An Isolate of Streptococcus mitis Displayed In Vitro Antimicrobial Activity and Deleterious Effect in a Preclinical Model of Lung Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, Growth Conditions
2.2. gDNA Extraction and Genome Analysis of S. mitis EM-371
2.3. Agar Well Diffusion Method
2.4. Animals and Procedures
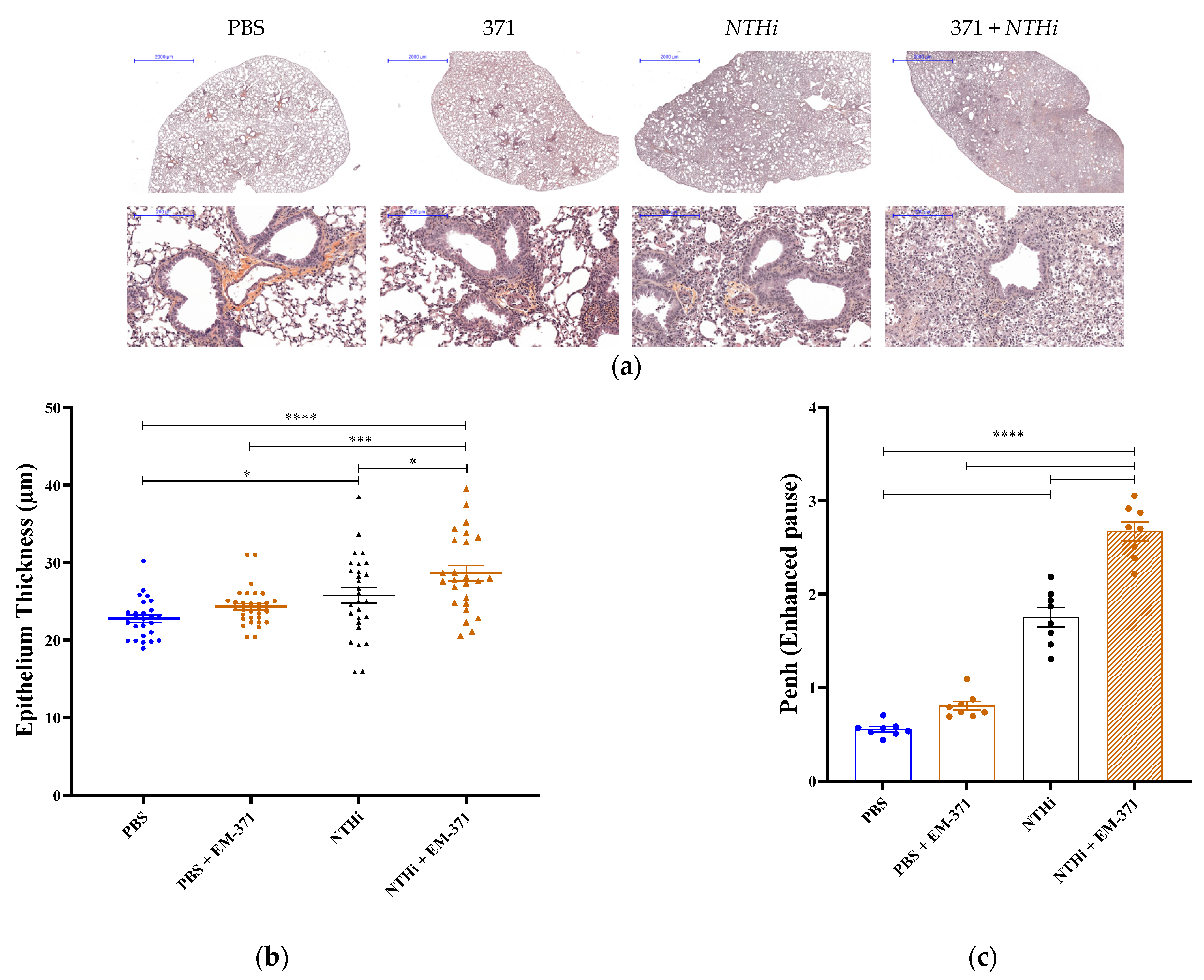
2.5. Plethysmography
2.6. Sample Collection
2.7. Histology
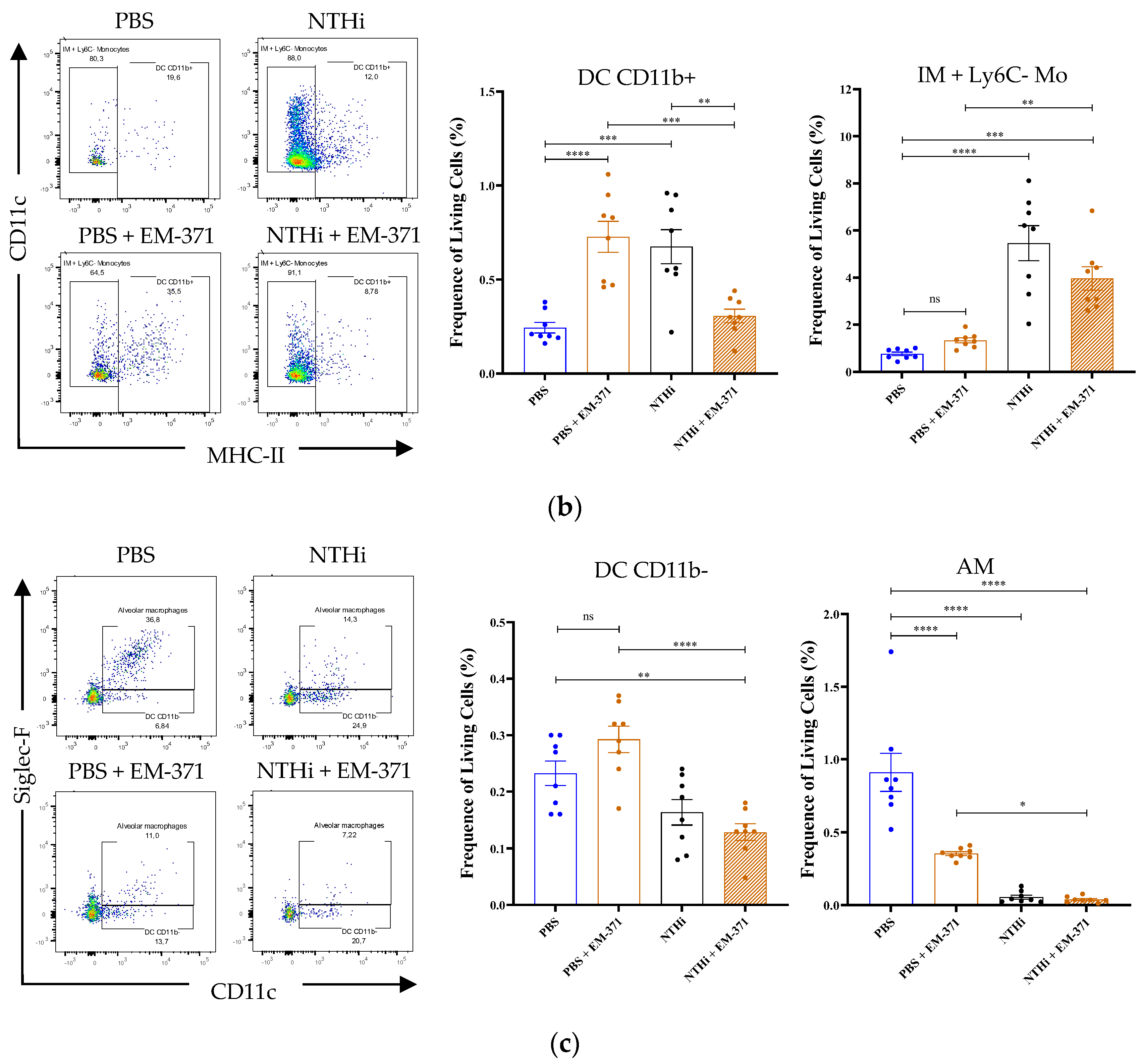
2.8. Flow Cytometry
2.9. SCFA Analysis
2.10. Statistical Analysis
3. Results
3.1. Probiotic Potential of S. mitis EM-371
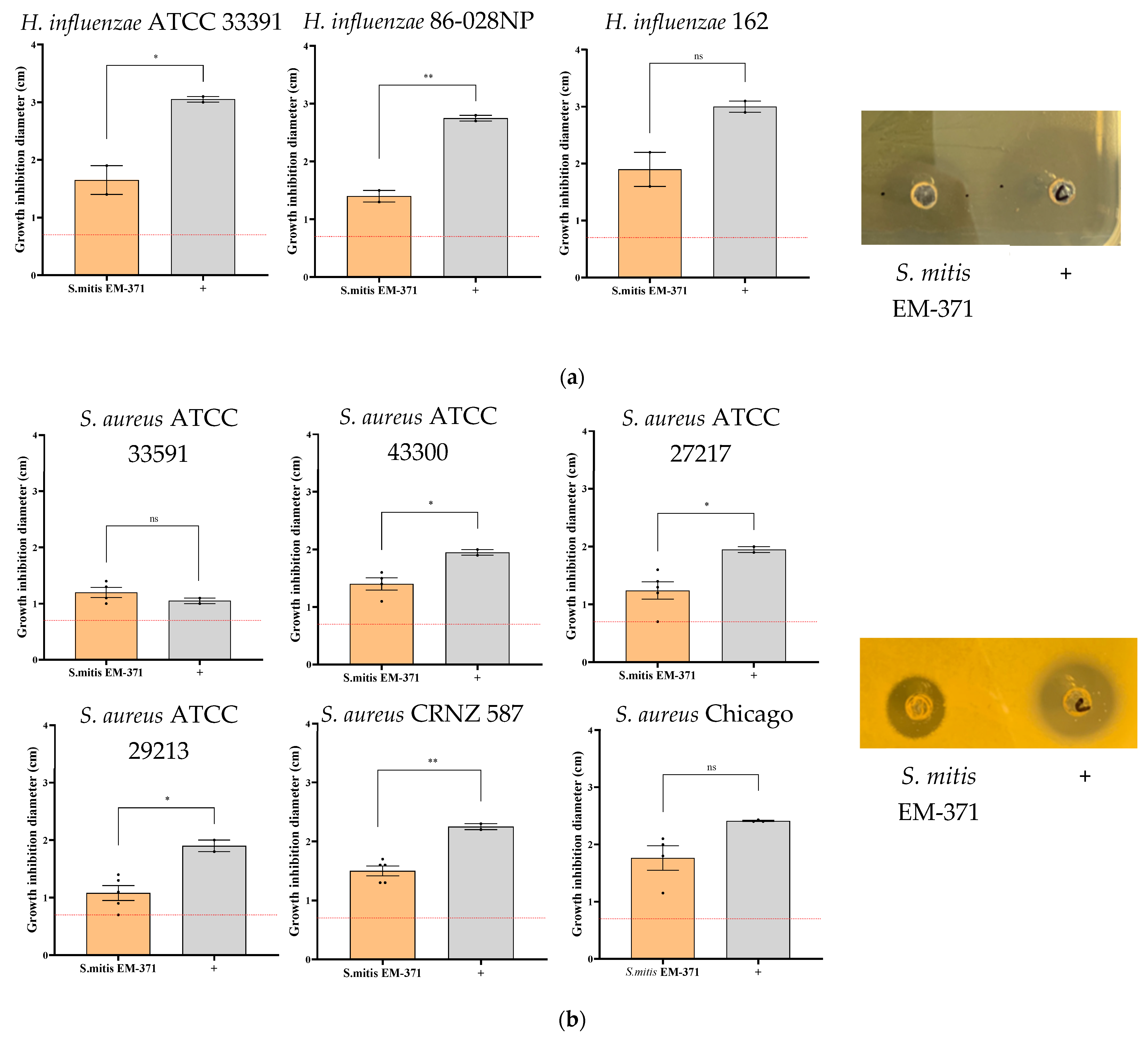
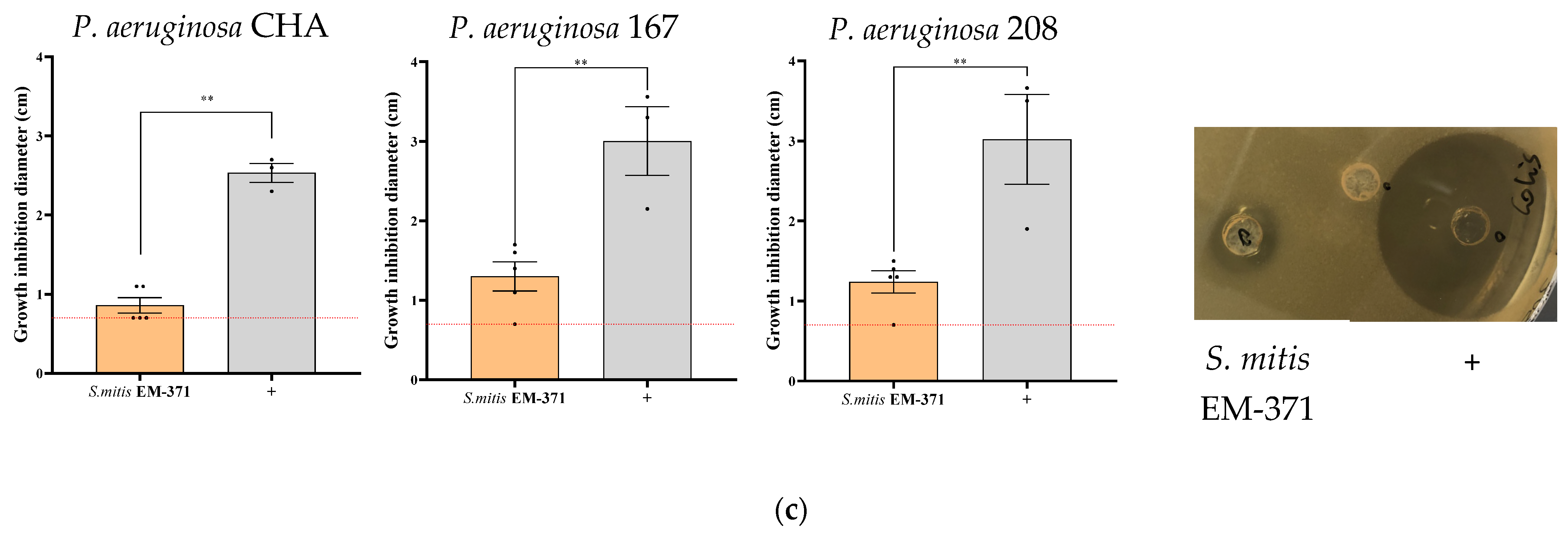
3.2. S. mitis EM-371 Inhibits the Growth of Bacterial Respiratory Pathogens In Vitro on Agar-Diffusion Test
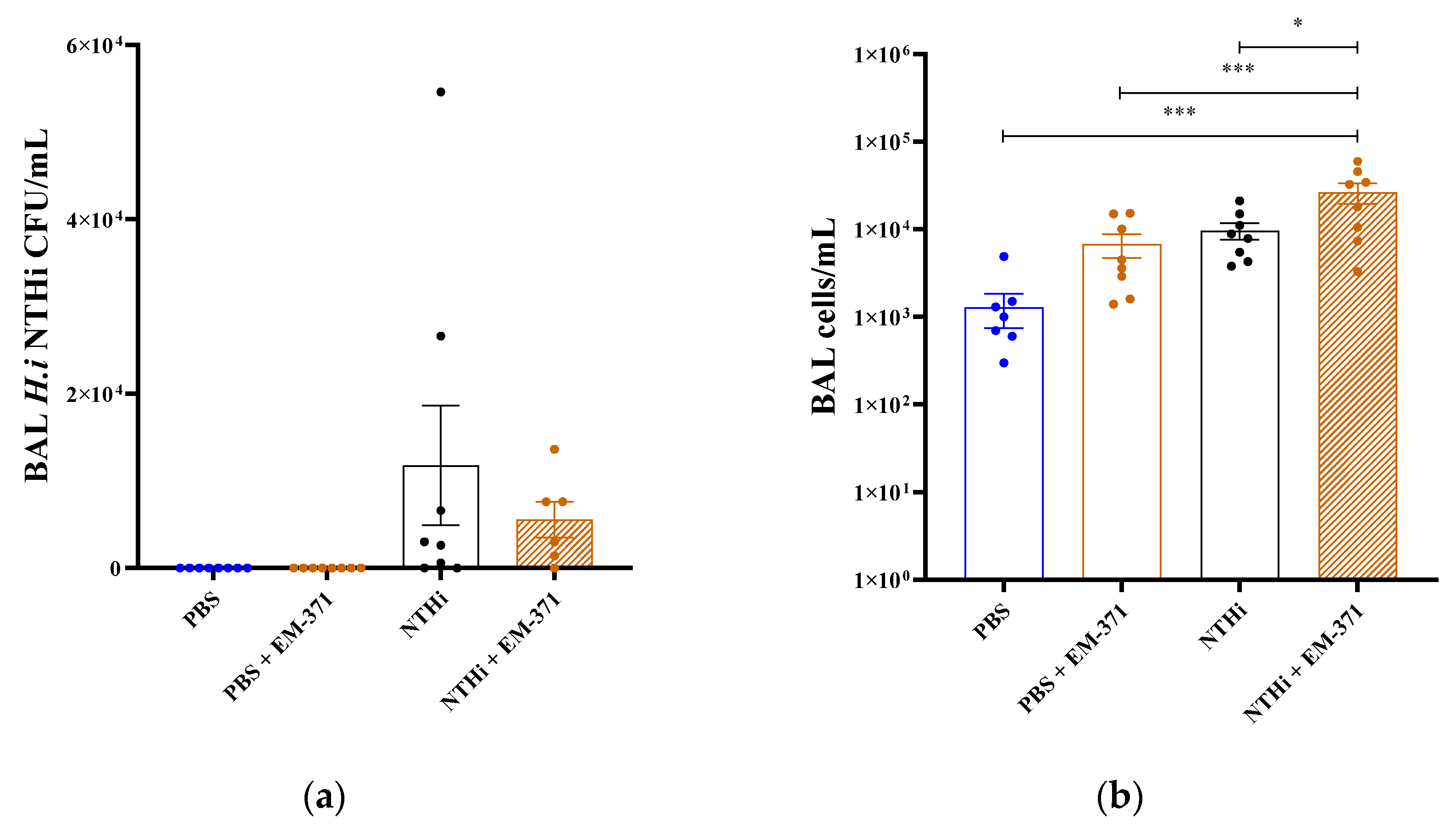
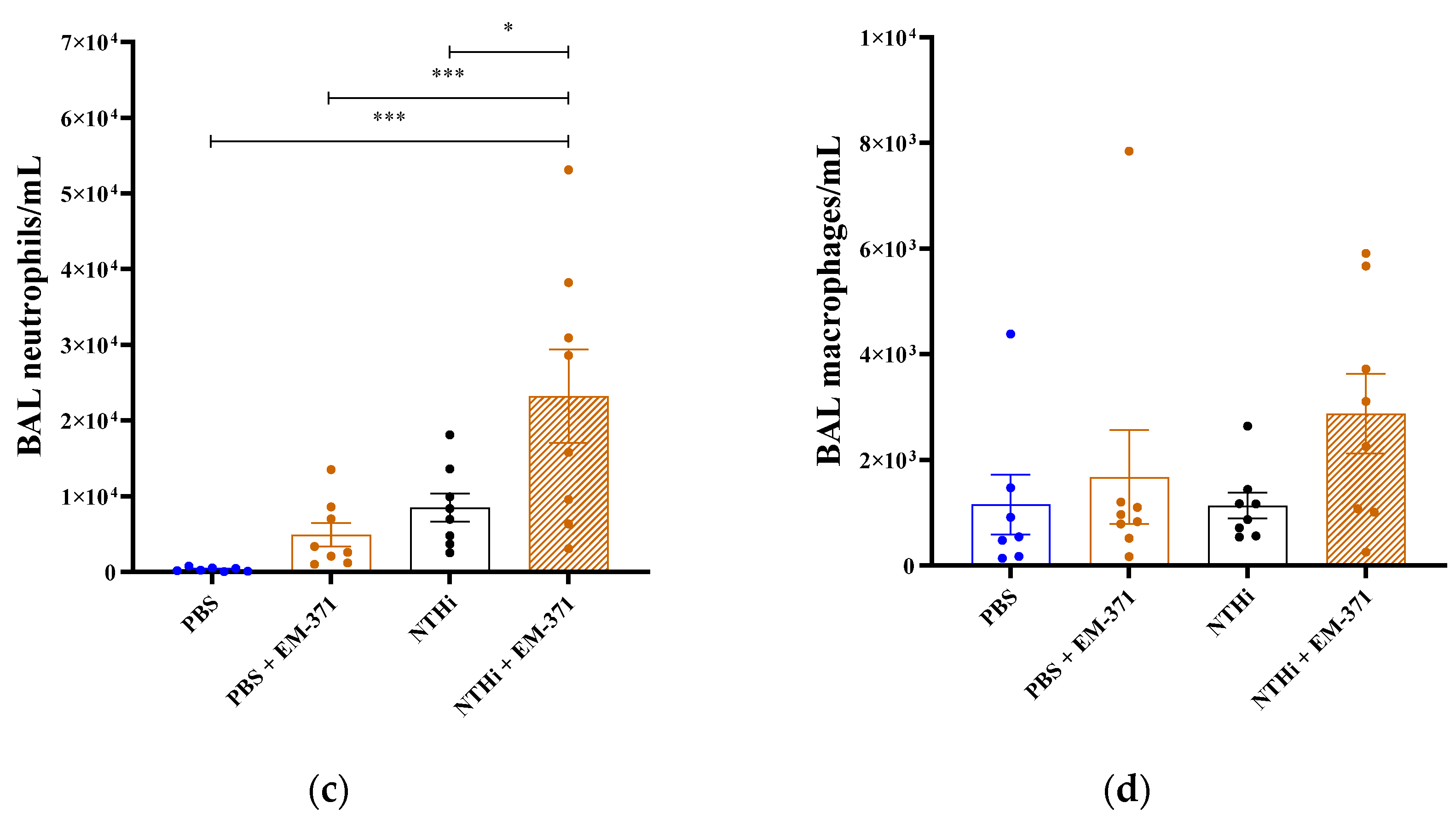
3.3. Preventive Treatment with S. mitis EM-371 Did Not Protect the Mice against Haemophilus influenzae (NTHi) Lung Infection in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, S.C.; Lin, C.-H.; Sung, C.T.; Fang, J.-Y. Antibacterial activities of bacteriocins: Application in foods and pharmaceuticals. Front. Microbiol. 2014, 5, 241. [Google Scholar] [PubMed]
- Lebeer, S.; Claes, I.; Tytgat, H.L.; Verhoeven, T.L.; Marien, E.; von Ossowski, I.; Reunanen, J.; Palva, A.; Vos, W.M.; Keersmaecker, S.C.; et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of Action of Probiotics. Adv. Nutr. 2019, 10 (Suppl. S1), S49–S66, Erratum in: Adv. Nutr. 2020, 11, 1054. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Beaumont, M.; Martín, R.; Langella, P.; Braesco, V.; Thomas, M. A proposed framework for an appropriate evaluation scheme for microorganisms as novel foods with a health claim in Europe. Microb. Cell Fact. 2015, 14, 48. [Google Scholar] [CrossRef]
