An Association between Decreased Small Intestinal RNA Modification and Disturbed Glucagon-like Peptide-1 Secretion under High-Fat Diet Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Studies and Procedures
2.2. RT-qPCR Analysis
2.3. Immunohistochemistry
2.4. LC-MS Analysis for m5C and m6A Level
2.5. ELISA for SAM Content Quantification
2.6. Statistical Analysis
3. Results
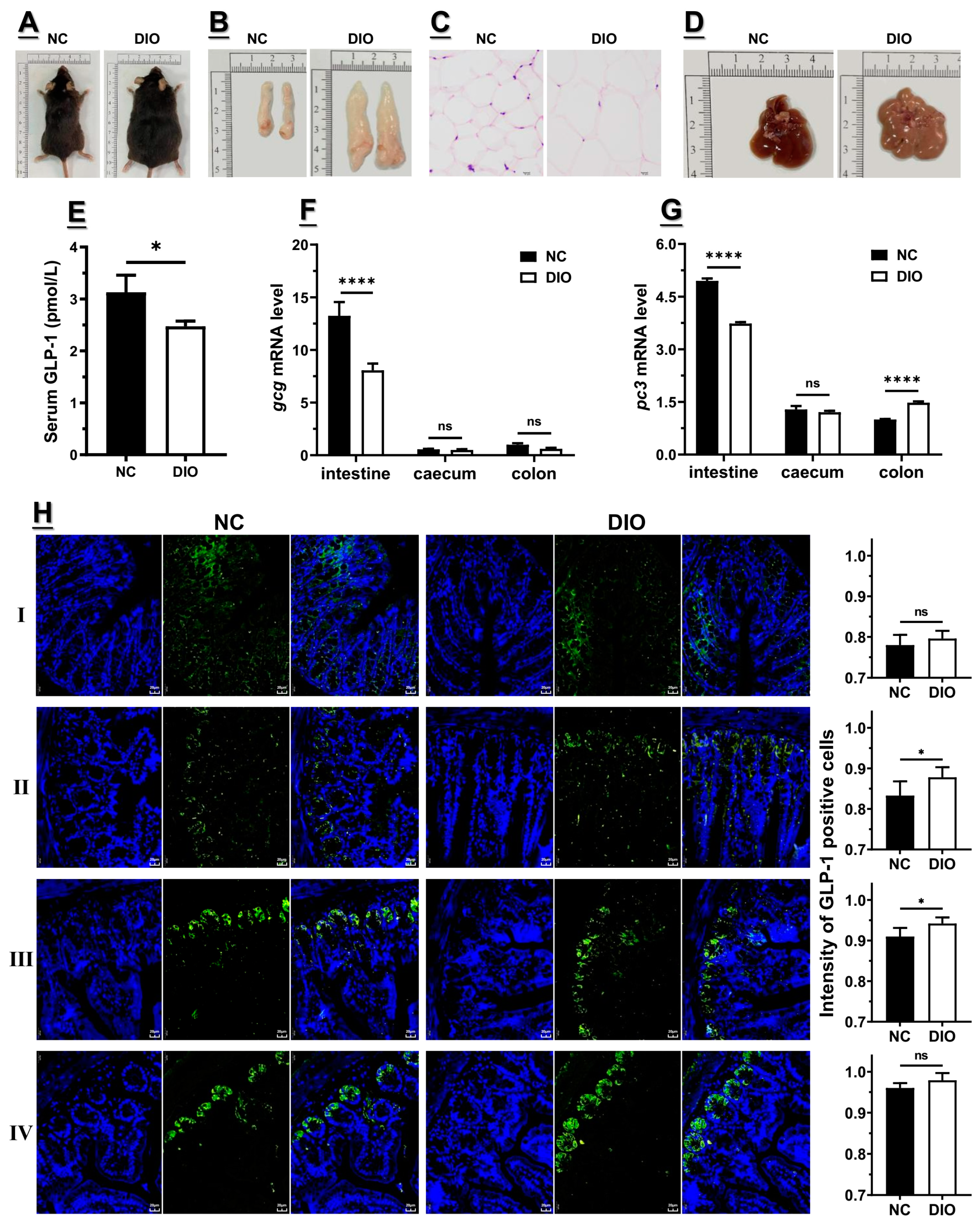
3.1. Decreased Expression of Gcg and Pc3 Is Consistent with the Lower GLP1 Level under High-Fat Diet
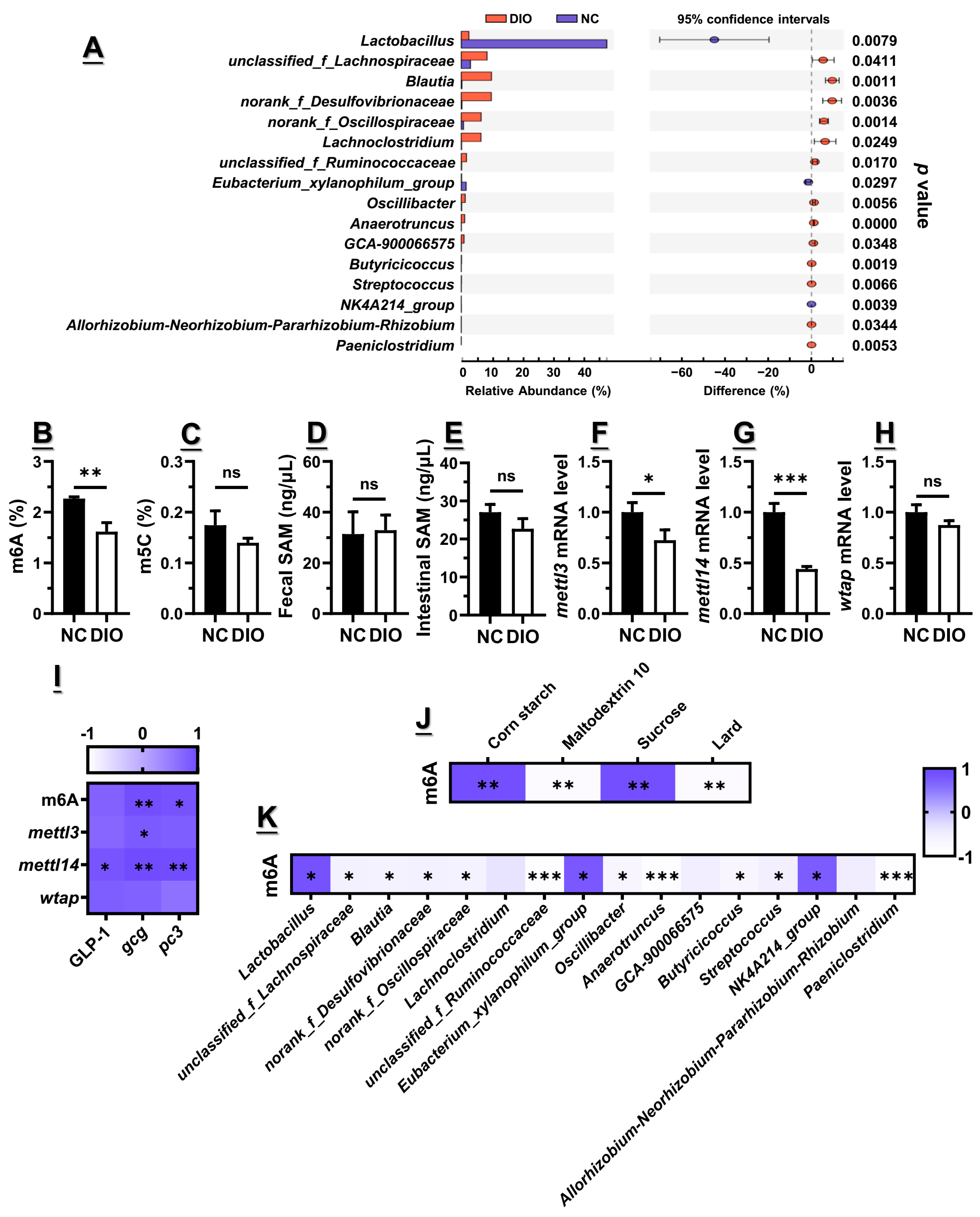
3.2. Decreased m6A Level Might Contribute to the Lower Gcg and Pc3 Expression Levels
3.3. Microbiota Dysbiosis Accompanied with Decreased GLP1, as Well as Gcg and Pc3 Expression Levels
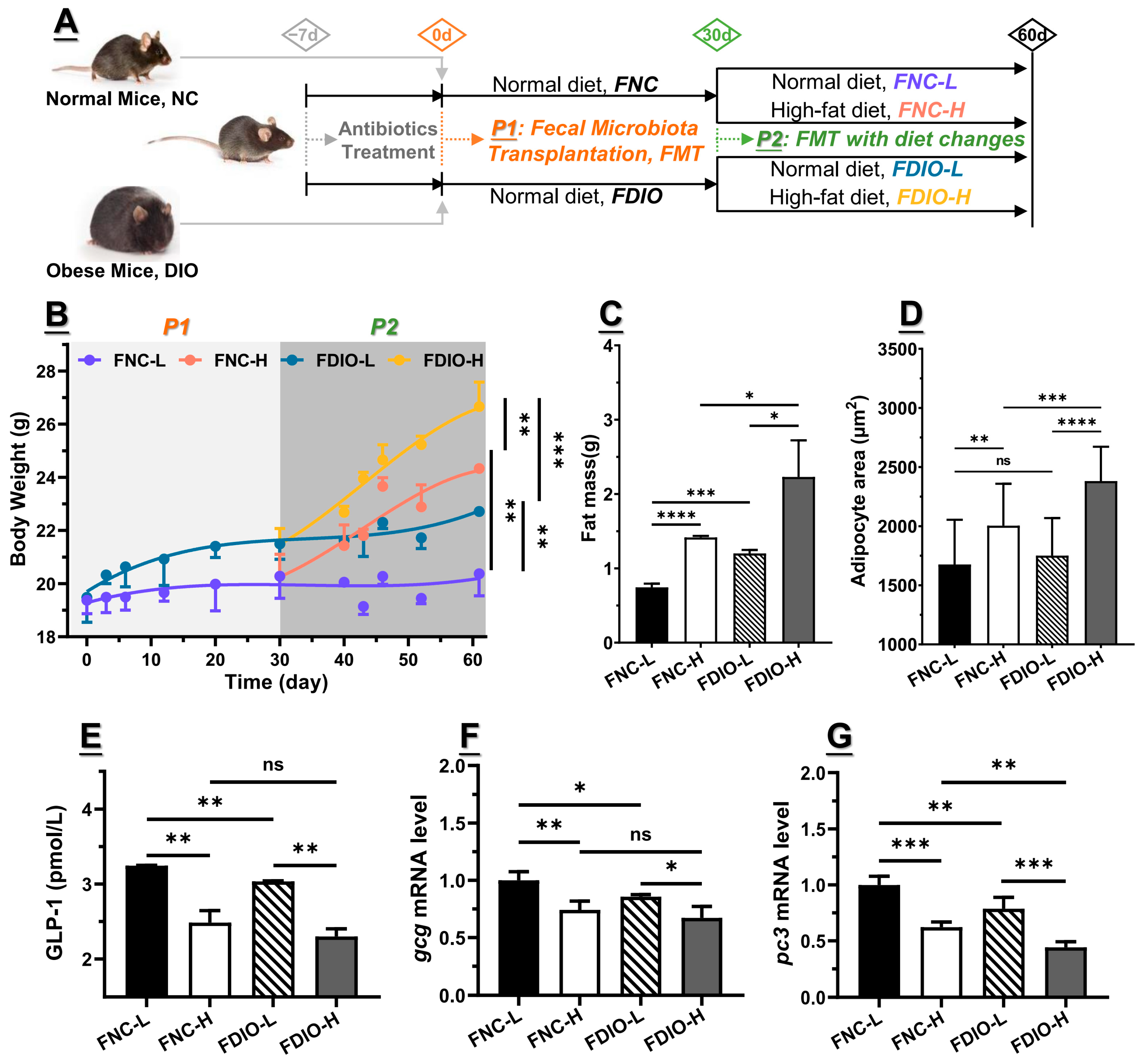
3.4. Feces from DIO Mice Decreased Obesity-Associated GLP1 Level
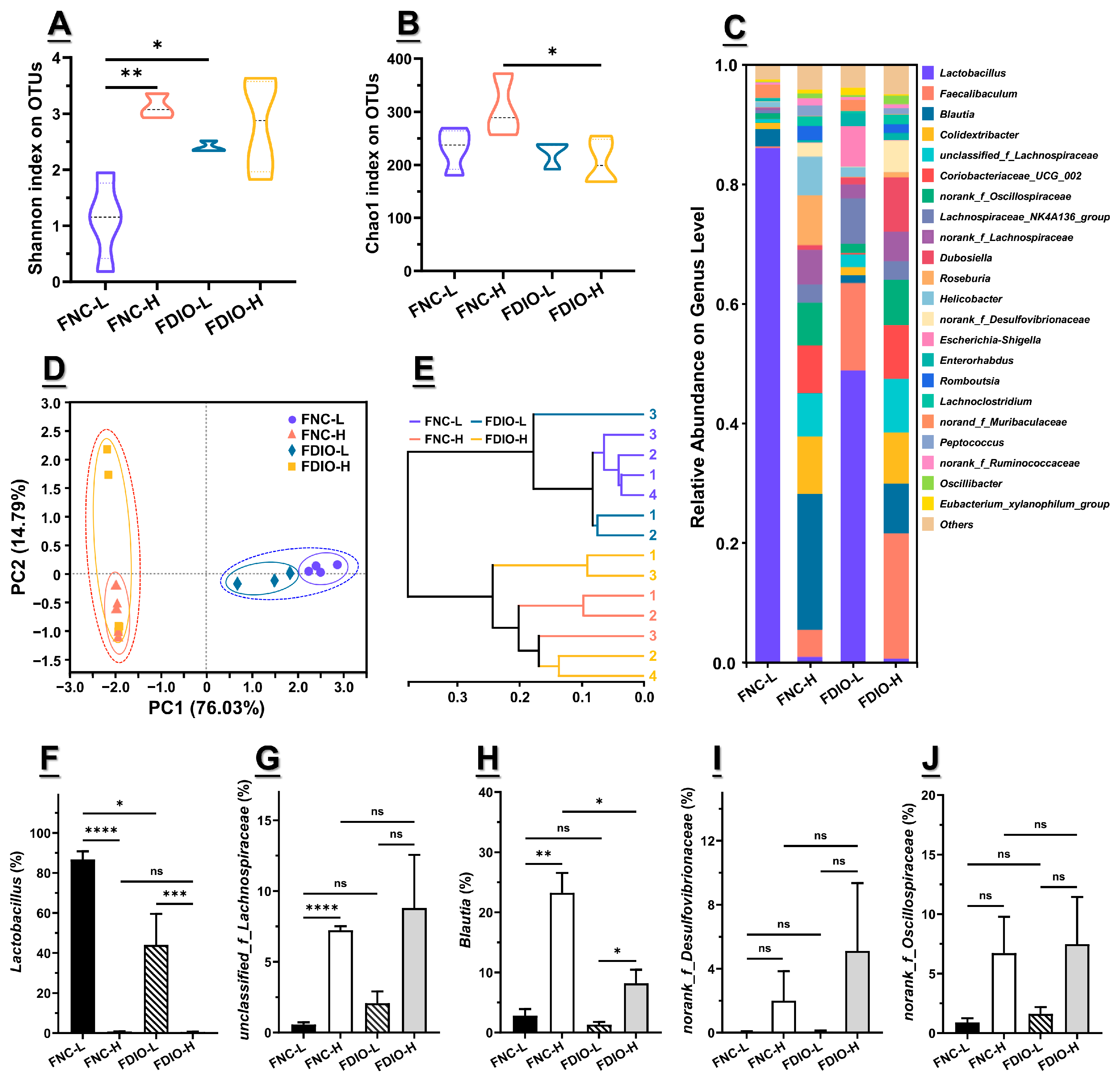
3.5. High-Fat Diet Aggravated Microbial Dysbiosis of Recipient Mice
3.6. High-Fat Diet Exacerbated the Decreased m6A Level and Epitranscriptome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; Naghavi, M.; et al. Health effects of overweight and obesity in 195 countries over 25 years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef]
- Tremmel, M.; Gerdtham, U.G.; Nilsson, P.M.; Saha, S. Economic burden of obesity: A systematic literature review. Int. J. Environ. Res. Public Health 2017, 14, 435. [Google Scholar] [CrossRef]
- Mizuno, T.M. Fat mass and obesity associated (fto) gene and hepatic glucose and lipid metabolism. Nutrients 2018, 10, 1600. [Google Scholar] [CrossRef]
