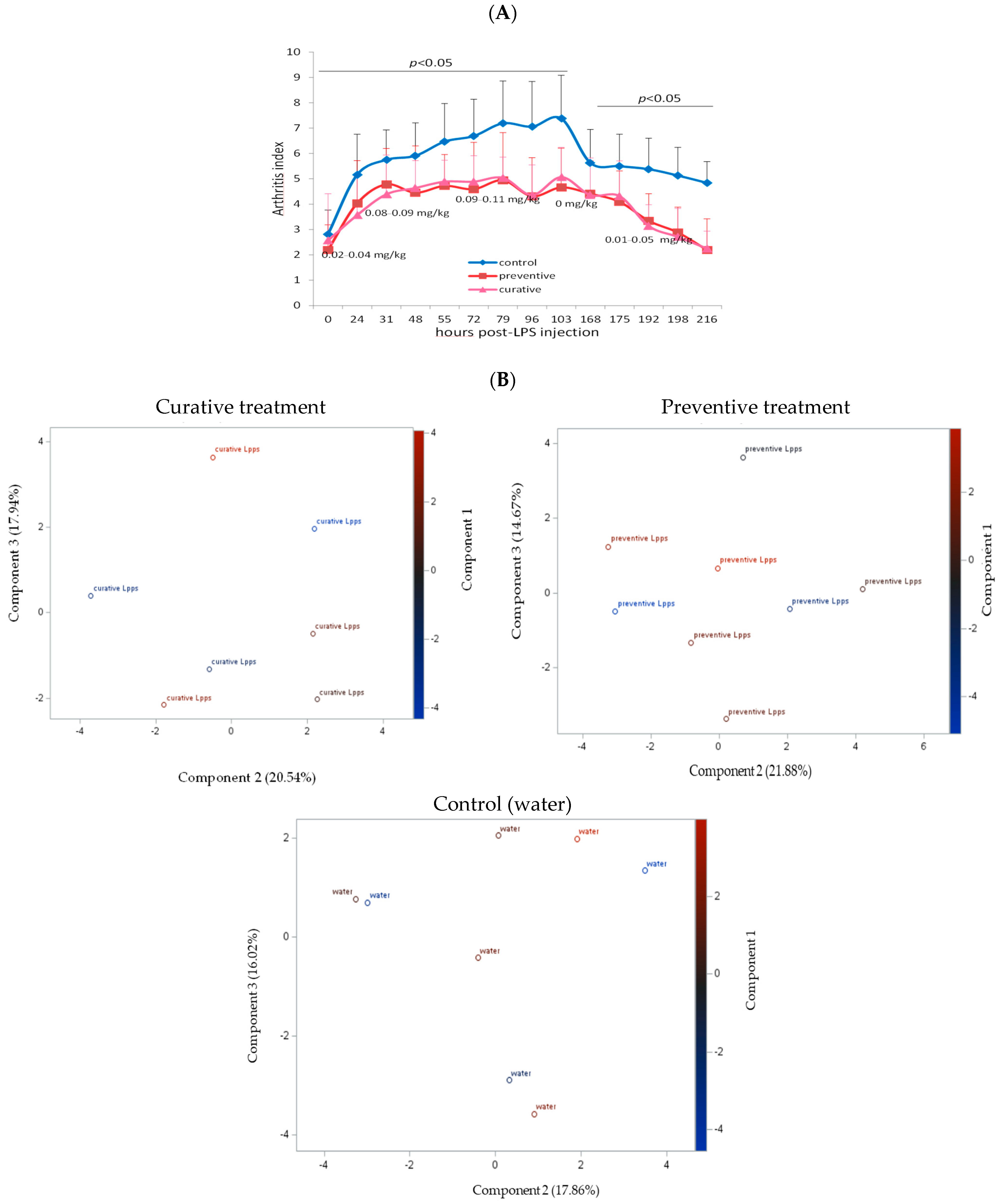
3.2. The Degree of Disease Progression Was Significantly Milder in Both Preventive and Therapeutic Lpps CAIA Groups
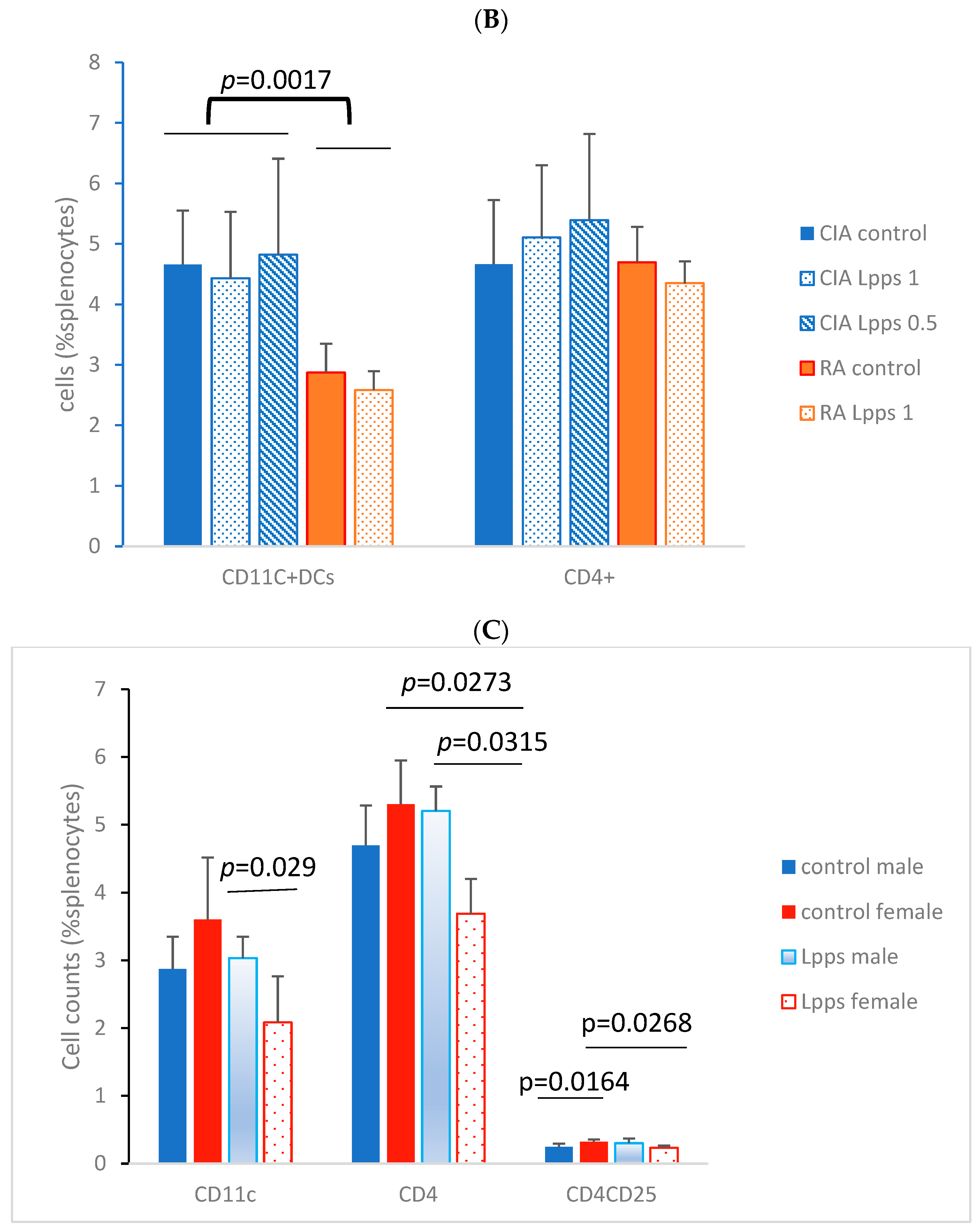
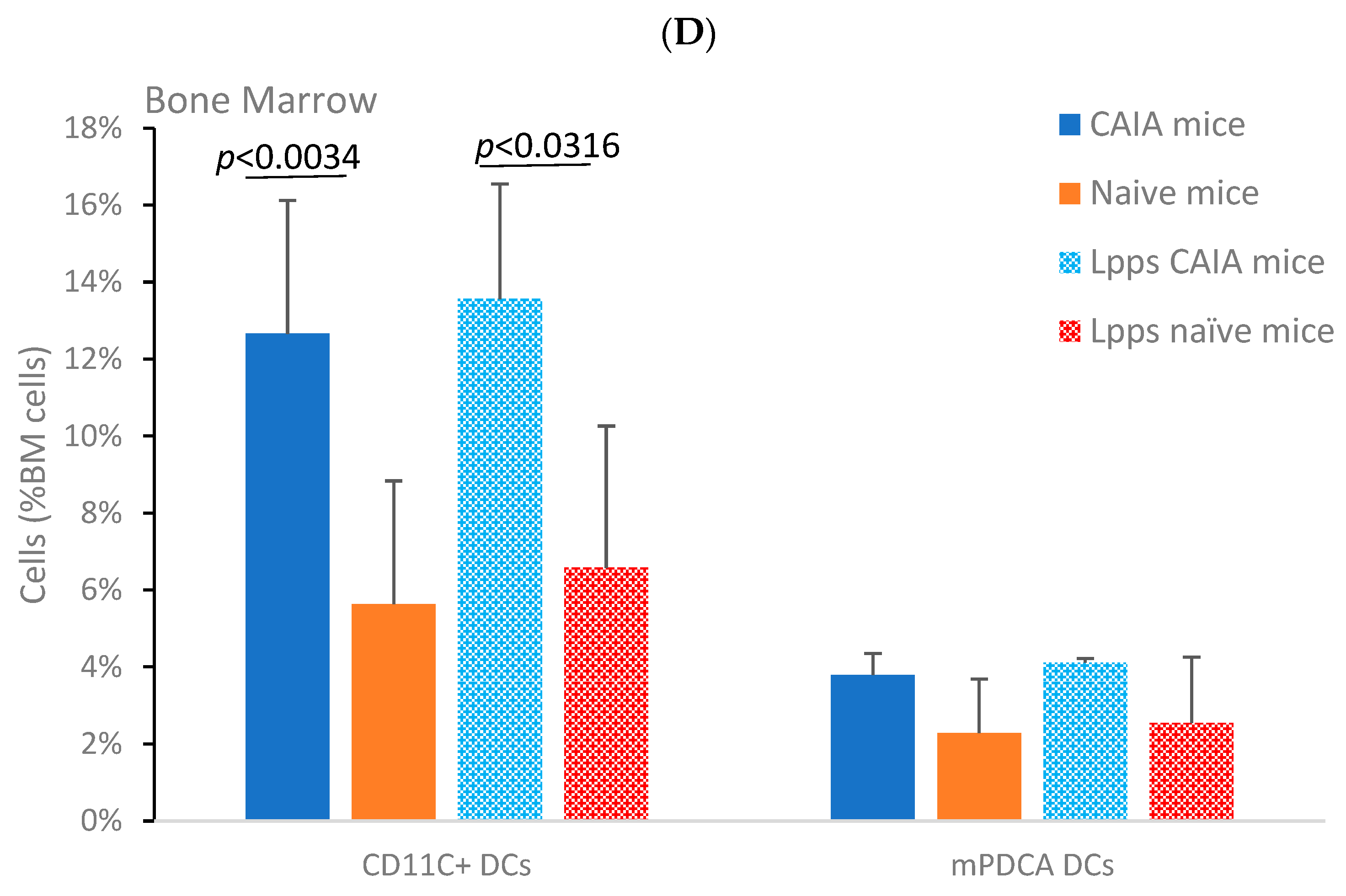
3.3. Higher IL-10 Splenocyte Response to LPS in CIA Lpps-Treated Mice Was Likely Related to CD11c+ DC Recruitment
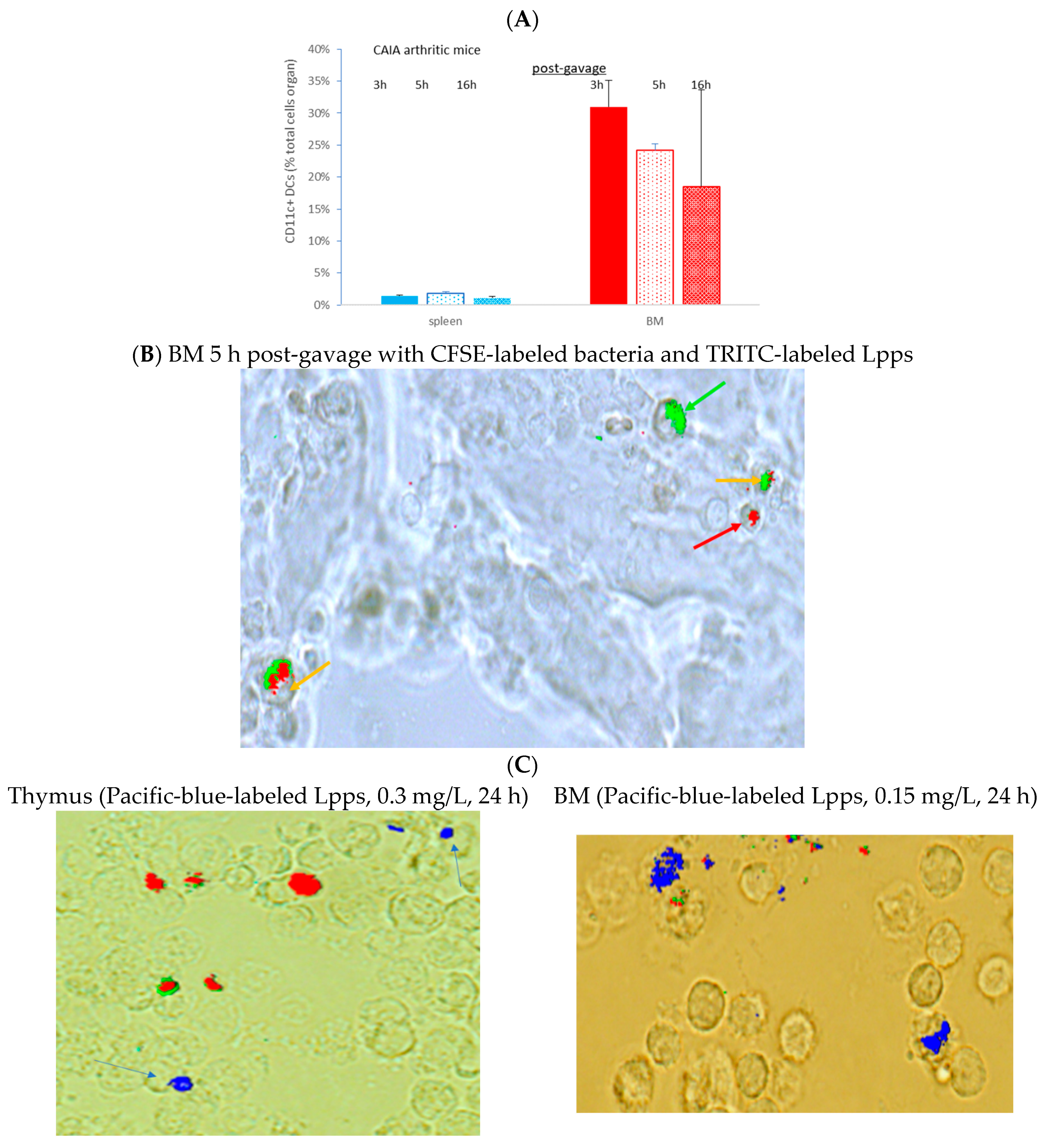
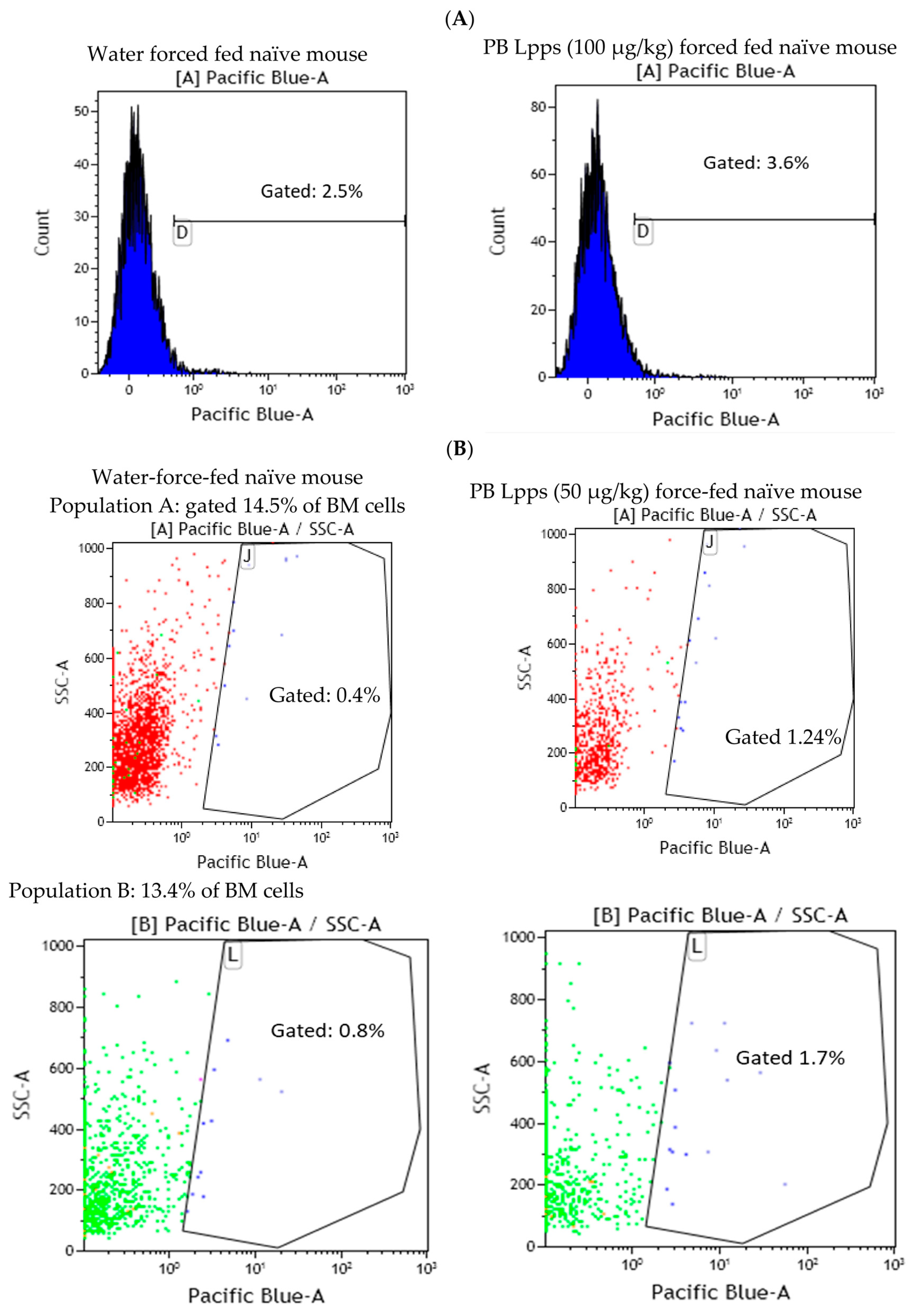
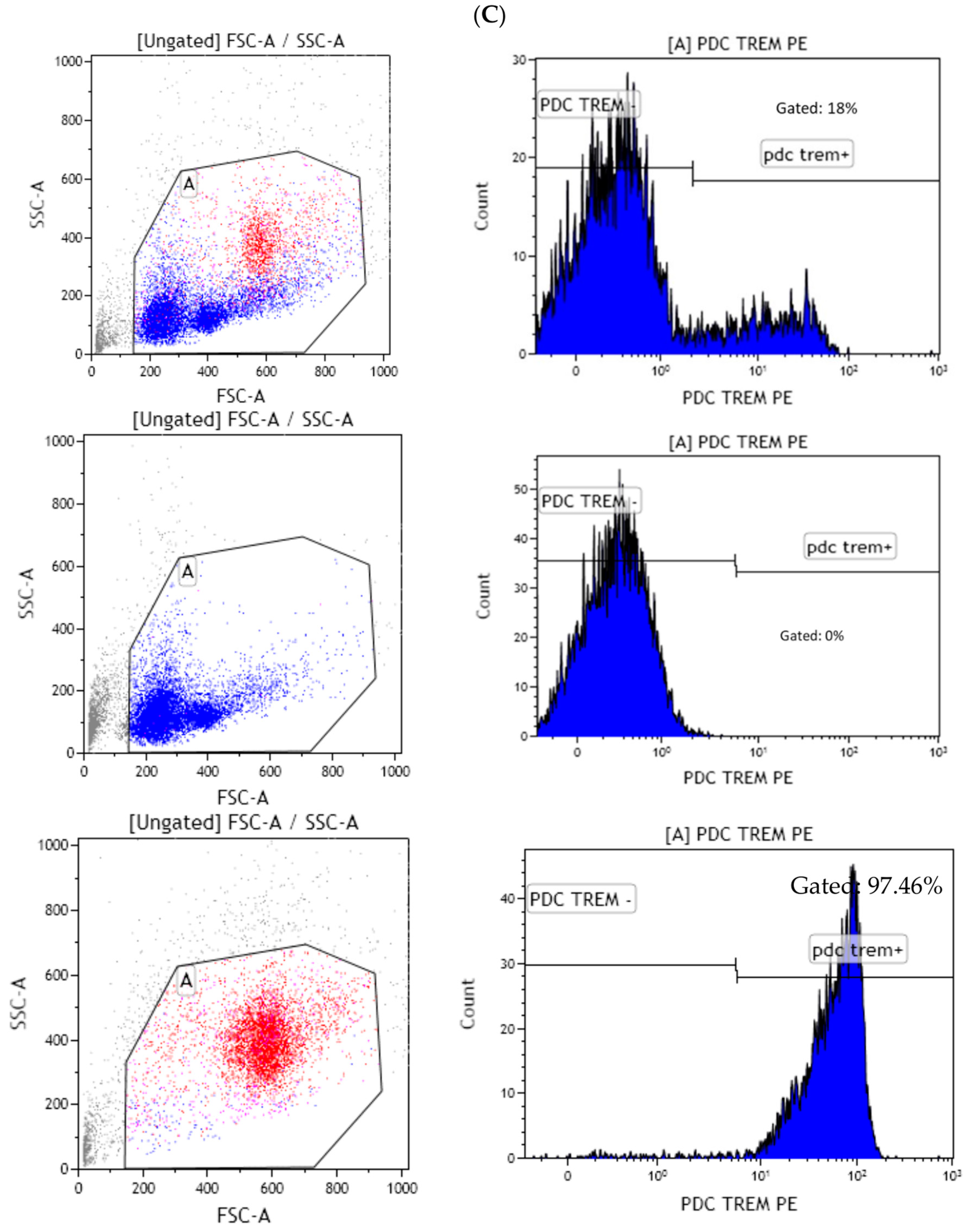
3.4. Lpps Was Primarily Detected in Bone Marrow Plasmacytoid DCs at a Late Post-Gavage Stage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Chen, B.D.; Zhao, L.D.; Li, H. The Gut Microbiota: Emerging Evidence in Autoimmune Diseases. Trends Mol. Med. 2020, 26, 862–873. [Google Scholar] [CrossRef]
- Ruiz-Limón, P.; Mena-Vázquez, N.; Moreno-Indias, I.; Manrique-Arija, S.; Lisbona-Montañez, J.M.; Cano-García, L.; Tinahones, F.J.; Fernández-Nebro, A. Collinsella is associated with cumulative inflammatory burden in an established rheumatoid arthritis cohort. Biomed. Pharmacother. 2022, 153, 113518. [Google Scholar] [CrossRef]
- Kishikawa, T.; Maeda, Y.; Nii, T.; Motooka, D.; Matsumoto, Y.; Matsushita, M.; Matsuoka, H.; Yoshimura, M.; Kawada, S.; Teshigawara, S.; et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum. Dis. 2020, 79, 103–111. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Jia, H.; Feng, Q.; Wang, D.; Liang, D.; Wu, X.; Li, J.; Tang, L.; Li, Y.; et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015, 21, 895–905. [Google Scholar] [CrossRef]
- Thompson, K.N.; Bonham, K.S.; Ilott, N.E.; Britton, G.J.; Colmenero, P.; Bullers, S.J.; McIver, L.J.; Ma, S.; Nguyen, L.H.; Filer, A.; et al. Alterations in the gut microbiome implicate key taxa and metabolic pathways across inflammatory arthritis phenotypes. Sci. Transl. Med. 2023, 15, eabn4722. [Google Scholar] [CrossRef]