- Van den Broek, M.F.L.; De Boeck, I.; Kiekens, F.; Boudewyns, A.; Vanderveken, O.M.; Lebeer, S. Translating Recent Microbiome Insights in Otitis Media into Probiotic Strategies. Clin. Microbiol. Rev. 2019, 32, e00010-18. [Google Scholar] [CrossRef]
- Forum of International Respiratory Societies. The Global Impact of Respiratory Disease, 2nd ed.; European Respiratory Society: Sheffield, UK, 2017. [Google Scholar]
- Su, Y.C.; Jalalvand, F.; Thegerström, J.; Riesbeck, K. The Interplay Between Immune Response and Bacterial Infection in COPD: Focus Upon Non-typeable Haemophilus influenzae. Front. Immunol. 2018, 9, 2530. [Google Scholar] [CrossRef]
- Murphy, T.F. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 2003, 16, 129–134. [Google Scholar] [CrossRef]
- Callaghan, M.; McClean, S. Bacterial host interactions in cystic fibrosis. Curr. Opin. Microbiol. 2012, 15, 71–77. [Google Scholar] [CrossRef]
- Cullen, L.; McClean, S. Bacterial Adaptation during Chronic Respiratory Infections. Pathogens 2015, 4, 66–89. [Google Scholar] [CrossRef]
- Planet, P.J. Pseudomonas aeruginosa. In Principles and Practice of Pediatric Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2018; pp. 866–870. [Google Scholar]
- Faure, E.; Kwong, K.; Nguyen, D. Pseudomonas aeruginosa in Chronic Lung Infections: How to Adapt Within the Host? Front. Immunol. 2018, 9, 2416. [Google Scholar] [CrossRef]