- Sarma, S.; Sockalingam, S.; Dash, S. Obesity as a multisystem disease: Trends in obesity rates and obesity-related complications. Diabetes Obes. Metab. 2021, 23, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Dicken, S.J.; Batterham, R.L. The role of diet quality in mediating the association between ultra-processed food intake, obesity and health-related outcomes: A review of prospective cohort studies. Nutrients 2022, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Monnier, L.; Schlienger, J.L.; Colette, C.; Bonnet, F. The obesity treatment dilemma: Why dieting is both the answer and the problem? A mechanistic overview. Diabetes Metab. 2021, 47, 101192. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Pedro, J.M.B.S. Targeting autophagy to counteract obesity-associated oxidative stress. Antioxidants 2021, 10, 102. [Google Scholar] [CrossRef]
- Zhou, Y.W.; Li, H.G.; Xia, N. The interplay between adipose tissue and vasculature: Role of oxidative stress in obesity. Front. Cardiovasc. Med. 2021, 8, 650214. [Google Scholar] [CrossRef]
- Lobstein, T.; Brownell, K.D. Endocrine-disrupting chemicals and obesity risk: A review of recommendations for obesity prevention policies. Obes. Rev. 2021, 22, e13332. [Google Scholar] [CrossRef]
- Zhou, M.; Johnston, L.J.; Wu, C.D.; Ma, X. Gut microbiota and its metabolites: Bridge of dietary nutrients and obesity-related diseases. Crit. Rev. Food Sci. Nutr. 2021, 63, 3236–3253. [Google Scholar] [CrossRef]
- Li, L.L.; Zhang, Y.B.; Speakman, J.R.; Hu, S.L.; Song, Y.P.; Qin, S. The gut microbiota and its products: Establishing causal relationships with obesity related outcomes. Obes. Rev. 2021, 22, e13341. [Google Scholar] [CrossRef] [PubMed]
- Weng, G.; Duan, Y.; Zhong, Y.; Song, B.; Zheng, J.; Zhang, S.; Yin, Y.; Deng, J. Plant extracts in obesity: A role of gut microbiota. Front. Nutr. 2021, 8, 727951. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.-X.; Zheng, K.-Y.; Wang, Y.-G. Gut microbiota-derived metabolites as key mucosal barrier modulators in obesity. World J. Gastroenterol. 2021, 27, 5555–5565. [Google Scholar] [CrossRef] [PubMed]