- Sköldstam, L.; Hagfors, L.; Johansson, G. An experimental study of a Mediterranean diet intervention for patients with rheumatoid arthritis. Ann. Rheum. Dis. 2003, 62, 208–214. [Google Scholar] [CrossRef]
- Häger, J.; Bang, H.; Hagen, M.; Frech, M.; Träger, P.; Sokolova, M.V.; Steffen, U.; Tascilar, K.; Sarter, K.; Schett, G.; et al. The Role of Dietary Fiber in Rheumatoid Arthritis Patients: A Feasibility Study. Nutrients 2019, 11, 2392. [Google Scholar] [CrossRef]
- Häupl, T.; Sörensen, T.; Smiljanovic, B.; Darcy, M.; Scheder-Bieschin, J.; Steckhan, N.; Hartmann, A.M.; Koppold, D.A.; Stuhlmüller, B.; Skriner, K.; et al. Intestinal Microbiota Reduction Followed by Fasting Discloses Microbial Triggering of Inflammation in Rheumatoid Arthritis. J. Clin. Med. 2023, 12, 4359. [Google Scholar] [CrossRef]
- Badsha, H. Role of Diet in Influencing Rheumatoid Arthritis Disease Activity. Open Rheumatol. J. 2018, 12, 19–28. [Google Scholar] [CrossRef]
- Peltonen, R.; Nenonen, M.; Helve, T.; Hänninen, O.; Toivanen, P.; Eerola, E. Faecal microbial flora and disease activity in rheumatoid arthritis during a vegan diet. Br. J. Rheumatol. 1997, 36, 64–68. [Google Scholar] [CrossRef]
- Kohashi, O.; Kohashi, Y.; Takahashi, T.; Ozawa, A.; Shigematsu, N. Suppressive effect of Escherichia coli on adjuvant-induced arthritis in germ-free rats. Arthritis Rheum. 1986, 29, 547–553. [Google Scholar] [CrossRef]
- Lehman, T.J.; Cremer, M.A.; Walker, S.M.; Dillon, A.M. The role of humoral immunity in Lactobacillus casei cell wall induced arthritis. J. Rheumatol. 1987, 14, 415–419. [Google Scholar]