- Fujitani, S.; Sun, H.Y.; Yu, V.L.; Weingarten, J.A. Pneumonia due to Pseudomonas aeruginosa: Part I: Epidemiology, clinical diagnosis, and source. Chest 2011, 139, 909–919. [Google Scholar] [CrossRef]
- Hauser, A.R.; Jain, M.; Bar-Meir, M.; McColley, S.A. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 2011, 24, 29–70. [Google Scholar] [CrossRef] [PubMed]
- Slack, M.P.E. A review of the role of Haemophilus influenzae in community-acquired pneumonia. Pneumonia 2015, 6, 26–43. [Google Scholar] [CrossRef] [PubMed]
- King, P.T.; Sharma, R. The Lung Immune Response to Nontypeable Haemophilus influenzae (Lung Immunity to NTHi). J. Immunol. Res. 2015, 2015, 706376. [Google Scholar] [CrossRef] [PubMed]
- Sadikot, R.T.; Blackwell, T.S.; Christman, J.W.; Prince, A.S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Prince, A. Staphylococcus aureus Infection in the Respiratory Tract. In Mucosal Immunology of Acute Bacterial Pneumonia; Columbia University: New York, NY, USA, 2013. [Google Scholar]
- Könönen, E. Development of oral bacterial flora in young children. Ann. Med. 2000, 32, 107–112. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of Oral Streptococci. Microbiol. Spectr. 2018. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Lung HIV Microbiome Project. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Beck, J.M.; Huffnagle, G.B.; Curtis, J.L. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann. Am. Thorac. Soc. 2015, 12, 821–830. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. Towards an ecology of the lung: New conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir. Med. 2014, 2, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, A.; Bassis, C.M.; Beck, J.M.; Young, V.B.; Curtis, J.L.; Huffnagle, G.B.; Schmidt, T.M. Application of a neutral community model to assess structuring of the human lung microbiome. mBio 2015, 6, e02284-14. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.; Dunne, E.M.; Wescombe, P.A.; Hale, J.D.F.; Mulholland, E.K.; Tagg, J.R.; Robins-Browne, R.M.; Satzke, S. Investigation of Streptococcus salivarius- mediated inhibition of pneumococcal adherence to pharyngeal epithelial cells. BMC Microbiol. 2016, 16, 225. [Google Scholar] [CrossRef] [PubMed]
- Ishijima, S.A.; Hayama, K.; Burton, J.P.; Reid, G.; Okada, M.; Matsushita, Y.; Abe, S. Effect of Streptococcus salivarius K12 on the in vitro growth of Candida albicans and its protective effect in an oral candidiasis model. Appl. Environ. Microbiol. 2012, 78, 2190–2199. [Google Scholar] [CrossRef]
- Shekhar, S.; Khan, R.; Schenck, K.; Petersen, F.C. Intranasal Immunization with the Commensal Streptococcus mitis Confers Protective Immunity against Pneumococcal Lung Infection. Appl. Environ. Microbiol. 2019, 85, e02235-18. [Google Scholar] [CrossRef]
- Mathieu, E.; MacPherson, C.W.; Belvis, J.; Mathieu, O.; Robert, V.; Saint-Criq, V.; Langella, P.; Tompkins, T.A.; Thomas, M. Oral Primo-Colonizing Bacteria Modulate Inflammation and Gene Expression in Bronchial Epithelial Cells. Microorganisms 2020, 8, 1094. [Google Scholar] [CrossRef]
