Predicting Cardiovascular Disease Mortality: Leveraging Machine Learning for Comprehensive Assessment of Health and Nutrition Variables
Abstract
:1. Introduction
1.1. Background
1.2. Overview
2. Materials and Methods
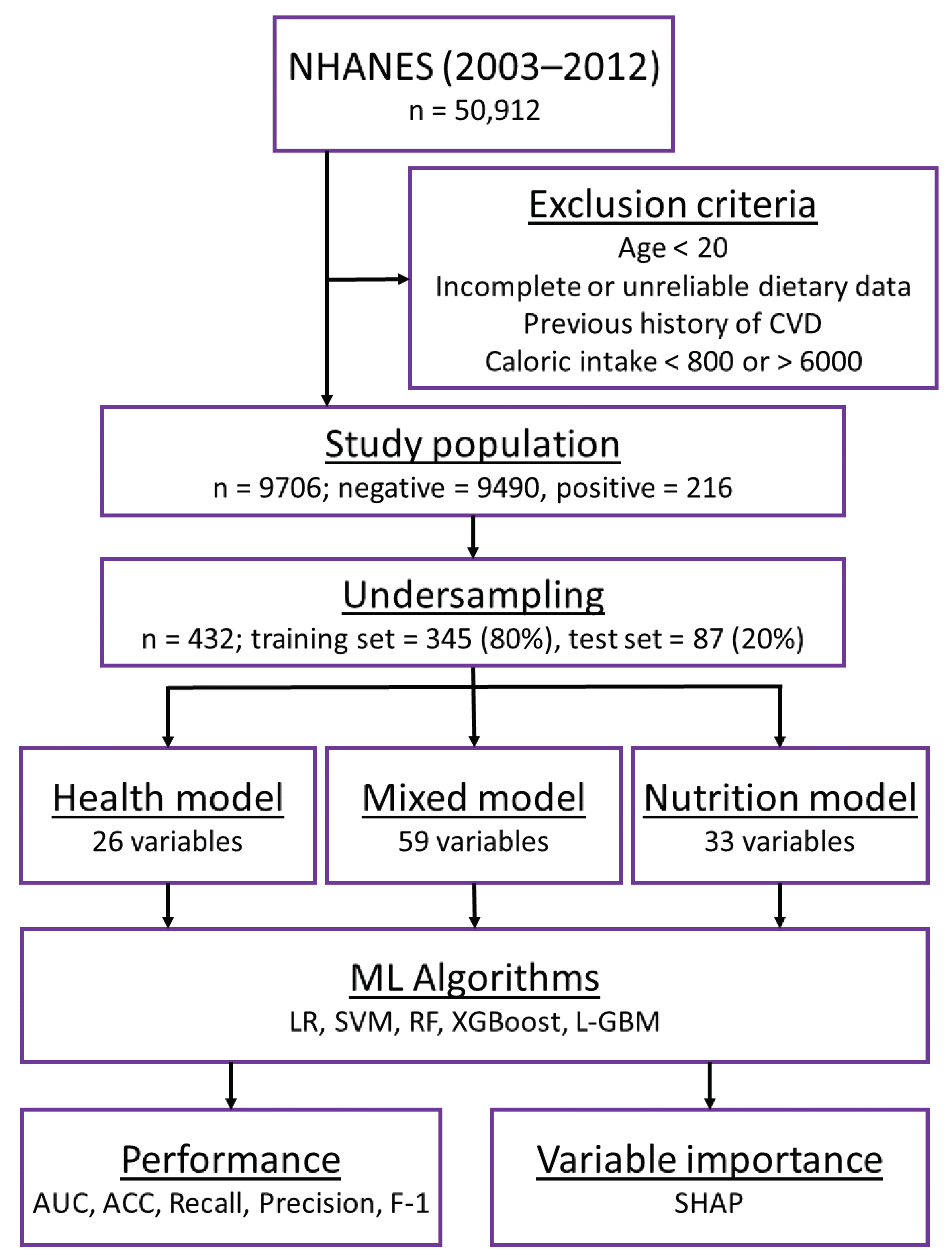
2.1. Study Design
2.2. Primary Outcome
2.3. Data Preprocessing and Variable Selection
2.4. Training of Machine Learning Models
2.5. Performance Metrics
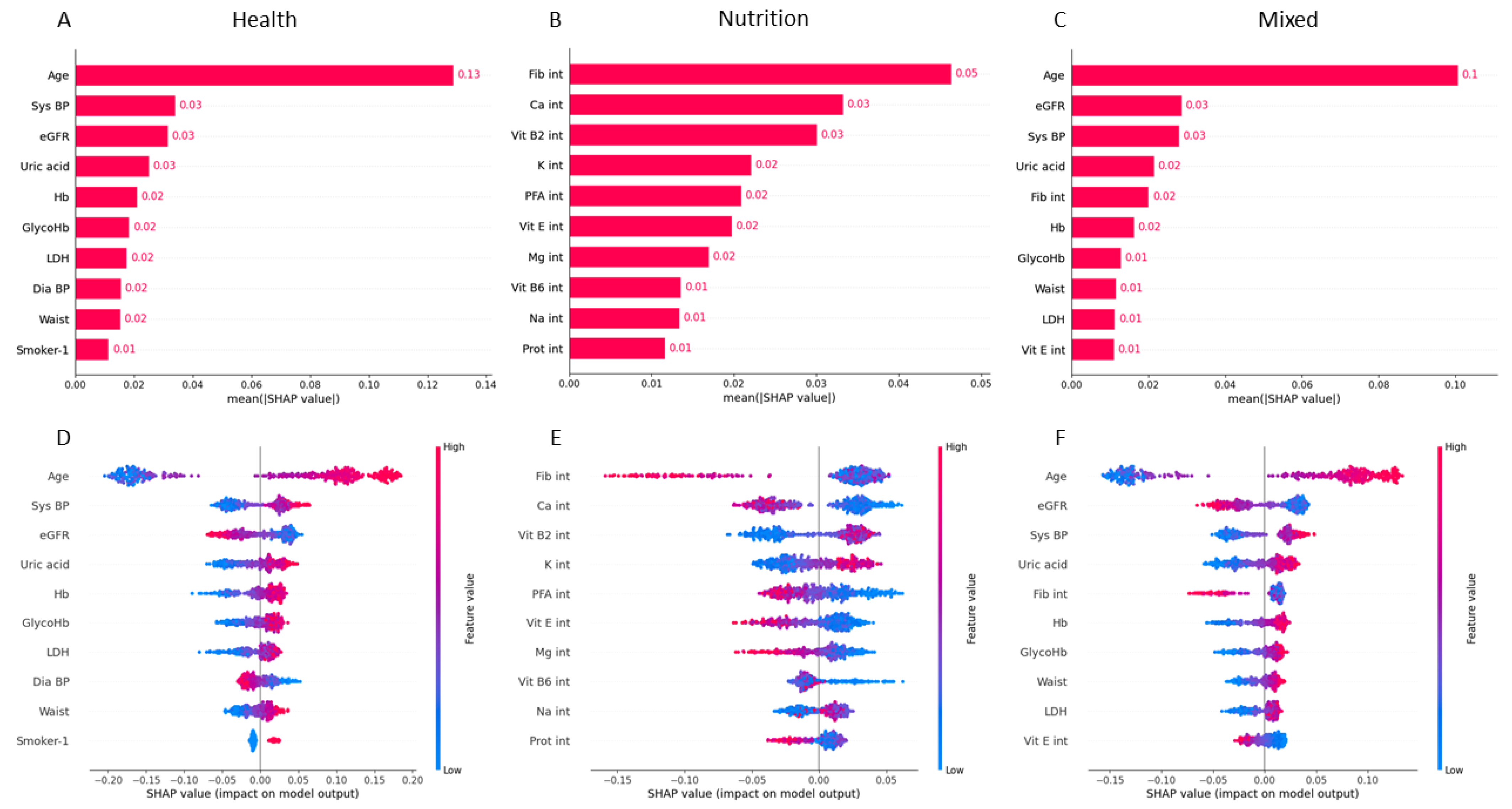
2.6. Variable Importance
3. Results
3.1. Variable Selection and General Characteristics
3.2. Comparison of Model Performance
3.3. Predictive Variable Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CVD | Cardiovascular disease |
| NHANES | National Health and Nutrition Examination Survey |
| ML | Machine learning |
| SHAP | Shapley additive explanation |
| AUC | Area under the curve |
| ACC | Accuracy |
| VIF | Variance inflation factor |
| NCHS | National Center for Health Statistics |
| LR | Logistic regression |
| SVM | Support vector machine |
| RF | Random forest |
| XGBoost | Extreme gradient boosting |
| L-GBM | Light gradient-boosting machine |
| NDI | National Death Index |
| BMI | Body mass index |
| HDL | High-density lipoprotein |
| LDL | Low-density lipoprotein |
| Estimated glomerular filtration rate | |
| TP | True positive |
| FN | False negative |
| TN | True negative |
| FP | False positive |
References
- Timmis, A.; Vardas, P.; Townsend, N.; Torbica, A.; Katus, H.; De Smedt, D.; Gale, C.P.; Maggioni, A.P.; Petersen, S.E.; Huculeci, R.; et al. European Society of Cardiology: Cardiovascular disease statistics 2021. Eur. Heart J. 2022, 43, 716–799. [Google Scholar] [CrossRef] [PubMed]
- Badimon, L.; Chagas, P.; Chiva-Blanch, G. Diet and cardiovascular disease: Effects of foods and nutrients in classical and emerging cardiovascular risk factors. Curr. Med. Chem. 2019, 26, 3639–3651. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Lichtenstein, A.H.; Wu, J.H.; Appel, L.J.; Creager, M.A.; Kris-Etherton, P.M.; Miller, M.; Rimm, E.B.; Rudel, L.L.; Robinson, J.G.; et al. Dietary fats and cardiovascular disease: A presidential advisory from the American Heart Association. Circulation 2017, 136, e1–e23. [Google Scholar] [CrossRef] [PubMed]