- Severijnen, A.J.; van Kleef, R.; Hazenberg, M.P.; van de Merwe, J.P. Cell wall fragments from major residents of the human intestinal flora induce chronic arthritis in rats. J. Rheumatol. 1989, 16, 1061–1068. [Google Scholar]
- Simelyte, E.; Rimpiläinen, M.; Lehtonen, L.; Zhang, X.; Toivanen, P. Bacterial cell wall-induced arthritis: Chemical composition and tissue distribution of four Lactobacillus strains. Infect. Immun. 2000, 68, 3535–3540. [Google Scholar] [CrossRef]
- Simelyte, E.; Rimpiläinen, M.; Zhang, X.; Toivanen, P. Role of peptidoglycan subtypes in the pathogenesis of bacterial cell wall arthritis. Ann. Rheum. Dis. 2003, 62, 976–982. [Google Scholar] [CrossRef]
- Kato, I.; Endo-Tanaka, K.; Yokokura, T. Suppressive effects of the oral administration of Lactobacillus casei on type II collagen-induced arthritis in DBA/1 mice. Life Sci. 1998, 63, 635–644. [Google Scholar] [CrossRef]
- Kano, H.; Kaneko, T.; Kaminogawa, S. Oral intake of Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1 prevents collagen-induced arthritis in mice. J. Food Prot. 2002, 65, 153–160. [Google Scholar] [CrossRef]
- Baharav, E.; Mor, F.; Halpern, M.; Weinberger, A. Lactobacillus GG bacteria ameliorate arthritis in Lewis rats. J. Nutr. 2004, 134, 1964–1969. [Google Scholar] [CrossRef]
- So, J.S.; Kwon, H.K.; Lee, C.G.; Yi, H.J.; Park, J.A.; Lim, S.Y.; Hwang, K.C.; Jeon, Y.H.; Im, S.H. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol. Immunol. 2008, 45, 2690–2699. [Google Scholar] [CrossRef]
- Fan, Z.; Ross, R.P.; Stanton, C.; Hou, B.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Lactobacillus casei CCFM1074 Alleviates Collagen-Induced Arthritis in Rats via Balancing Treg/Th17 and Modulating the Metabolites and Gut Microbiota. Front. Immunol. 2021, 12, 680073. [Google Scholar] [CrossRef]
