Dietary Flaxseed and Flaxseed Oil Differentially Modulate Aspects of the Microbiota Gut–Brain Axis Following an Acute Lipopolysaccharide Challenge in Male C57Bl/6 Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Flaxseed Nutrient Composition, Oil Extraction, and Oil Fatty Acid Composition
2.2. Experimental Diet Preparation
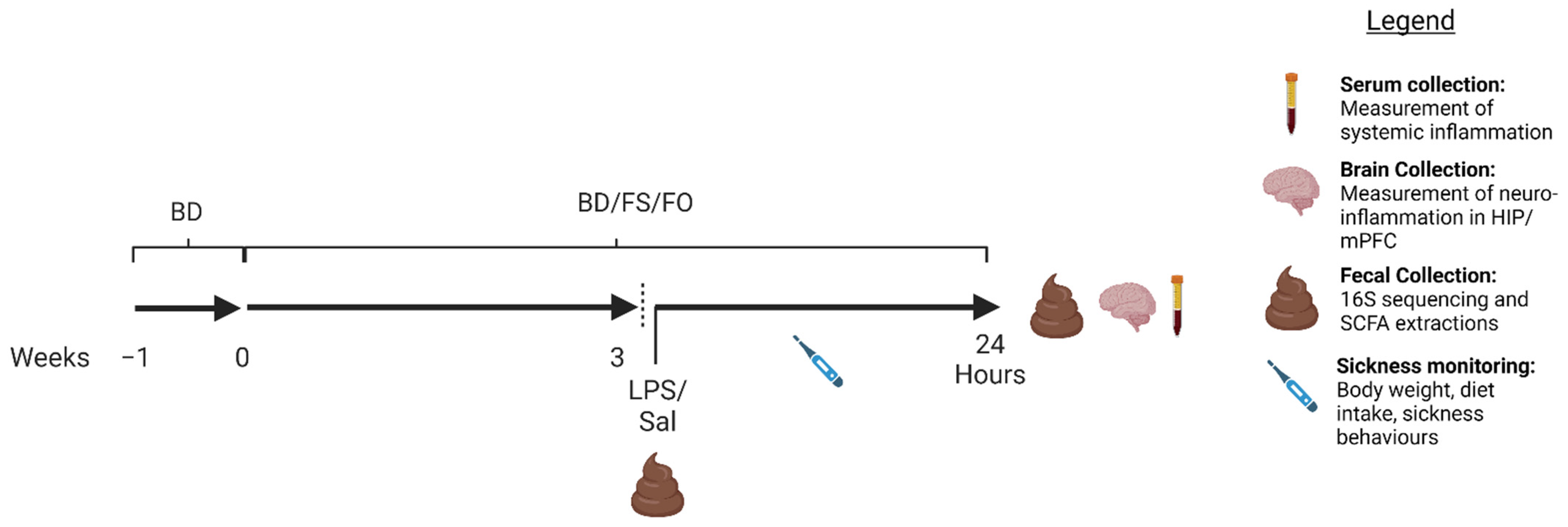
2.3. Animal and Experimental Design
2.4. LPS Preparation
2.5. Well-Being Measures Post-LPS/SAL Injection
2.5.1. Diet Intake (DI) and Body Weight (BW)
2.5.2. Analysis of Sickness Behaviours
2.5.3. Nest Quality Scoring
2.6. Assessment of Systemic Inflammation
2.7. Hippocampus (HIP) and Medial Prefrontal Cortex (mPFC) Isolation and Gene Expression of Inflammatory Cytokines
2.8. Fecal DNA Extraction and 16S rRNA Gene Sequencing