- Shivappa, N.; Godos, J.; Hébert, J.R.; Wirth, M.D.; Piuri, G.; Speciani, A.F.; Grosso, G. Dietary inflammatory index and cardiovascular risk and mortality—A meta-analysis. Nutrients 2018, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Salas-Salvadó, J.; Becerra-Tomás, N.; García-Gavilán, J.F.; Bullo, M.; Barrubes, L. Mediterranean diet and cardiovascular disease prevention: What do we know? Prog. Cardiovasc. Dis. 2018, 61, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Kirk, D.; Kok, E.; Tufano, M.; Tekinerdogan, B.; Feskens, E.J.; Camps, G. Machine Learning in Nutrition Research. Adv. Nutr. 2022, 13, 2573–2589. [Google Scholar] [CrossRef]
- Morgenstern, J.D.; Rosella, L.C.; Costa, A.P.; de Souza, R.J.; Anderson, L.N. Perspective: Big data and machine learning could help advance nutritional epidemiology. Adv. Nutr. 2021, 12, 621–631. [Google Scholar] [CrossRef]
- Liu, J.; Chou, E.L.; Lau, K.K.; Woo, P.Y.; Li, J.; Chan, K.H.K. Machine learning algorithms identify demographics, dietary features, and blood biomarkers associated with stroke records. J. Neurol. Sci. 2022, 440, 120335. [Google Scholar] [CrossRef]
- Morgenstern, J.D.; Rosella, L.C.; Costa, A.P.; Anderson, L.N. Development of machine learning prediction models to explore nutrients predictive of cardiovascular disease using Canadian linked population-based data. Appl. Physiol. Nutr. Metab. 2022, 47, 529–546. [Google Scholar] [CrossRef]
- Rigdon, J.; Basu, S. Machine learning with sparse nutrition data to improve cardiovascular mortality risk prediction in the USA using nationally randomly sampled data. BMJ Open 2019, 9, e032703. [Google Scholar] [CrossRef]