- Pan, H.; Guo, R.; Ju, Y.; Wang, Q.; Zhu, J.; Xie, Y.; Zheng, Y.; Li, T.; Liu, Z.; Lu, L.; et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome 2019, 7, 107. [Google Scholar] [CrossRef]
- Jhun, J.; Min, H.K.; Ryu, J.; Lee, S.Y.; Ryu, J.G.; Choi, J.W.; Na, H.S.; Lee, S.Y.; Jung, Y.; Park, S.J.; et al. Lactobacillus sakei suppresses collagen-induced arthritis and modulates the differentiation of T helper 17 cells and regulatory B cells. J. Transl. Med. 2020, 18, 317. [Google Scholar] [CrossRef]
- Yamashita, M.; Matsumoto, K.; Endo, T.; Ukibe, K.; Hosoya, T.; Matsubara, Y.; Nakagawa, H.; Sakai, F.; Miyazaki, T. Preventive Effect of Lactobacillus helveticus SBT2171 on Collagen-Induced Arthritis in Mice. Front. Microbiol. 2017, 8, 1159. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Zou, Q.; Zhong, B.; Wang, H.; Mou, F.; Wu, L.; Fang, Y. Lactobacillus salivarius Isolated from Patients with Rheumatoid Arthritis Suppresses Collagen-Induced Arthritis and Increases Treg Frequency in Mice. J. Interferon Cytokine Res. 2016, 36, 706–712. [Google Scholar] [CrossRef]
- Esvaran, M.; Conway, P.L. Lactobacillus fermentum PC1 has the Capacity to Attenuate Joint Inflammation in Collagen-Induced Arthritis in DBA/1 Mice. Nutrients 2019, 11, 785. [Google Scholar] [CrossRef]
- Eerola, E.; Möttönen, T.; Hannonen, P.; Luukkainen, R.; Kantola, I.; Vuori, K.; Tuominen, J.; Toivanen, P. Intestinal flora in early rheumatoid arthritis. Br. J. Rheumatol. 1994, 33, 1030–1038. [Google Scholar] [CrossRef]
- Zhao, Q.; Ren, H.; Yang, N.; Xia, X.; Chen, Q.; Zhou, D.; Liu, Z.; Chen, X.; Chen, Y.; Huang, W.; et al. Bifidobacterium pseudocatenulatum-Mediated Bile Acid Metabolism to Prevent Rheumatoid Arthritis via the Gut-Joint Axis. Nutrients 2023, 15, 255. [Google Scholar] [CrossRef]