- Siggins, M.K.; Gill, S.; Langford, P.R.; Li, Y.; Ladhani, S.N.; Tregoning, J.S. PHiD-CV induces anti-Protein D antibodies but does not augment pulmonary clearance of nontypeable Haemophilus influenzae in mice. Vaccine 2015, 33, 38. [Google Scholar] [CrossRef][Green Version]
- Sencio, V.; Barthelemy, A.; Tavares, L.P.; Machado, M.G.; Soulard, D.; Cuinat, C.; Queiroz-Junior, C.M.; Noordine, M.L.; Salome-Desnoulez, S.; Deryuter, L. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 2020, 30, 2934–2947.e2936. [Google Scholar] [CrossRef]
- O’Toole, P.; Marchesi, J.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Denapaite, D.; Brückner, R.; Nuhn, M.; Reichmann, P.; Henrich, B.; Maurer, P.; Schähle, Y.; Selbmann, P.; Zimmermann, W.; Wambutt, R.; et al. The genome of Streptococcus mitis B6--what is a commensal? PLoS ONE 2010, 5, e9426. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J. Streptococcus mitis: Walking the line between commensalism and pathogenesis. Mol. Oral Microbiol. 2011, 26, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, N.; Wrzosek, L.; Radziwill-Bienkowska, J.M.; Ringot-Destrez, B.; Duviau, M.P.; Noordine, M.L.; Laroute, V.; Robert, V.; Cherbuy, C.; Daveran-Mingot, M.L.; et al. Characterization of Mucus-Related Properties of Streptococcus thermophilus: From Adhesion to Induction. Front. Physiol. 2018, 9, 980. [Google Scholar] [CrossRef] [PubMed]
- Turpin, W.; Humblot, C.; Noordine, M.L.; Thomas, M.; Guyot, J.P. Lactobacillaceae and cell adhesion: Genomic and functional screening. PLoS ONE 2012, 7, e38034. [Google Scholar] [CrossRef]