- Zhao, Y.; Naumova, E.N.; Bobb, J.F.; Claus Henn, B.; Singh, G.M. Joint associations of multiple dietary components with cardiovascular disease risk: A machine-learning approach. Am. J. Epidemiol. 2021, 190, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4768–4777. [Google Scholar]
- Van Camp, G. Cardiovascular disease prevention. Acta Clin. Belg. 2014, 69, 407–411. [Google Scholar] [CrossRef]
- Ortega, F.B.; Lavie, C.J.; Blair, S.N. Obesity and cardiovascular disease. Circ. Res. 2016, 118, 1752–1770. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Tanaka, A.; Node, K.; Kobayashi, Y. Uric acid and cardiovascular disease: A clinical review. J. Cardiol. 2021, 78, 51–57. [Google Scholar] [CrossRef]
- Martinez, M.; Santamarina, J.; Pavesi, A.; Musso, C.; Umpierrez, G.E. Glycemic variability and cardiovascular disease in patients with type 2 diabetes. BMJ Open Diabetes Res. Care 2021, 9, e002032. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, M.; Coppolino, G.; Faga, T.; Garofalo, C.; Serra, R.; Andreucci, M. Epidemiology of cardiovascular risk in chronic kidney disease patients: The real silent killer. Rev. Cardiovasc. Med. 2019, 20, 209–220. [Google Scholar] [PubMed]
- Skeaff, C.M.; Miller, J. Dietary Fat and Coronary Heart Disease. Ann. Nutr. Metab. 2009, 55, 173–201. [Google Scholar] [CrossRef]
- Delgado-Lista, J.; Perez-Martinez, P.; Lopez-Miranda, J.; Perez-Jimenez, F. Long chain omega-3 fatty acids and cardiovascular disease: A systematic review. Br. J. Nutr. 2012, 107, S201–S213. [Google Scholar] [CrossRef]
- Mazidi, M.; Rezaie, P.; Banach, M. Effect of magnesium supplements on serum C-reactive protein: A systematic review and meta-analysis. Arch. Med. Sci. 2018, 14, 707–716. [Google Scholar] [CrossRef]
- Soliman, G.A. Dietary fiber, atherosclerosis, and cardiovascular disease. Nutrients 2019, 11, 1155. [Google Scholar] [CrossRef] [PubMed]
- Casas, R.; Castro-Barquero, S.; Estruch, R.; Sacanella, E. Nutrition and cardiovascular health. Int. J. Mol. Sci. 2018, 19, 3988. [Google Scholar] [CrossRef] [PubMed]