- Li, B.; Ding, M.; Liu, X.; Zhao, J.; Ross, R.P.; Stanton, C.; Yang, B.; Chen, W. Bifidobacterium breve CCFM1078 Alleviates Collagen-Induced Arthritis in Rats via Modulating the Gut Microbiota and Repairing the Intestinal Barrier Damage. J. Agric. Food Chem. 2022, 70, 14665–14678. [Google Scholar] [CrossRef]
- Jeong, Y.; Jhun, J.; Lee, S.Y.; Na, H.S.; Choi, J.; Cho, K.H.; Lee, S.Y.; Lee, A.R.; Park, S.J.; You, H.J.; et al. Therapeutic Potential of a Novel Bifidobacterium Identified Through Microbiome Profiling of RA Patients with Different RF Levels. Front. Immunol. 2021, 12, 736196. [Google Scholar] [CrossRef]
- Alipour, B.; Homayouni-Rad, A.; Vaghef-Mehrabany, E.; Sharif, S.K.; Vaghef-Mehrabany, L.; Asghari-Jafarabadi, M.; Nakhjavani, M.R.; Mohtadi-Nia, J. Effects of Lactobacillus casei supplementation on disease activity and inflammatory cytokines in rheumatoid arthritis patients: A randomized double-blind clinical trial. Int. J. Rheum. Dis. 2014, 17, 519–527. [Google Scholar] [CrossRef]
- Campagne, J.; Guichard, J.F.; Moulhade, M.C.; Kawski, H.; Maurier, F. Lactobacillus endocarditis: A case report in France and literature review. IDCases 2020, 21, e00811. [Google Scholar] [CrossRef]
- Grazioli-Gauthier, L.; Rigamonti, E.; Leo, L.A.; Martinetti Lucchini, G.; Lo Priore, E.; Bernasconi, E. Lactobacillus jensenii mitral valve endocarditis: Case report, literature review and new perspectives. IDCases 2022, 27, e01401. [Google Scholar] [CrossRef]
- Pasala, S.; Singer, L.; Arshad, T.; Roach, K. Lactobacillus endocarditis in a healthy patient with probiotic use. IDCases 2020, 22, e00915. [Google Scholar] [CrossRef]
- Mikucka, A.; Deptuła, A.; Bogiel, T.; Chmielarczyk, A.; Nurczyńska, E.; Gospodarek-Komkowska, E. Bacteraemia Caused by Probiotic Strains of Lacticaseibacillus rhamnosus-Case Studies Highlighting the Need for Careful Thought before Using Microbes for Health Benefits. Pathogens 2022, 11, 977. [Google Scholar] [CrossRef]