16S rRNA Data Processing and Bioinformatics Analyses
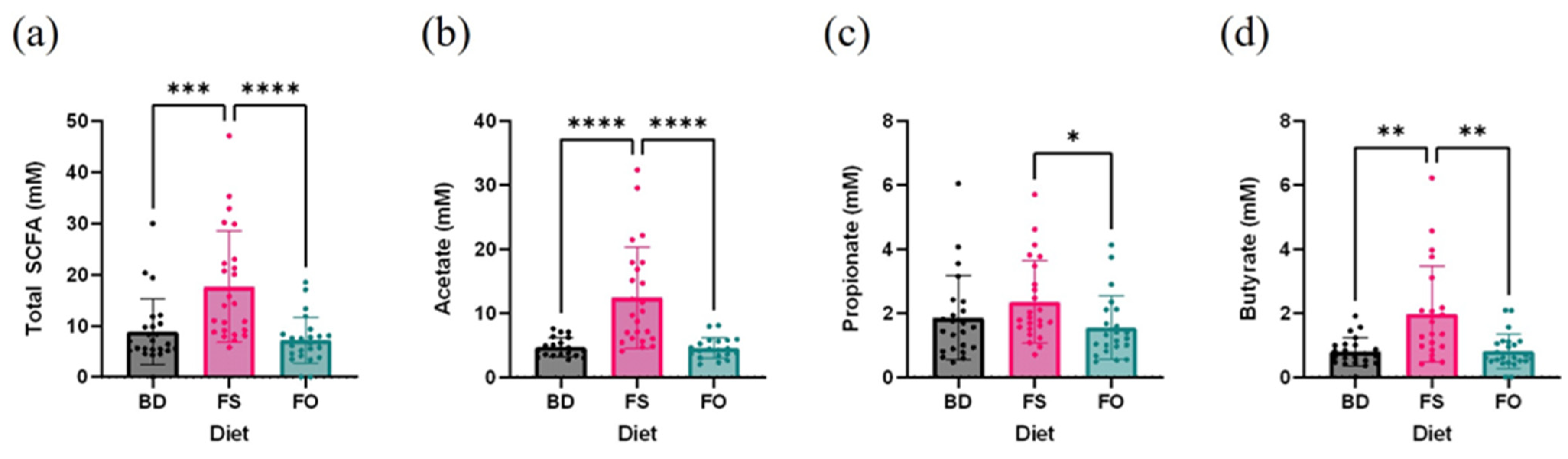
2.9. Fecal Short-Chain Fatty Acid Analyses
2.10. Statistical Analyses
3. Results
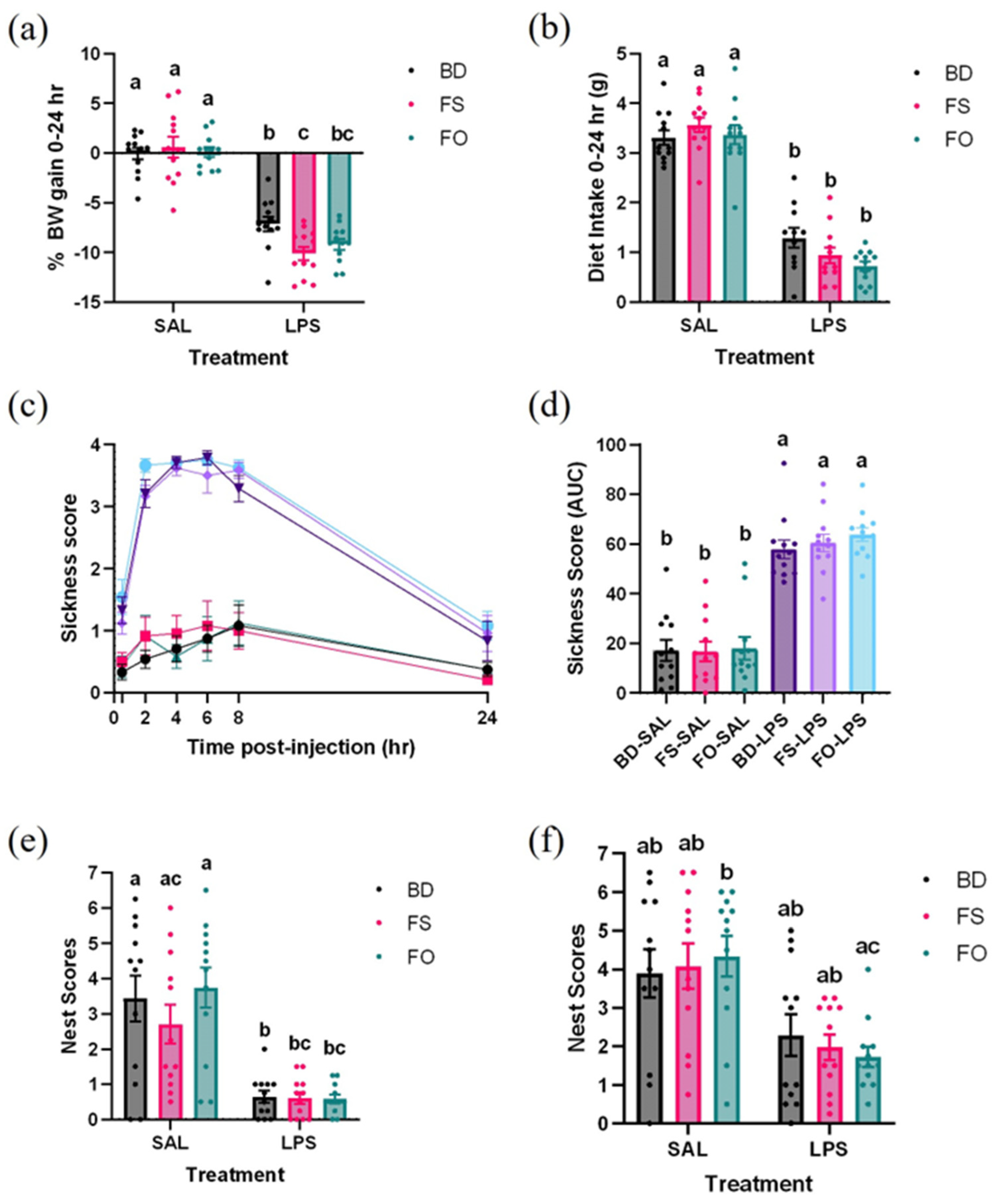
3.1. LPS Induces Body Weight Loss and Appetite Suppression, Which Is Not Attenuated by Flaxseed or Flaxseed-Oil Supplemented Diets
3.2. LPS-Induced Decline in Well-Being Was Not Improved by FS or FO Diet Supplementation
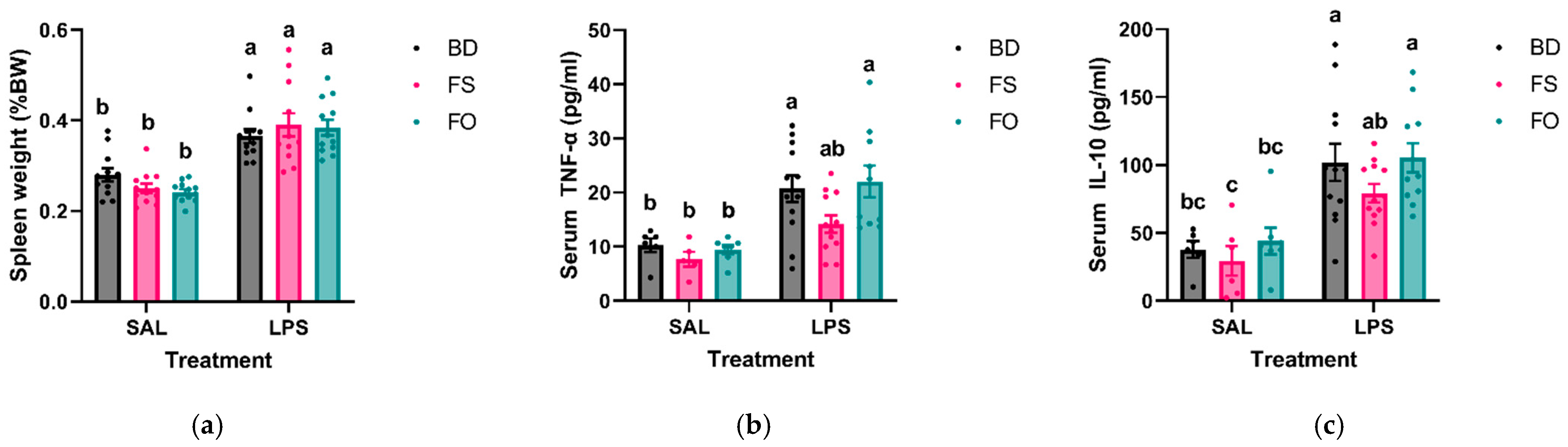
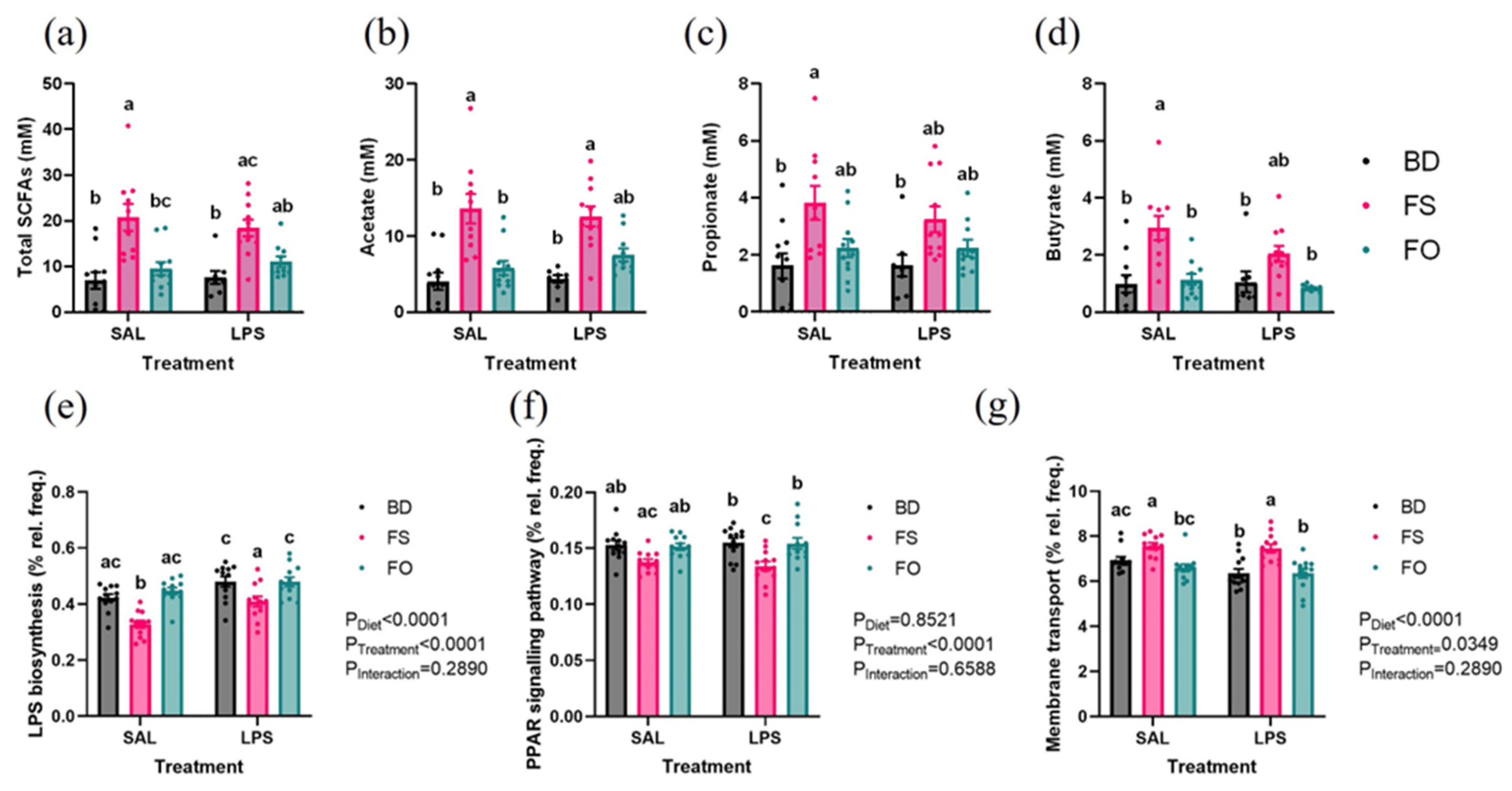
3.3. Systemic Inflammation Induced by LPS Was Positively Impacted by FS but Not FO Diet Supplementation
3.4. LPS-Induced Neuroinflammation Was Partially Attenuated by FS Supplementation
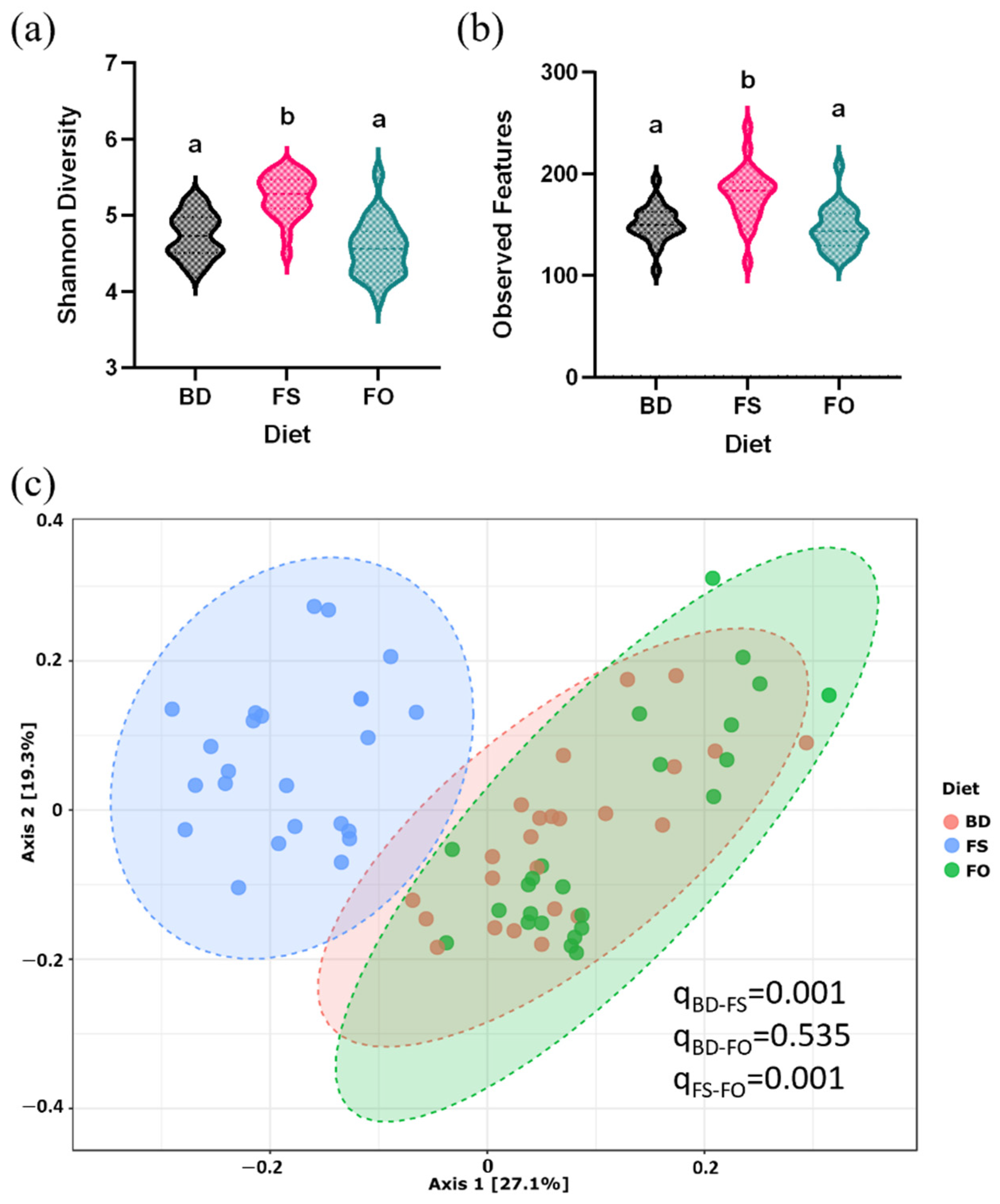
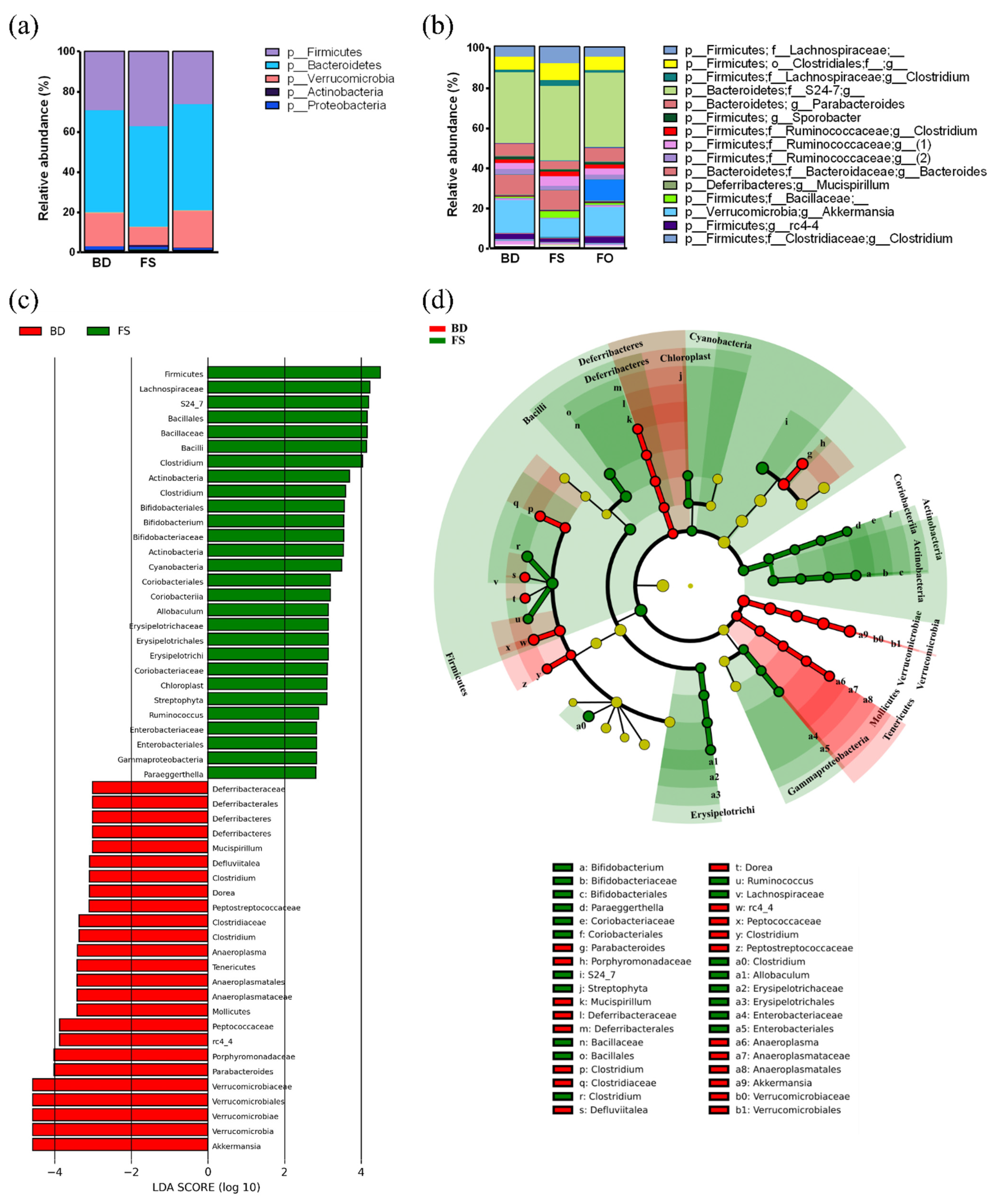