- Colangeli, L.; Escobar Marcillo, D.I.; Simonelli, V.; Iorio, E.; Rinaldi, T.; Sbraccia, P.; Fortini, P.; Guglielmi, V. The crosstalk between gut microbiota and white adipose tissue mitochondria in obesity. Nutrients 2023, 15, 1723. [Google Scholar] [CrossRef]
- Theilade, S.; Christensen, M.B.; Vilsboll, T.; Knop, F.K. An overview of obesity mechanisms in humans: Endocrine regulation of food intake, eating behaviour and common determinants of body weight. Diabetes Obes. Metab. 2021, 23, 17–35. [Google Scholar] [CrossRef]
- Lafferty, R.A.; O’Harte, F.P.M.; Irwin, N.; Gault, V.A.; Flatt, P.R. Proglucagon-derived peptides as therapeutics. Front. Endocrinol. 2021, 12, 689678. [Google Scholar] [CrossRef]
- Le, J.; Zhang, X.; Jia, W.; Zhang, Y.; Luo, J.; Sun, Y.; Ye, J. Regulation of microbiota-GLP1 axis by sennoside A in diet-induced obese mice. Acta Pharm. Sin. B 2019, 9, 758–768. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 2019, 15, 226–237. [Google Scholar] [CrossRef]
- Andersen, A.; Lund, A.; Knop, F.K.; Vilsboll, T. Glucagon-like peptide 1 in health and disease. Nat. Rev. Endocrinol. 2018, 14, 390–403. [Google Scholar] [CrossRef]
- Davis, E.M.; Sandoval, D.A. Glucagon-like peptide-1: Actions and influence on pancreatic hormone function. Compr. Physiol. 2020, 10, 577–595. [Google Scholar] [CrossRef]
- Wu, R.F.; Wang, X.X. Epigenetic regulation of adipose tissue expansion and adipogenesis by N(6)-methyladenosine. Obes. Rev. 2020, 22, e13124. [Google Scholar] [CrossRef]
- Gao, W.; Liu, J.L.; Lu, X.; Yang, Q. Epigenetic regulation of energy metabolism in obesity. J. Mol. Cell Biol. 2021, 13, 480–499. [Google Scholar] [CrossRef]