- Romond, M.B.; Ais, A.; Guillemot, F.; Bounouader, R.; Cortot, A.; Romond, C. Cell-free whey from milk fermented with Bifidobacterium breve C50 used to modify the colonic microflora of healthy subjects. J. Dairy Sci. 1998, 81, 1229–1235. [Google Scholar] [CrossRef]
- Scuotto, A.; Djorie, S.; Colavizza, M.; Romond, P.C.; Romond, M.B. Bifidobacterium breve C50 secretes lipoprotein with CHAP domain recognized in aggregated form by TLR2. Biochimie 2014, 107 Pt B, 367–375. [Google Scholar] [CrossRef]
- Scuotto, A.; Romond, P.C.; Djorie, S.; Alric, M.; Romond, M.B. In silico mining and characterization of bifidobacterial lipoprotein with CHAP domain secreted in an aggregated form. Int. J. Biol. Macromol. 2016, 82, 653–662. [Google Scholar] [CrossRef]
- Sane, F.; Piva, F.; Romond, M.B. Free lipoproteins from Bifidobacterium longum alleviate osteoarthritis through modulation of the gut microbiome. Microbiome Res. Rep. 2023, 2, 18. [Google Scholar] [CrossRef]
- Brand, D.D.; Latham, K.A.; Rosloniec, E.F. Collagen-induced arthritis. Nat. Protoc. 2007, 2, 1269–1275. [Google Scholar] [CrossRef]
- Sarkar, S.; Cooney, L.A.; White, P.; Dunlop, D.B.; Endres, J.; Jorns, J.M.; Wasco, M.J.; Fox, D.A. Regulation of pathogenic IL-17 responses in collagen-induced arthritis: Roles of endogenous interferon-gamma and IL-4. Arthritis Res. Ther. 2009, 11, R158. [Google Scholar] [CrossRef]
- El Kfoury, K.A.; Romond, M.B.; Scuotto, A.; Alidjinou, E.K.; Daboussi, F.; Hamze, M.; Engelmann, I.; Sane, F.; Hober, D. Bifidobacteria-derived lipoproteins inhibit the infection with coxsackievirus B4 in vitro. Int. J. Antimicrob. Agents. 2017, 50, 177–185. [Google Scholar] [CrossRef]