- Kirasich, K.; Smith, T.; Sadler, B. Random forest vs logistic regression: Binary classification for heterogeneous datasets. SMU Data Sci. Rev. 2018, 1, 9. [Google Scholar]
- Dinh, A.; Miertschin, S.; Young, A.; Mohanty, S.D. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med. Inform. Decis. Mak. 2019, 19, 211. [Google Scholar] [CrossRef] [PubMed]
- Panaretos, D.; Koloverou, E.; Dimopoulos, A.C.; Kouli, G.M.; Vamvakari, M.; Tzavelas, G.; Pitsavos, C.; Panagiotakos, D.B. A comparison of statistical and machine-learning techniques in evaluating the association between dietary patterns and 10-year cardiometabolic risk (2002–2012): The ATTICA study. Br. J. Nutr. 2018, 120, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.; Kim, D.; Lee, S.; Won, C.; Kim, S.; Yang, J.S.; Yu, J.; Kim, B.; Lee, J. Machine learning-based diagnosis and risk factor analysis of cardiocerebrovascular disease based on KNHANES. Sci. Rep. 2022, 12, 2250. [Google Scholar] [CrossRef]
- Abdalrada, A.S.; Abawajy, J.; Al-Quraishi, T.; Islam, S.M.S. Machine learning models for prediction of co-occurrence of diabetes and cardiovascular diseases: A retrospective cohort study. J. Diabetes Metab. Disord. 2022, 21, 251–261. [Google Scholar] [CrossRef]
- Pan, A.; Sun, Q.; Bernstein, A.M.; Schulze, M.B.; Manson, J.E.; Stampfer, M.J.; Willett, W.C.; Hu, F.B. Red meat consumption and mortality: Results from 2 prospective cohort studies. Arch. Intern. Med. 2012, 172, 555–563. [Google Scholar]
- Aaron, K.J.; Sanders, P.W. Role of dietary salt and potassium intake in cardiovascular health and disease: A review of the evidence. Mayo Clin. Proc. 2013, 88, 987–995. [Google Scholar] [CrossRef]
- Garg, A.; Lee, J.C.Y. Vitamin E: Where are we now in vascular diseases? Life 2022, 12, 310. [Google Scholar] [CrossRef]
| Variable | Control (n = 9490) | CVD Death (n = 216) | p-Value |
|---|---|---|---|
| Health model variables | |||
| Age, years | 43.0 [31.0, 57.0] | 68.0 [60.0, 75.0] | <0.001 |
| Sex | <0.001 | ||
| Male | 4600 (48.5) | 148 (68.5) | |
| Female | 4890 (51.5) | 68 (31.5) | |
| Health Ins | <0.001 | ||