- Sun, M.L.; Zhang, X.A. Epigenetic regulation of N6-methyladenosine modifications in obesity. J. Diabetes Investig. 2021, 12, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Bi, Z.; Liu, Y.H.; Zhao, Y.L.; Yao, Y.X.; Wu, R.F.; Liu, Q.; Wang, Y.Z.; Wang, X.X. A dynamic reversible RNA N-6-methyladenosine modification: Current status and perspectives. J. Cell. Physiol. 2019, 234, 7948–7956. [Google Scholar] [CrossRef]
- Kan, R.L.; Chen, J.J.; Sallam, T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022, 38, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Tatekawa, S.; Ofusa, K.; Chijimatsu, R.; Vecchione, A.; Tamari, K.; Ogawa, K.; Ishii, H. Methylosystem for cancer sieging strategy. Cancers 2021, 13, 5088. [Google Scholar] [CrossRef]
- Peng, L.; Long, T.; Li, F.; Xie, Q. Emerging role of m(6)A modification in cardiovascular diseases. Cell Biol. Int. 2022, 46, 711–722. [Google Scholar] [CrossRef]
- Azzam, S.K.; Alsafar, H.; Sajini, A.A. FTO m6A demethylase in obesity and cancer: Implications and underlying molecular mechanisms. Int. J. Mol. Sci. 2022, 23, 3800. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Jaffrey, S.R. Hidden codes in mRNA: Control of gene expression by m6A. Mol. Cell 2022, 82, 2236–2251. [Google Scholar] [CrossRef]
- He, L.E.; Li, H.Y.; Wu, A.Q.; Peng, Y.L.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef]
- Mentch, S.J.; Locasale, J.W. One-carbon metabolism and epigenetics: Understanding the specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98. [Google Scholar] [CrossRef]
- Bekdash, R.A. Neuroprotective effects of choline and other methyl donors. Nutrients 2019, 11, 2995. [Google Scholar] [CrossRef]
- Jabs, S.; Biton, A.; Becavin, C.; Nahori, M.A.; Ghozlane, A.; Pagliuso, A.; Spano, G.; Guerineau, V.; Touboul, D.; Giai Gianetto, Q.; et al. Impact of the gut microbiota on the m(6)A epitranscriptome of mouse cecum and liver. Nat. Commun. 2020, 11, 1344. [Google Scholar] [CrossRef]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes 2022, 14, 2022407. [Google Scholar] [CrossRef]
