Dynamics and Crosstalk between Gut Microbiota, Metabolome, and Fecal Calprotectin in Very Preterm Infants: Insights into Feeding Intolerance
Abstract
:1. Introduction
2. Materials and Methods
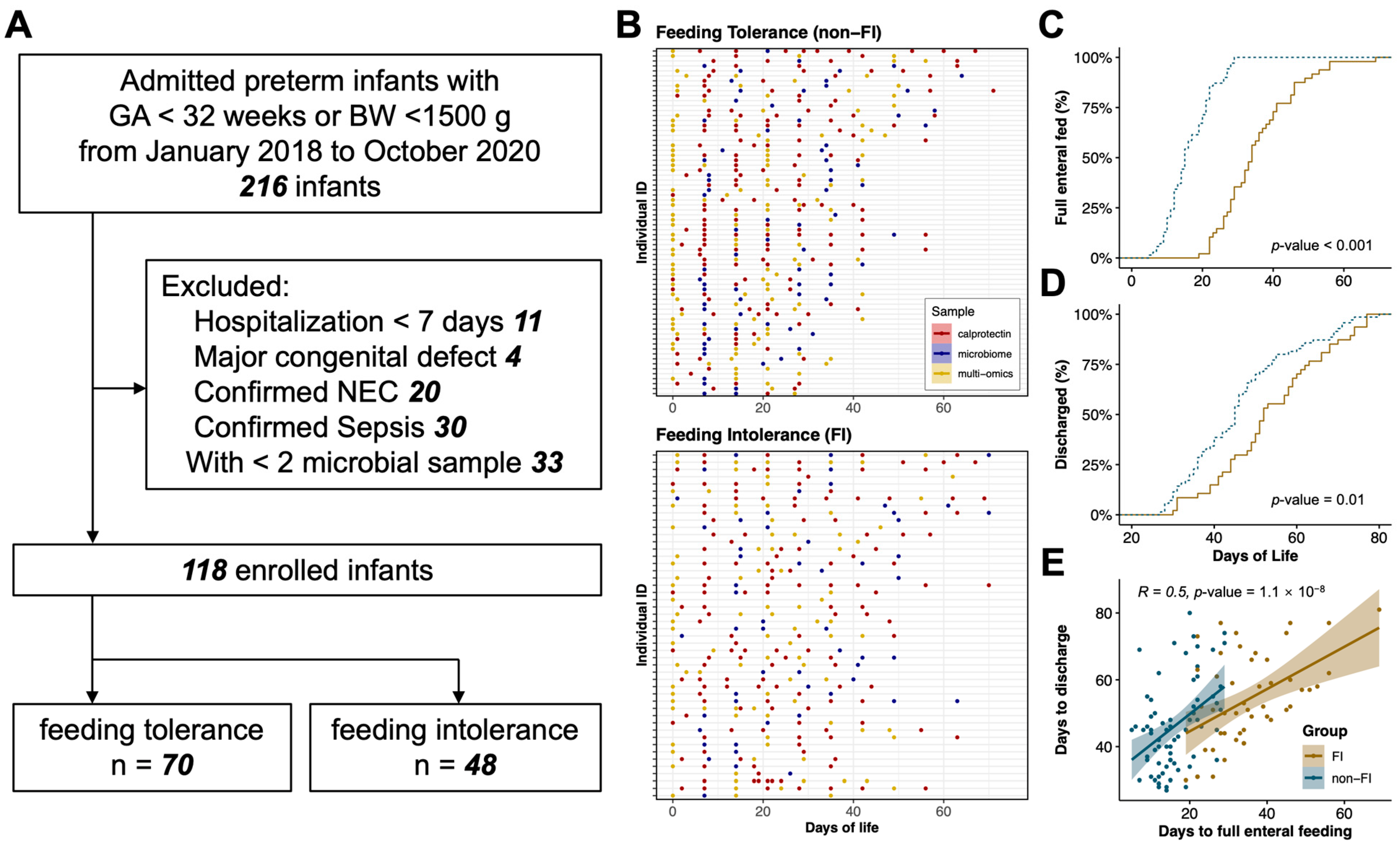
2.1. Study Design and Participants
2.2. Definitions
2.3. Fecal Sample Collection, Preparation, and FC Measurement
2.4. Stool 16S rRNA Gene Sequencing and Quantitative Microbial Profiling
2.5. Broad Range Metabolome by Liquid Chromatography–Mass Spectrometry
2.6. Statistical Analysis and Data Integration
3. Results
3.1. Infants, Maternal, and Sample Characteristics
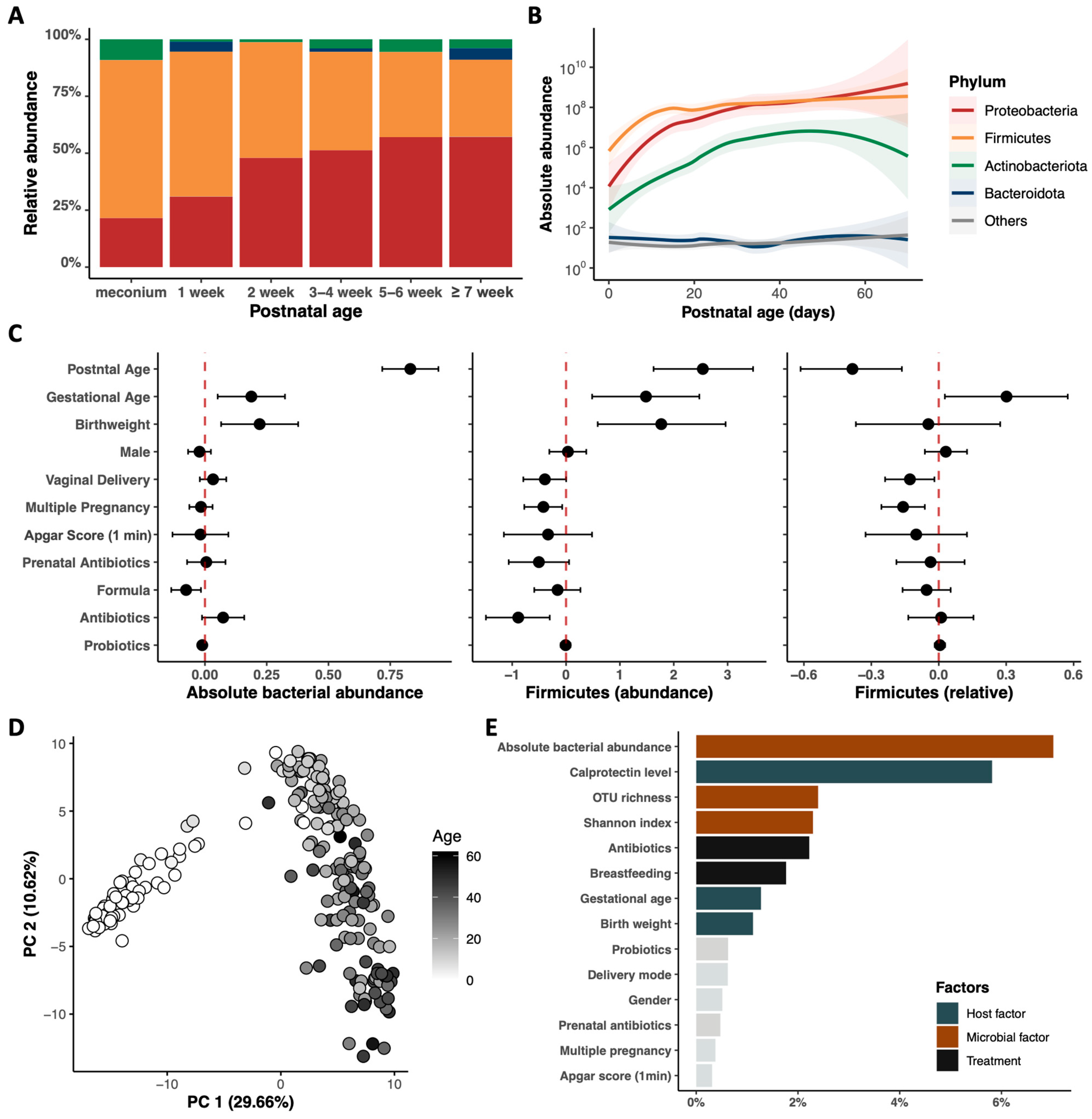
3.2. Postnatal Dynamics of the Gut Microbiome and Metabolome in Very Preterm Infants
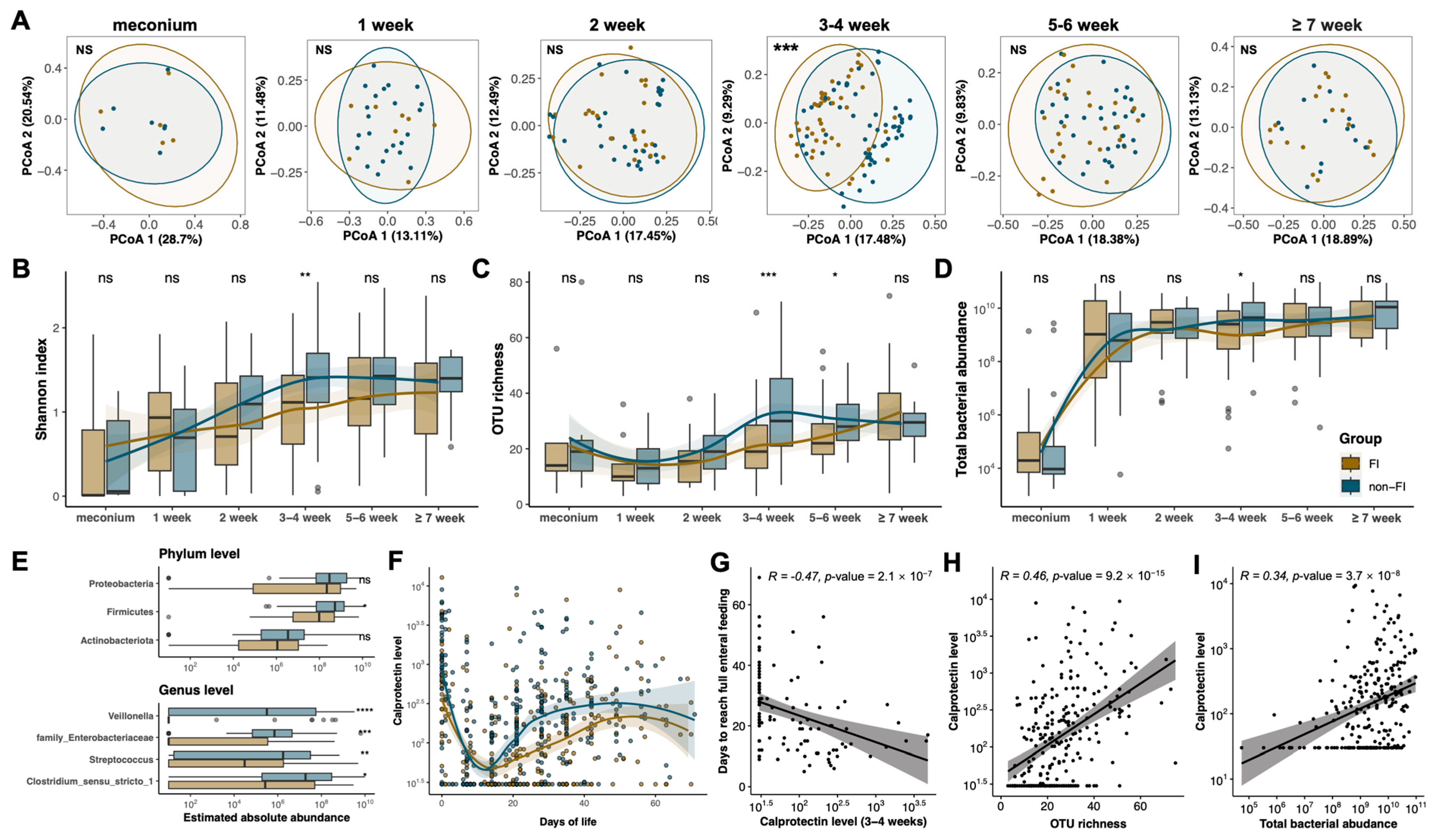
3.3. Compromised Developmental Trajectory of the Gut Microbiome Is Associated with Altered Calprotectin Levels in Preterm Infants with Feeding Intolerance
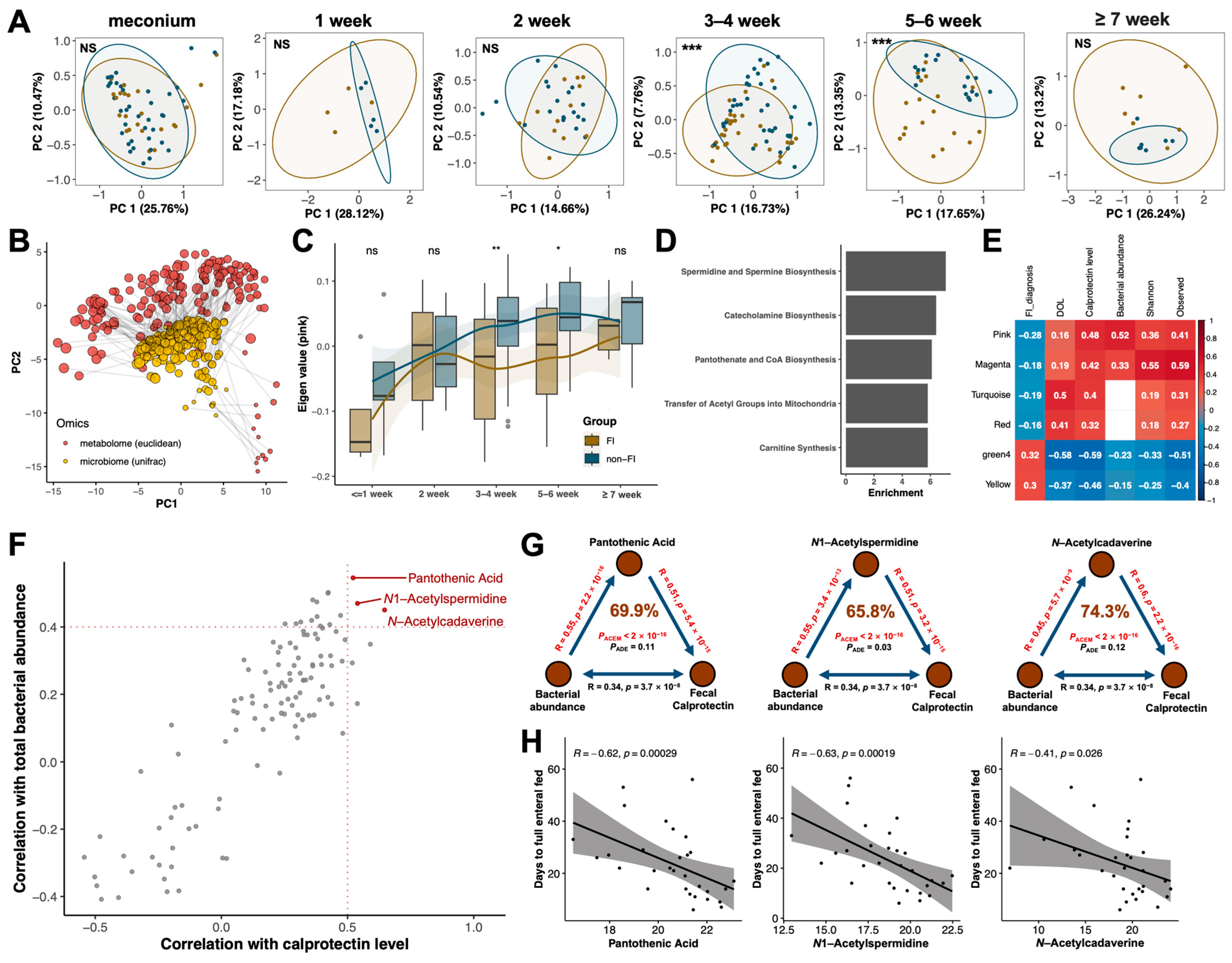
3.4. Multi-Omics Analysis Identified Pantothenic Acid and Polyamine Metabolites as Potential Key Mediators in the Crosstalk between Gut Microbiota and Host