- Santagati, M.; Scillato, M.; Patanè, F.; Aiello, C.; Stefani, S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol. Med. Microbiol. 2012, 65, 23–31. [Google Scholar] [CrossRef]
- Ikryannikova, L.N.; Malakhova, M.V.; Lominadze, G.G.; Karpova, I.Y.; Kostryukova, E.S.; Mayansky, N.A.; Kruglov, A.N.; Klimova, E.A.; Lisitsina, E.S.; Ilina, E.N.; et al. Inhibitory effect of streptococci on the growth of M. catarrhalis strains and the diversity of putative bacteriocin-like gene loci in the genomes of S. pneumoniae and its relatives. AMB Express. 2017, 7, 218. [Google Scholar] [CrossRef]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The influence of the microbiome on respiratory health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef]
- Sencio, V.; Gallerand, A.; Gomes Machado, M.; Deruyter, L.; Heumel, S.; Soulard, D.; Barthelemy, J.; Cuinat, C.; Vieira, A.T.; Barthelemy, A.; et al. Influenza Virus Infection Impairs the Gut’s Barrier Properties and Favors Secondary Enteric Bacterial Infection through Reduced Production of Short-Chain Fatty Acids. Infect. Immun. 2021, 89, e0073420. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, S.; Sencio, V.; Raise, A.; Huillet, E.; Layec, S.; Deruyter, L.; Heumel, S.; Auger, S.; Robert, V.; Langella, P.; et al. Description of a Newly Isolated Blautia faecis Strain and Its Benefit in Mouse Models of Post-Influenza Secondary Enteric and Pulmonary Infections. Nutrients 2022, 14, 1478. [Google Scholar] [CrossRef]
- Ahearn, C.P.; Gallo, M.C.; Murphy, T.F. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog. Dis. 2017, 75, ftx042. [Google Scholar] [CrossRef]