- Miro-Blanch, J.; Yanes, O. Epigenetic regulation at the interplay between gut microbiota and host metabolism. Front. Genet. 2019, 10, 638. [Google Scholar] [CrossRef]
- Shock, T.; Badang, L.; Ferguson, B.; Martinez-Guryn, K. The interplay between diet, gut microbes, and host epigenetics in health and disease. J. Nutr. Biochem. 2021, 95, 108631. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate production by probiotic bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Wang, Z.L.; Cai, H.B.; Zhou, C.L. Progress in the microbial production of S-adenosyl-L-methionine. World J. Microb. Biotechnol. 2016, 32, 153. [Google Scholar] [CrossRef] [PubMed]
- Krautkramer, K.A.; Kreznar, J.H.; Romano, K.A.; Vivas, E.I.; Barrett-Wilt, G.A.; Rabaglia, M.E.; Keller, M.P.; Attie, A.D.; Rey, F.E.; Denu, J.M. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 2016, 64, 982–992. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Chen, J.; Yi, K.; Peng, L.; Xie, J.; Gou, X.; Peng, T.; Tang, L. Phlorizin ameliorates obesity-associated endotoxemia and insulin resistance in high-fat diet-fed mice by targeting the gut microbiota and intestinal barrier integrity. Gut Microbes 2020, 12, 1842990. [Google Scholar] [CrossRef]
- Chukwuma, C.I.; Mopuri, R.; Nagiah, S.; Chuturgoon, A.A.; Islam, M.S. Erythritol reduces small intestinal glucose absorption, increases muscle glucose uptake, improves glucose metabolic enzymes activities and increases expression of Glut-4 and IRS-1 in type 2 diabetic rats. Eur. J. Nutr. 2018, 57, 2431–2444. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.S.; Lin, Z.X.; Liu, Y.J.; Yang, S.M.; Zhang, Y.; Yu, X.Y. Ultra performance liquid chromatography-tandem mass spectrometry assay for the quantification of RNA and DNA methylation. J. Pharmaceut. Biomed. 2021, 197, 113969. [Google Scholar] [CrossRef]