3.5. Dietary Supplementation with FS, but Not FO, Alters the Baseline Fecal Microbiota Profile and Activity, Prior to LPS/SAL Administration
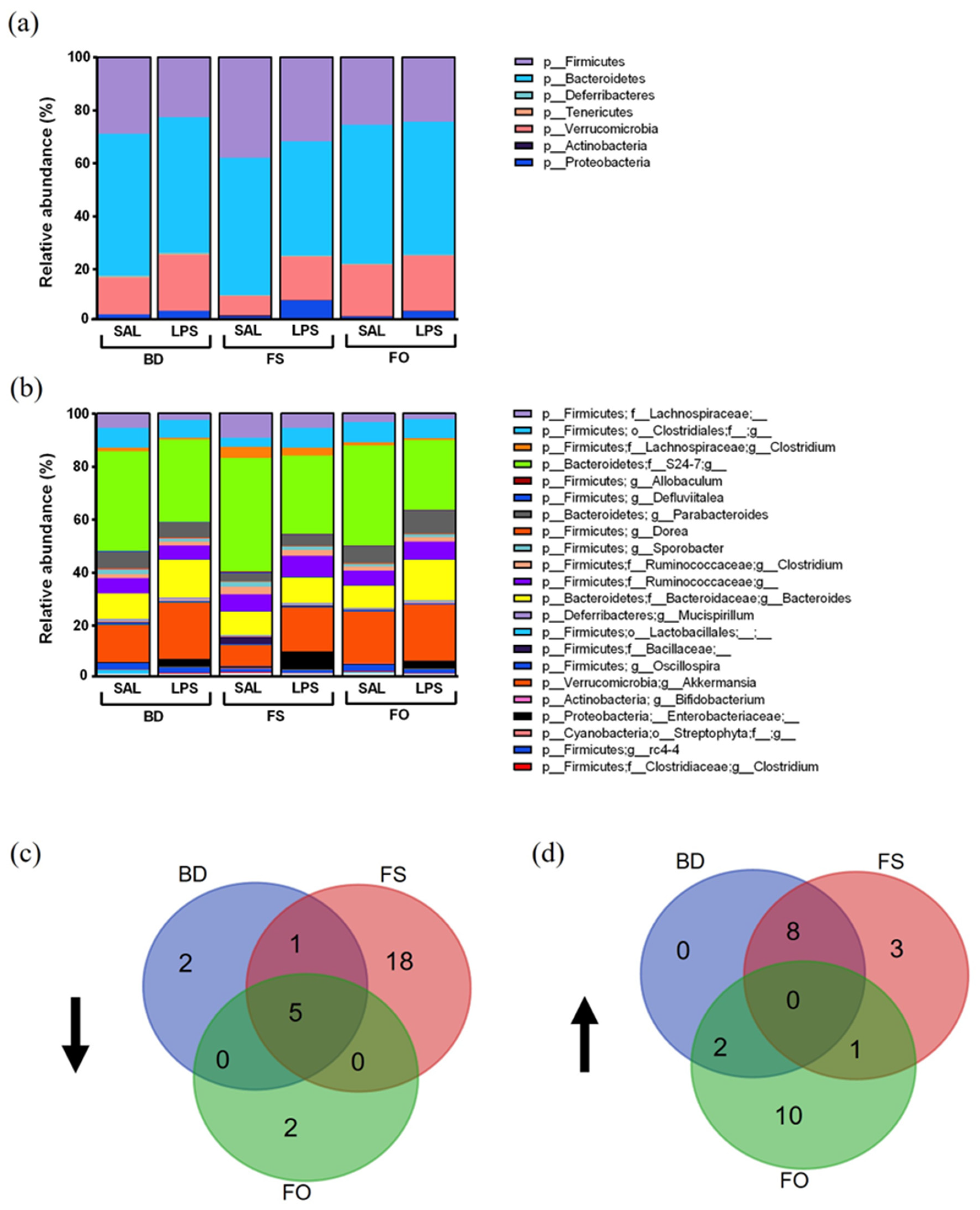
3.6. LPS-Challenged Mice Display Altered Fecal Microbiota Diversity and Composition in a Diet Dependent Manner
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cryan, J.F.; O’Mahony, S.M. The Microbiome-Gut-Brain Axis: From Bowel to Behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.G.; Cryan, J.F.; Mayer, E.A. The Microbiota-Gut-Brain Axis: From Motility to Mood. Gastroenterology 2021, 160, 1486–1501. [Google Scholar] [CrossRef] [PubMed]
- Quigley, E.M.M. Microbiota-Brain-Gut Axis and Neurodegenerative Diseases. Curr. Neurol. Neurosci. Rep. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, V.L.; Smith, M.R.B.; Hall, L.J.; Cleare, A.J.; Stone, J.M.; Young, A.H. Perturbations in Gut Microbiota Composition in Psychiatric Disorders: A Review and Meta-Analysis. JAMA Psychiatry 2021, 78, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Zhang, H.; Chen, X.; Zhang, Y.; Wu, J.; Zhao, L.; Wang, D.; Pu, J.; Ji, P.; et al. Toward a Deeper Understanding of Gut Microbiome in Depression: The Promise of Clinical Applicability. Adv. Sci. 2022, 9, 2203707. [Google Scholar] [CrossRef]
- Jensen, S.B.; Sheikh, M.A.; Akkouh, I.A.; Szabo, A.; O’Connell, K.S.; Lekva, T.; Engh, J.A.; Agartz, I.; Elvsåshagen, T.; Ormerod, M.B.E.G. Elevated Systemic Levels of Markers Reflecting Intestinal Barrier Dysfunction and Inflammasome Activation Are Correlated in Severe Mental Illness. Schizophr. Bull. 2023, 49, 635–645. [Google Scholar] [CrossRef]
- Wu, H.; Wang, J.; Teng, T.; Yin, B.; He, Y.; Jiang, Y.; Liu, X.; Yu, Y.; Li, X.; Zhou, X. Biomarkers of Intestinal Permeability and Blood-Brain Barrier Permeability in Adolescents with Major Depressive Disorder. J. Affect. Disord. 2023, 323, 659–666. [Google Scholar] [CrossRef]
- Kelly, J.R.; Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G.; Hyland, N.P. Breaking down the Barriers: The Gut Microbiome, Intestinal Permeability and Stress-Related Psychiatric Disorders. Front. Cell Neurosci. 2015, 9, 392. [Google Scholar] [CrossRef]
- Pengal, R.A.; Ganesan, L.P.; Wei, G.; Fang, H.; Ostrowski, M.C.; Tridandapani, S. Lipopolysaccharide-Induced Production of Interleukin-10 Is Promoted by the Serine/Threonine Kinase Akt. Mol. Immunol. 