| Yes | 7525 (79.3) | 199 (92.1) | |
| No | 1965 (20.7) | 17 (7.9) | |
| Education | <0.001 | ||
| Less than 9th grade | 2004 (21.1) | 72 (33.3) | |
| High school grade | 2049 (21.6) | 65 (30.1) | |
| Some college, AA degree | 2817 (29.7) | 40 (18.5) | |
| College grad | 2620 (27.6) | 39 (18.1) | |
| Diabetic | <0.001 | ||
| No | 8646 (91.1) | 161 (74.5) | |
| Yes | 844 (8.9) | 55 (25.5) | |
| Smoker | <0.001 | ||
| Smoking | 1782 (18.8) | 60 (27.8) | |
| Quit | 2207 (23.3) | 77 (35.6) | |
| Never | 5501 (58.0) | 79 (36.6) | |
| Waist, cm | 97.8 (15.3) | 104.8 (14.9) | <0.001 |
| Sys BP, mmHg | 119.0 [110.0, 128.3] | 131.0 [122.0, 144.0] | <0.001 |
| GlycoHb, % | 5.4 [5.2, 5.7] | 5.7 [5.5, 6.1] | <0.001 |
| K, mmol/L | 3.9 [3.8, 4.1] | 4.0 [3.9, 4.2] | <0.001 |
| LDH, U/L | 127.3 (21.3) | 138.5 (20.5) | <0.001 |
| Na, mmol/L | 139.0 [138.0, 140.0] | 139.7 [139.0, 140.5] | <0.001 |
| Uric acid, mg/dL | 5.3 [4.4, 6.1] | 5.8 [5.3, 6.7] | <0.001 |
| , mL/min/1.73 m2 | 103.3 (26.4) | 81.4 (21.9) | <0.001 |
| Nutrition model variables | |||
| Ca int, mg | 882.8 [630.5, 1214.4] | 726.8 [516.9, 959.0] | <0.001 |
| Cu int, mg | 1.2 [1.0, 1.6] | 1.1 [0.9, 1.4] | <0.001 |
| Fib int, g | 16.2 [11.7, 22.4] | 13.9 [10.7, 18.0] | <0.001 |
| Fol int, µg | 389.0 [289.5, 528.0] | 344.2 [270.2, 447.2] | <0.001 |
| Mg int, mg | 289.0 [225.0, 370.5] | 260.0 [200.0, 312.6] | <0.001 |
| Nia int, mg | 24.1 [18.3, 31.5] | 21.2 [16.5, 27.0] | <0.001 |
| PFA int, g | 16.3 [11.6, 22.4] | 14.0 [9.7, 18.4] | <0.001 |
| Prot int, g | 80.9 [62.8, 102.9] | 73.4 [58.8, 86.6] | <0.001 |
| Na int, mg | 3335.5 [2548.5, 4307.4] | 2981.0 [2393.9, 3766.9] | <0.001 |
| Sug int, g | 107.8 [74.6, 150.5] | 87.9 [61.3, 130.9] | <0.001 |
| Vit B1 int, mg | 1.6 [1.2, 2.1] | 1.4 [1.1, 1.9] | <0.001 |
| Vit B6 int, mg | 1.9 [1.4, 2.6] | 1.7 [1.2, 2.3] | <0.001 |
| Vit E int, µg | 6.9 [4.9, 9.9] | 5.6 [4.2, 7.8] | <0.001 |
| Model | Algorithms | ACC | AUC | Recall | Precision | F1-Score |
|---|---|---|---|---|---|---|