Immunity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Mauro, A.; Neu, J.; Riezzo, G.; Raimondi, F.; Martinelli, D.; Francavilla, R.; Indrio, F. Gastrointestinal function development and microbiota. Ital. J. Pediatr. 2013, 39, 15. [Google Scholar] [CrossRef] [PubMed]
- Indrio, F.; Riezzo, G.; Cavallo, L.; Di Mauro, A.; Francavilla, R. Physiological basis of food intolerance in VLBW. J. Matern.-Fetal Neonatal Med. 2011, 24 (Suppl. S1), 64–66. [Google Scholar] [CrossRef]
- Weeks, C.L.; Marino, L.V.; Johnson, M.J. A systematic review of the definitions and prevalence of feeding intolerance in preterm infants. Clin. Nutr. 2021, 40, 5576–5586. [Google Scholar] [CrossRef] [PubMed]
- Walls Castellanos, M.; Claud, E.C. The microbiome, guard or threat to infant health. Trends Mol. Med. 2021, 27, 1175–1186. [Google Scholar] [CrossRef] [PubMed]
- Healy, D.B.; Ryan, C.A.; Ross, R.P.; Stanton, C.; Dempsey, E.M. Clinical implications of preterm infant gut microbiome development. Nat. Microbiol. 2022, 7, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.L.; Lagomarcino, A.J.; Schibler, K.R.; Taft, D.H.; Yu, Z.; Wang, B.; Altaye, M.; Wagner, M.; Gevers, D.; Ward, D.V.; et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 2013, 1, 13. [Google Scholar] [CrossRef]