- Yuan, B.F. Liquid chromatography-mass spectrometry for analysis of rna adenosine methylation. RNA Methylation Methods Protoc. 2017, 1562, 33–42. [Google Scholar] [CrossRef]
- Aranias, T.; Grosfeld, A.; Poitou, C.; Omar, A.A.; Le Gall, M.; Miquel, S.; Garbin, K.; Ribeiro, A.; Bouillot, J.L.; Bado, A.; et al. Lipid-rich diet enhances L-cell density in obese subjects and in mice through improved L-cell differentiation. J. Nutr. Sci. 2015, 4, e22. [Google Scholar] [CrossRef] [PubMed]
- Aranias, T.; Gilet, J.; Guilmeau, S.; Grosfeld, A.; Le Gall, M.; Brot-Laroche, E.; Leturque, A.; Serradas, P. High fat diet increases enteroendocrine L cell number in the jejunum of mice in conditions of insulin resistance. Diabetologia 2011, 54, S210–S211. [Google Scholar]
- Reimann, F.; Habib, A.M.; Tolhurst, G.; Parker, H.E.; Rogers, G.J.; Gribble, F.M. Glucose sensing in l cells: A primary cell study. Cell Metab. 2008, 8, 532–539. [Google Scholar] [CrossRef]
- Van der Wielen, N.; ten Klooster, J.P.; Muckenschnabl, S.; Pieters, R.; Hendriks, H.F.J.; Witkamp, R.F.; Meijerink, J. The noncaloric sweetener rebaudioside a stimulates glucagon-like peptide 1 release and increases enteroendocrine cell numbers in 2-dimensional mouse organoids derived from different locations of the intestine. J. Nutr. 2016, 146, 2429–2435. [Google Scholar] [CrossRef]
- Jorsal, T.; Rhee, N.A.; Pedersen, J.; Wahlgren, C.D.; Mortensen, B.; Jepsen, S.L.; Jelsing, J.; Dalboge, L.S.; Vilmann, P.; Hassan, H.; et al. Enteroendocrine K and L cells in healthy and type 2 diabetic individuals. Diabetologia 2018, 61, 284–294. [Google Scholar] [CrossRef]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Panaro, B.L.; Yusta, B.; Matthews, D.; Koehler, J.A.; Song, Y.M.; Sandoval, D.A.; Drucker, D.J. Intestine -selective reduction of Gcg expression reveals the importance of the distal gut for GLP-1 secretion. Mol. Metab. 