| Health | Logistic Regression | 0.75 | 0.86 | 0.75 | 0.75 | 0.75 |
| Random Forest | 0.8 | 0.87 | 0.8 | 0.8 | 0.8 | |
| ThunderSVM | 0.79 | 0.88 | 0.79 | 0.79 | 0.79 | |
| XGBoost | 0.78 | 0.86 | 0.78 | 0.78 | 0.78 | |
| LightGBM | 0.76 | 0.86 | 0.76 | 0.76 | 0.76 | |
| Nutrition | Logistic Regression | 0.68 | 0.7 | 0.68 | 0.68 | 0.68 |
| Random Forest | 0.7 | 0.7 | 0.7 | 0.7 | 0.7 | |
| ThunderSVM | 0.63 | 0.67 | 0.63 | 0.64 | 0.63 | |
| XGBoost | 0.66 | 0.69 | 0.66 | 0.66 | 0.66 | |
| LightGBM | 0.66 | 0.7 | 0.66 | 0.66 | 0.66 | |
| Mixed | Logistic Regression | 0.76 | 0.86 | 0.76 | 0.76 | 0.76 |
| Random Forest | 0.82 | 0.88 | 0.82 | 0.82 | 0.82 | |
| ThunderSVM | 0.77 | 0.87 | 0.77 | 0.77 | 0.77 | |
| XGBoost | 0.8 | 0.87 | 0.8 | 0.8 | 0.8 | |
| LightGBM | 0.79 | 0.87 | 0.79 | 0.79 | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin-Morales, A.; Yamamoto, M.; Inoue, M.; Vu, T.; Dawadi, R.; Araki, M. Predicting Cardiovascular Disease Mortality: Leveraging Machine Learning for Comprehensive Assessment of Health and Nutrition Variables. Nutrients 2023, 15, 3937. https://doi.org/10.3390/nu15183937
Martin-Morales A, Yamamoto M, Inoue M, Vu T, Dawadi R, Araki M. Predicting Cardiovascular Disease Mortality: Leveraging Machine Learning for Comprehensive Assessment of Health and Nutrition Variables. Nutrients. 2023; 15(18):3937. https://doi.org/10.3390/nu15183937
Chicago/Turabian StyleMartin-Morales, Agustin, Masaki Yamamoto, Mai Inoue, Thien Vu, Research Dawadi, and Michihiro Araki. 2023. "Predicting Cardiovascular Disease Mortality: Leveraging Machine Learning for Comprehensive Assessment of Health and Nutrition Variables" Nutrients 15, no. 18: 3937. https://doi.org/10.3390/nu15183937
APA StyleMartin-Morales, A., Yamamoto, M., Inoue, M., Vu, T., Dawadi, R., & Araki, M. (2023). Predicting Cardiovascular Disease Mortality: Leveraging Machine Learning for Comprehensive Assessment of Health and Nutrition Variables. Nutrients, 15(18), 3937. https://doi.org/10.3390/nu15183937