- Claud, E.C.; Keegan, K.P.; Brulc, J.M.; Lu, L.; Bartels, D.; Glass, E.; Chang, E.B.; Meyer, F.; Antonopoulos, D.A. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 2013, 1, 20. [Google Scholar] [CrossRef]
- Aguilar-Lopez, M.; Dinsmoor, A.M.; Ho, T.T.B.; Donovan, S.M. A systematic review of the factors influencing microbial colonization of the preterm infant gut. Gut Microbes 2021, 13, 1884514. [Google Scholar] [CrossRef]
- Beghetti, I.; Barone, M.; Brigidi, P.; Sansavini, A.; Corvaglia, L.; Aceti, A.; Turroni, S. Early-life gut microbiota and neurodevelopment in preterm infants: A narrative review. Front. Nutr. 2023, 10, 1241303. [Google Scholar] [CrossRef]
- Iliodromiti, Z.; Triantafyllou, A.R.; Tsaousi, M.; Pouliakis, A.; Petropoulou, C.; Sokou, R.; Volaki, P.; Boutsikou, T.; Iacovidou, N. Gut Microbiome and Neurodevelopmental Disorders: A Link Yet to Be Disclosed. Microorganisms 2023, 11, 487. [Google Scholar] [CrossRef]
- Liu, X.C.; Sun, Q.; Ji, Y.C.; Fu, L.Z.; Wang, Z.L.; He, Y.; Li, L.Q. Differences in the Gut Microbiota Composition and Metabolites Associated with Feeding Intolerance in VLBW Infants with a Gestational Age of ≤30 Weeks: A Pilot Study. Front. Cell Infect. Microbiol. 2022, 12, 726322. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ao, D.; Cai, X.; Huang, P.; Cai, N.; Lin, S.; Wu, B. Early gut microbiota in very low and extremely low birth weight preterm infants with feeding intolerance: A prospective case-control study. J. Microbiol. 2022, 60, 1021–1031. [Google Scholar] [CrossRef] [PubMed]
- Ruoss, J.L.; Bazacliu, C.; Russell, J.T.; Cruz, D.; Li, N.; Gurka, M.J.; Filipp, S.L.; Polin, R.A.; Triplett, E.W.; Neu, J. Routine Early Antibiotic Use in SymptOmatic Preterm Neonates: A Pilot Randomized Controlled Trial. J. Pediatr. 2021, 229, 294–298.e3. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chang, Y.; Wang, Z.; Bao, W.; Li, Z. Altered gut microbiota is associated with feeding intolerance in preterm infants. Turk. J. Pediatr. 2021, 63, 206–217. [Google Scholar] [CrossRef]
- Yuan, Z.; Yan, J.; Wen, H.; Deng, X.; Li, X.; Su, S. Feeding intolerance alters the gut microbiota of preterm infants. PLoS ONE 2019, 14, e0210609. [Google Scholar] [CrossRef] [PubMed]