- Moghaddam, S.J.; Clement, C.G.; De la Garza, M.M.; Zou, X.; Travis, E.L.; Young, H.W.; Evans, C.M.; Tuvim, M.J.; Dickey, B.F. Haemophilus influenzae lysate induces aspects of the chronic obstructive pulmonary disease phenotype. Am. J. Respir. Cell Mol. Biol. 2008, 38, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Schyns, J.; Bureau, F.; Marichal, T. Lung Interstitial Macrophages: Past, Present, and Future. J. Immunol. Res. 2018, 2018, 5160794. [Google Scholar] [CrossRef] [PubMed]
- Collin, M.; Bigley, V. Human dendritic cell subsets: An update. Immunology 2018, 154, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.O.; Zhang, W.; Du, J.Y.; Yu, Q. BDCA1-positive dendritic cells (DCs) represent a unique human myeloid DC subset that induces innate and adaptive immune responses to Staphylococcus aureus Infection. Infect. Immun. 2014, 82, 4466–4476. [Google Scholar] [CrossRef]



| Analysis | S. mitis EM-371 |
|---|---|
| Genome Analysis Presence of genes potentially coding for adhesion sites and receptors (*) | Choline-binding protein: CbpD, lytC, pce Fibronectin-binding protein: pavA Plasminogen-binding protein: GAPDH Laminin-binding protein: lmb Other surface proteins: NanA, Eno, ZmpB, PsaA |
| Genome Analysis Presence of genes potentially associated to bacteriocins production (*) | blpT, blpZ, comE, blpR, pncP, tsaD, blpH, comA, cibA, pncG, comB, srtA, blpL and comD |
| Modulation of Cytokines/Chemokines— in comparison to untreated BEAS-2B cells (**) | Pro-inflammatory mediators not modulated: TNFα, IL-6 and IL-8 Mediators up-regulated: CCL25, Basic FGF, MIF |