- Piva, F.; Romond, M.B. Bifidobactéries: De la bactérie commensale aux probiotiques et métabiotiques/postbiotiques. In Génie des Procédés Enzymatiques, Fermentaires et D’encapsulation Appliqués Aux Industries Agro-Alimentaires; Ghoul, M., Ed.; ISTE Group: London, UK, 2023; Volume 3, pp. 154–196. [Google Scholar]
- Romond, M.B.; Ais, A.; Yazourh, A.; Romond, C. Cell-free wheys from bifidobacteria fermented milks exert a regulatory effect on the intestinal microflora of mice and humans. Anaerobe 1997, 3, 137–143. [Google Scholar] [CrossRef]
- Piva, F.; Romond, M.B. Bifidobacteria: From commensal bacteria to probiotics and metabiotics/postbiotics. In Implementation of Enzymatic Processes and Lactic Acid Bacteria in Food Industry; Ghoul, M., Ed.; ISTE: London, UK, 2023; Chapter 8; pp. 189–225. [Google Scholar]
- Mullié, C.; Yazourh, A.; Singer, E.; Lecroix, F.; Blareau, J.P.; Romond, M.B.; Romond, C. Partial Characterization of Bifidobacterium breve C50 Cell-Free Whey Compounds Inducing Modifications to the Intestinal Microflora. J. Dairy Sci. 2002, 85, 1383–1389. [Google Scholar] [CrossRef]
- Tolve, R.; Cela, N.; Condelli, N.; Di Cairano, M.; Caruso, M.C.; Galgano, F. Microencapsulation as a Tool for the Formulation of Functional Foods: The Phytosterols’ Case Study. Foods 2020, 9, 470. [Google Scholar] [CrossRef]
- Dresch, C.; Ackermann, M.; Vogt, B.; de Andrade Pereira, B.; Shortman, K.; Fraefel, C. Thymic but not splenic CD8(+) DCs can efficiently cross-prime T cells in the absence of licensing factors. Eur. J. Immunol. 2011, 41, 2544–2555. [Google Scholar] [CrossRef]
- Jimenez-Boj, E.; Redlich, K.; Türk, B.; Hanslik-Schnabel, B.; Wanivenhaus, A.; Chott, A.; Smolen, J.S.; Schett, G. Interaction between Synovial Inflammatory Tissue and Bone Marrow in Rheumatoid Arthritis. J. Immunol. 2005, 175, 2579–2588. [Google Scholar] [CrossRef]