- Jukic, A.; Bakiri, L.; Wagner, E.F.; Tilg, H.; Adolph, T.E. Calprotectin: From biomarker to biological function. Gut 2021, 70, 1978–1988. [Google Scholar] [CrossRef] [PubMed]
- Ricciuto, A.; Griffiths, A.M. Clinical value of fecal calprotectin. Crit. Rev. Clin. Lab. Sci. 2019, 56, 307–320. [Google Scholar] [CrossRef]
- Willers, M.; Ulas, T.; Völlger, L.; Vogl, T.; Heinemann, A.S.; Pirr, S.; Pagel, J.; Fehlhaber, B.; Halle, O.; Schöning, J.; et al. S100A8 and S100A9 Are Important for Postnatal Development of Gut Microbiota and Immune System in Mice and Infants. Gastroenterology 2020, 159, 2130–2145. [Google Scholar] [CrossRef]
- Hong, L.; Huang, Y.; Jiang, S.; Han, J.; Li, S.; Zhang, L.; Zhou, Q.; Cao, X.; Yu, W.; Yang, Y.; et al. Postnatal Dynamics and Clinical Associations of Fecal Calprotectin in Very Preterm Infants: Implications for Necrotizing Enterocolitis and Feeding Intolerance. Clin. Transl. Gastroenterol. 2023, 14, e00604. [Google Scholar] [CrossRef]
- Clinical guidelines for the diagnosis and treatment of feeding intolerance in preterm infants (2020). Zhongguo Dang Dai Er Ke Za Zhi (Chin. J. Contemp. Pediatr.) 2020, 22, 1047–1055.