2020, 37, 100990. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.M.; Zhao, Y.; Wang, X.; Kong, L.C.; Johnston, L.J.; Lu, L.; Ma, X. Dietary nutrients shape gut microbes and intestinal mucosa via epigenetic modifications. Crit. Rev. Food Sci. 2022, 62, 783–797. [Google Scholar] [CrossRef]
- Gribble, F.M.; Diakogiannaki, E.; Reimann, F. Gut hormone regulation and secretion via FFA1 and FFA4. Handb. Exp. Pharmacol. 2017, 236, 181–203. [Google Scholar] [CrossRef] [PubMed]
- Boque, N.; de la Iglesia, R.; de la Garza, A.L.; Milagro, F.I.; Olivares, M.; Banuelos, O.; Soria, A.C.; Rodriguez-Sanchez, S.; Martinez, J.A.; Campion, J. Prevention of diet-induced obesity by apple polyphenols in Wistar rats through regulation of adipocyte gene expression and DNA methylation patterns. Mol. Nutr. Food Res. 2013, 57, 1473–1478. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Yang, S.; Lu, J.; Jin, X.; Wu, M. Diet-gut microbiota-epigenetics in metabolic diseases: From mechanisms to therapeutics. Biomed. Pharmacother. 2022, 153, 113290. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Martin, M.A. Impact of diet on gut microbiota. Curr. Opin. Food Sci. 2021, 37, 83–90. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Li, M.; Zhang, Y.; Sun, M.; Wang, L.; Lin, J.; Cui, Y.; Chen, Q.; Jin, C.; et al. Fusobacterium nucleatum reduces METTL3-mediated m(6)A modification and contributes to colorectal cancer metastasis. Nat. Commun. 2022, 13, 1248. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]

| FNC-L | FNC-H | FDIO-L | FDIO-H | |
|---|---|---|---|---|
| FBG (mM) | 4.40 ± 0.40 | 5.57 ± 0.12 ** | 5.3 ± 0.44 | 6.60 ± 0.40 #§ |