- Nehmar, R.; Alsaleh, G.; Voisin, B.; Flacher, V.; Mariotte, A.; Saferding, V.; Puchner, A.; Niederreiter, B.; Vandamme, T.; Schabbauer, G.; et al. Therapeutic modulation of plasmacytoid dendritic cells in experimental Arthritis. Arthritis Rheum. 2017, 69, 2124–2135. [Google Scholar] [CrossRef]
- Kool, M.; GeurtsVanKesse, C.; Muskens, F.; Branco Madeira, F.; van Nimwegen, M.; Kuipers, H.; Thielemans, K.; Hoogsteden, H.C.; Hammad, H.; Lambrech, B.N. Facilitated antigen uptake and timed exposure to TLR ligands dictate the antigen presenting potential of plasmacytoid DCs. J. Leuk. Biol. 2011, 90, 1170–1190. [Google Scholar] [CrossRef]
- Swiecki, M.; Colonna, M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015, 15, 471–485. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piva, F.; Gervois, P.; Karrout, Y.; Sané, F.; Romond, M.-B. Gut–Joint Axis: Impact of Bifidobacterial Cell Wall Lipoproteins on Arthritis Development. Nutrients 2023, 15, 4861. https://doi.org/10.3390/nu15234861
Piva F, Gervois P, Karrout Y, Sané F, Romond M-B. Gut–Joint Axis: Impact of Bifidobacterial Cell Wall Lipoproteins on Arthritis Development. Nutrients. 2023; 15(23):4861. https://doi.org/10.3390/nu15234861
Chicago/Turabian StylePiva, Frank, Philippe Gervois, Youness Karrout, Famara Sané, and Marie-Bénédicte Romond. 2023. "Gut–Joint Axis: Impact of Bifidobacterial Cell Wall Lipoproteins on Arthritis Development" Nutrients 15, no. 23: 4861. https://doi.org/10.3390/nu15234861
APA StylePiva, F., Gervois, P., Karrout, Y., Sané, F., & Romond, M.-B. (2023). Gut–Joint Axis: Impact of Bifidobacterial Cell Wall Lipoproteins on Arthritis Development. Nutrients, 15(23), 4861. https://doi.org/10.3390/nu15234861