- Bell, M.J.; Ternberg, J.L.; Feigin, R.D.; Keating, J.P.; Marshall, R.; Barton, L.; Brotherton, T. Neonatal necrotizing enterocolitis. Therapeutic decisions based upon clinical staging. Ann. Surg. 1978, 187, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiang, S.; Sun, J.; Hei, M.; Wang, L.; Zhang, H.; Ma, X.; Wu, H.; Li, X.; Sun, H.; et al. Assessment of Neonatal Intensive Care Unit Practices, Morbidity, and Mortality among Very Preterm Infants in China. JAMA Netw. Open 2021, 4, e2118904. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, L.; Yan, W.; Li, S.; Han, J.; Zhou, Q.; Yang, Y.; Lee, S.K.; Cao, Y.; Group, R.-E.S. Antibiotic Use in Neonatal Intensive Care Units in China: A Multicenter Cohort Study. J. Pediatr. 2021, 239, 136–142.e4. [Google Scholar] [CrossRef]
- Expert consensus on the diagnosis and management of neonatal sepsis (version 2019). Zhonghua Er Ke Za Zhi (Chin. J. Pediatr.) 2019, 57, 252–257. [CrossRef]
- Lutsar, I.; Chazallon, C.; Carducci, F.I.C.; Trafojer, U.; Abdelkader, B.; de Cabre, V.M.; Esposito, S.; Giaquinto, C.; Heath, P.T.; Ilmoja, M.-L.; et al. Current management of late onset neonatal bacterial sepsis in five European countries. Eur. J. Pediatr. 2014, 173, 997–1004. [Google Scholar] [CrossRef]
- Jiang, S.; Hong, L.; Gai, J.; Shi, J.; Yang, Y.; Lee, S.K.; Cao, Y.; Group, R.-E.S. Early-onset Sepsis among Preterm Neonates in China, 2015 to 2018. Pediatr. Infect. Dis. J. 2019, 38, 1236–1241. [Google Scholar] [CrossRef]
- Ballard, J.L.; Novak, K.K.; Driver, M. A simplified score for assessment of fetal maturation of newly born infants. J. Pediatr. 1979, 95, 769–774. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, R.; Zhang, S.; Shi, W.; Yan, W.; Wang, X.; Lyu, Q.; Liu, L.; Zhou, Q.; Qiu, Q.; et al. Chinese neonatal birth weight curve for different gestational age. Zhonghua Er Ke Za Zhi 2015, 53, 97–103. [Google Scholar]
- Dorling, J.; Abbott, J.; Berrington, J.; Bosiak, B.; Bowler, U.; Boyle, E.; Embleton, N.; Hewer, O.; Johnson, S.; Juszczak, E.; et al. Controlled Trial of Two Incremental Milk-Feeding Rates in Preterm Infants. N. Engl. J. Med. 2019, 381, 1434–1443. [Google Scholar] [CrossRef]
- Hong, L.; Zhang, L.; Zhou, Q.; Li, S.; Han, J.; Jiang, S.; Han, X.; Yang, Y.; Hong, S.; Cao, Y. Impacts of Enriched Human Milk Cells on Fecal Metabolome and Gut Microbiome of Premature Infants with Stage I Necrotizing Enterocolitis: A Pilot Study. Mol. Nutr. Food Res. 2022, 66, e2100342. [Google Scholar] [CrossRef]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.; Schmidt, T.M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015, 43, D593–D598. [Google Scholar] [CrossRef] [PubMed]
- Jian, C.; Luukkonen, P.; Yki-Jarvinen, H.; Salonen, A.; Korpela, K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 2020, 15, e0227285. [Google Scholar] [CrossRef] [PubMed]
- Cassir, N.; Simeoni, U.; La Scola, B. Gut microbiota and the pathogenesis of necrotizing enterocolitis in preterm neonates. Future Microbiol. 2016, 11, 273–292. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Tingley, D.; Yamamoto, T.; Hirose, K.; Keele, L.; Imai, K. Mediation: R Package for Causal Mediation Analysis. J. Stat. Softw. 2014, 59, 1–38. [Google Scholar] [CrossRef]
- Rao, J.N.; Xiao, L.; Wang, J.-Y. Polyamines in Gut Epithelial Renewal and Barrier Function. Physiology 2020, 35, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Ooga, T.; Matsumoto, M. Intestinal luminal putrescine is produced by collective biosynthetic pathways of the commensal microbiome. Gut Microbes 2019, 10, 159–171. [Google Scholar] [CrossRef]