| FSI (mIU/L) | 1.28 ± 0.07 | 1.47 ± 0.01 ** | 1.45 ± 0.06 & | 1.67 ± 0.06 ##§§ |
| HOMA-IR | 0.25 ± 0.01 | 0.32 ± 0.02 ** | 0.32 ± 0.02 && | 0.42 ± 0.03 #§§ |
| HOMA-IS | 0.18 ± 0.01 | 0.14 ± 0.01 ** | 0.14 ± 0.01 && | 0.11 ± 0.01 #§ |
| TC (mM) | 2.10 ± 0.14 | 3.13 ± 0.65 * | 2.38 ± 0.11 & | 3.76 ± 0.51 ## |
| TG (mM) | 0.45 ± 0.03 | 0.69 ± 0.03 *** | 0.67 ± 0.08 && | 0.74 ± 0.09 |
| LDL-C (mM) | 0.18 ± 0.02 | 0.24 ± 0.04 | 0.20 ± 0.03 | 0.28 ± 0.06 |
| HDL-C (mM) | 3.18 ± 0.27 | 1.92 ± 0.05 **** | 2.43 ± 0.38 & | 1.65 ± 0.22 # |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Deng, L.-L.; Xiao, X.-L.; Long, S.-Y.; Deng, Y.; Peng, T.; Xie, J.; Zhang, X.-Y. An Association between Decreased Small Intestinal RNA Modification and Disturbed Glucagon-like Peptide-1 Secretion under High-Fat Diet Stress. Nutrients 2023, 15, 3707. https://doi.org/10.3390/nu15173707
Chen J, Deng L-L, Xiao X-L, Long S-Y, Deng Y, Peng T, Xie J, Zhang X-Y. An Association between Decreased Small Intestinal RNA Modification and Disturbed Glucagon-like Peptide-1 Secretion under High-Fat Diet Stress. Nutrients. 2023; 15(17):3707. https://doi.org/10.3390/nu15173707
Chicago/Turabian StyleChen, Jiang, Lin-Ling Deng, Xing-Lin Xiao, Shi-Yuan Long, Yuan Deng, Tong Peng, Jie Xie, and Xiao-Yu Zhang. 2023. "An Association between Decreased Small Intestinal RNA Modification and Disturbed Glucagon-like Peptide-1 Secretion under High-Fat Diet Stress" Nutrients 15, no. 17: 3707. https://doi.org/10.3390/nu15173707
APA StyleChen, J., Deng, L.-L., Xiao, X.-L., Long, S.-Y., Deng, Y., Peng, T., Xie, J., & Zhang, X.-Y. (2023). An Association between Decreased Small Intestinal RNA Modification and Disturbed Glucagon-like Peptide-1 Secretion under High-Fat Diet Stress. Nutrients, 15(17), 3707. https://doi.org/10.3390/nu15173707