| Modulation of Short-chain fatty acid (SCFA)— in culture supernatant (***) | Acetate (4 mM) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathieu, E.; Marquant, Q.; Chain, F.; Bouguyon, E.; Saint-Criq, V.; Le-Goffic, R.; Descamps, D.; Langella, P.; Tompkins, T.A.; Binda, S.; et al. An Isolate of Streptococcus mitis Displayed In Vitro Antimicrobial Activity and Deleterious Effect in a Preclinical Model of Lung Infection. Nutrients 2023, 15, 263. https://doi.org/10.3390/nu15020263
Mathieu E, Marquant Q, Chain F, Bouguyon E, Saint-Criq V, Le-Goffic R, Descamps D, Langella P, Tompkins TA, Binda S, et al. An Isolate of Streptococcus mitis Displayed In Vitro Antimicrobial Activity and Deleterious Effect in a Preclinical Model of Lung Infection. Nutrients. 2023; 15(2):263. https://doi.org/10.3390/nu15020263
Chicago/Turabian StyleMathieu, Elliot, Quentin Marquant, Florian Chain, Edwige Bouguyon, Vinciane Saint-Criq, Ronan Le-Goffic, Delphyne Descamps, Philippe Langella, Thomas A. Tompkins, Sylvie Binda, and et al. 2023. "An Isolate of Streptococcus mitis Displayed In Vitro Antimicrobial Activity and Deleterious Effect in a Preclinical Model of Lung Infection" Nutrients 15, no. 2: 263. https://doi.org/10.3390/nu15020263
APA StyleMathieu, E., Marquant, Q., Chain, F., Bouguyon, E., Saint-Criq, V., Le-Goffic, R., Descamps, D., Langella, P., Tompkins, T. A., Binda, S., & Thomas, M. (2023). An Isolate of Streptococcus mitis Displayed In Vitro Antimicrobial Activity and Deleterious Effect in a Preclinical Model of Lung Infection. Nutrients, 15(2), 263. https://doi.org/10.3390/nu15020263