- Nakamura, A.; Kurihara, S.; Takahashi, D.; Ohashi, W.; Nakamura, Y.; Kimura, S.; Onuki, M.; Kume, A.; Sasazawa, Y.; Furusawa, Y.; et al. Symbiotic polyamine metabolism regulates epithelial proliferation and macrophage differentiation in the colon. Nat. Commun. 2021, 12, 2105. [Google Scholar] [CrossRef]
- Milovic, V. Polyamines in the gut lumen: Bioavailability and biodistribution. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Uda, K.; Tsujikawa, T.; Fujiyama, Y.; Bamba, T. Rapid absorption of luminal polyamines in a rat small intestine ex vivo model. J. Gastroenterol. Hepatol. 2003, 18, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Dufour, C.; Dandrifosse, G.; Forget, P.; Vermesse, F.; Romain, N.; Lepoint, P. Spermine and spermidine induce intestinal maturation in the rat. Gastroenterology 1988, 95, 112–116. [Google Scholar] [CrossRef]
- Luk, G.D. Polyamines in intestinal growth. Biochem. Soc. Trans. 1990, 18, 1090–1091. [Google Scholar] [CrossRef]
- Buts, J.P.; De Keyser, N.; Kolanowski, J.; Sokal, E.; Van Hoof, F. Maturation of villus and crypt cell functions in rat small intestine. Role of dietary polyamines. Dig. Dis. Sci. 1993, 38, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Capano, G.; Bloch, K.J.; Schiffrin, E.J.; Dascoli, J.A.; Israel, E.J.; Harmatz, P.R. Influence of the polyamine, spermidine, on intestinal maturation and dietary antigen uptake in the neonatal rat. J. Pediatr. Gastroenterol. Nutr. 1994, 19, 34–42. [Google Scholar] [CrossRef]
- Kaouass, M.; Deloyer, P.; Wery, I.; Dandrifosse, G. Analysis of structural and biochemical events occurring in the small intestine after dietary polyamine ingestion in suckling rats. Dig. Dis. Sci. 1996, 41, 1434–1444. [Google Scholar] [CrossRef]
- ter Steege, J.C.; Buurman, W.A.; Forget, P.P. Spermine induces maturation of the immature intestinal immune system in neonatal mice. J. Pediatr. Gastroenterol. Nutr. 1997, 25, 332–340. [Google Scholar] [CrossRef]
- Capano, G.; Bloch, K.J.; Carter, E.A.; Dascoli, J.A.; Schoenfeld, D.; Harmatz, P.R. Polyamines in human and rat milk influence intestinal cell growth in vitro. J. Pediatr. Gastroenterol. Nutr. 1998, 27, 281–286. [Google Scholar] [CrossRef]
- van Wettere, W.H.; Willson, N.L.; Pain, S.J.; Forder, R.E. Effect of oral polyamine supplementation pre-weaning on piglet growth and intestinal characteristics. Animal 2016, 10, 1655–1659. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, Z.; Hu, H.; Duan, W.; Wang, A.; Dong, Y.; Gao, W.; Deng, S.; Cheng, B.; Li, J.; et al. Differentiation and Proliferation of Intestinal Stem Cells and its Underlying Regulated Mechanisms during Weaning. Curr. Protein Pept. Sci. 2019, 20, 690–695. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.X.; Cai, L.; Zhao, X.M.; Jiang, X.R.; Li, X.L. Effects of Spermidine on Cell Proliferation, Migration, and Inflammatory Response in Porcine Enterocytes. Front. Biosci. (Landmark Ed.) 2022, 27, 194. [Google Scholar] [CrossRef]
- Ma, L.; Ni, Y.; Wang, Z.; Tu, W.; Ni, L.; Zhuge, F.; Zheng, A.; Hu, L.; Zhao, Y.; Zheng, L.; et al. Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 2020, 12, 1832857. [Google Scholar] [CrossRef]
- Pérez-Cano, F.J.; González-Castro, A.; Castellote, C.; Franch, A.; Castell, M. Influence of breast milk polyamines on suckling rat immune system maturation. Dev. Comp. Immunol. 2010, 34, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Niechcial, A.; Schwarzfischer, M.; Wawrzyniak, M.; Atrott, K.; Laimbacher, A.; Morsy, Y.; Katkeviciute, E.; Häfliger, J.; Westermann, P.; Akdis, C.A.; et al. Spermidine ameliorates colitis via induction of anti-inflammatory macrophages and prevention of intestinal dysbiosis. J. Crohns Colitis 2023, 17, 1489–1503. [Google Scholar] [CrossRef]
- Sabui, S.; Kapadia, R.; Ghosal, A.; Schneider, M.; Lambrecht, N.W.G.; Said, H.M. Biotin and pantothenic acid oversupplementation to conditional SLC5A6 KO mice prevents the development of intestinal mucosal abnormalities and growth defects. Am. J. Physiol. Cell Physiol. 2018, 315, C73–C79. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, S.; Ravcheev, D.; de Crécy-Lagard, V.; Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 2015, 6, 148. [Google Scholar] [CrossRef]
- Uebanso, T.; Shimohata, T.; Mawatari, K.; Takahashi, A. Functional Roles of B-Vitamins in the Gut and Gut Microbiome. Mol. Nutr. Food Res. 2020, 64, e2000426. [Google Scholar] [CrossRef]
| Variable | FI (n = 48) | Non-FI (n = 70) | p Value |
|---|---|---|---|
| Infant characteristics | |||
| Gestational age (weeks), mean (SD) | 29.8 (1.5) | 30.2 (1.7) | 0.208 |
| Birth weight (g), mean (SD) | 1265.2 (177.2) | 1335.1 (202.3) | 0.055 |
| Male, n/N (%) | 25 (52.1) | 29 (41.4) | 0.341 |
| Small for gestational age, n/N (%) | 3 (6.2) | 6 (8.6) | 0.909 |
| Apgar score at 1 min, mean (SD) | 7.6 (1.8) | 7.9 (1.6) | 0.318 |
| Maternal characteristics | |||
| Multiple pregnancy, n/N (%) | 22 (45.8) | 28 (40.0) | 0.660 |
| Vaginal delivery, n/N (%) | 16 (33.3) | 23 (32.9) | 1.000 |
| PPROM a, n/N (%) | 11 (22.9) | 17 (24.3) | 1.000 |
| Use of prenatal antibiotics, n/N (%) | 11 (22.9) | 10 (14.3) | 0.337 |
| Gestational diabetes mellitus, n/N (%) | 12 (25.0) | 13 (18.6) | 0.542 |
| Gestational hypertension, n/N (%) | 6 (12.5) | 7 (10.0) | 0.899 |
| Treatment | |||
| Early antibiotics use b (days), mean (SD) | 4.2 (2.2) | 3.6 (2.2) | 0.128 |
| Exclusive breastmilk feeding, n/N (%) | 40 (81.4) | 57 (91.6) | 0.656 |
| Probiotics use c (days), mean (SD) | 6.2 (12.0) | 5.3 (8.4) | 0.656 |
| Drugs for PDA d, n/N (%) | 8 (16.7) | 6 (8.6) | 0.296 |
| Proton pump inhibitors, n/N (%) | 1 (2.1) | 2 (2.9) | 1.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, L.; Huang, Y.; Han, J.; Li, S.; Zhang, L.; Jiang, S.; Zhou, Q.; Cao, X.; Yu, W.; Yang, Y.; et al. Dynamics and Crosstalk between Gut Microbiota, Metabolome, and Fecal Calprotectin in Very Preterm Infants: Insights into Feeding Intolerance. Nutrients 2023, 15, 4849. https://doi.org/10.3390/nu15224849
Hong L, Huang Y, Han J, Li S, Zhang L, Jiang S, Zhou Q, Cao X, Yu W, Yang Y, et al. Dynamics and Crosstalk between Gut Microbiota, Metabolome, and Fecal Calprotectin in Very Preterm Infants: Insights into Feeding Intolerance. Nutrients. 2023; 15(22):4849. https://doi.org/10.3390/nu15224849
Chicago/Turabian StyleHong, Luyang, Yihuang Huang, Junyan Han, Shujuan Li, Lan Zhang, Siyuan Jiang, Qi Zhou, Xincheng Cao, Weiyin Yu, Yi Yang, and et al. 2023. "Dynamics and Crosstalk between Gut Microbiota, Metabolome, and Fecal Calprotectin in Very Preterm Infants: Insights into Feeding Intolerance" Nutrients 15, no. 22: 4849. https://doi.org/10.3390/nu15224849
APA StyleHong, L., Huang, Y., Han, J., Li, S., Zhang, L., Jiang, S., Zhou, Q., Cao, X., Yu, W., Yang, Y., Hong, S., Zhou, Y., Yan, W., & Cao, Y. (2023). Dynamics and Crosstalk between Gut Microbiota, Metabolome, and Fecal Calprotectin in Very Preterm Infants: Insights into Feeding Intolerance. Nutrients, 15(22), 4849. https://doi.org/10.3390/nu15224849