2006, 43, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Kell, D.B.; Pretorius, E. The Role of Lipopolysaccharide-Induced Cell Signalling in Chronic Inflammation. Chronic Stress 2022, 6, 24705470221076390. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Dauphinee, S.M.; Karsan, A. Lipopolysaccharide Signaling in Endothelial Cells. Lab. Investig. 2006, 86, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Lively, S.; Schlichter, L.C. Microglia Responses to Pro-Inflammatory Stimuli (LPS, IFNγ+TNFα) and Reprogramming by Resolving Cytokines (IL-4, IL-10). Front. Cell Neurosci. 2018, 12, 215. [Google Scholar] [CrossRef] [PubMed]
- Gorina, R.; Font-Nieves, M.; Márquez-Kisinousky, L.; Santalucia, T.; Planas, A.M. Astrocyte TLR4 Activation Induces a Proinflammatory Environment through the Interplay between MyD88-dependent NFκB Signaling, MAPK, and Jak1/Stat1 Pathways. Glia 2011, 59, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Batista, C.R.A.; Gomes, G.F.; Candelario-Jalil, E.; Fiebich, B.L.; De Oliveira, A.C.P. Lipopolysaccharide-Induced Neuroinflammation as a Bridge to Understand Neurodegeneration. Int. J. Mol. Sci. 2019, 20, 2293. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bi, W.; Xiao, S.; Lan, X.; Cheng, X.; Zhang, J.; Lu, D.; Wei, W.; Wang, Y.; Li, H.; et al. Neuroinflammation Induced by Lipopolysaccharide Causes Cognitive Impairment in Mice. Sci. Rep. 2019, 9, 5790. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.-Y.; Lee, J.W.; Lin, G.; Lee, Y.K.; Lee, Y.H.; Choi, I.S.; Han, S.B.; Jung, J.K.; Kim, Y.H.; Kim, K.H. Obovatol Attenuates LPS-Induced Memory Impairments in Mice via Inhibition of NF-ΚB Signaling Pathway. Neurochem. Int. 2012, 60, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Rolls, A.; Shechter, R.; London, A.; Ziv, Y.; Ronen, A.; Levy, R.; Schwartz, M. Toll-like Receptors Modulate Adult Hippocampal Neurogenesis. Nat. Cell Biol. 2007, 9, 1081–1088. [Google Scholar] [CrossRef]
- Valero, J.; Mastrella, G.; Neiva, I.; Sánchez, S.; Malva, J.O. Long-Term Effects of an Acute and Systemic Administration of LPS on Adult Neurogenesis and Spatial Memory. Front. Neurosci. 2014, 8, 83. [Google Scholar] [CrossRef]
- de Gomes, M.G.; Souza, L.C.; Goes, A.R.; Del Fabbro, L.; Filho, C.B.; Donato, F.; Prigol, M.; Luchese, C.; Roman, S.S.; Puntel, R.L.; et al. Fish Oil Ameliorates Sickness Behavior Induced by Lipopolysaccharide in Aged Mice through the Modulation of Kynurenine Pathway. J. Nutr. Biochem. 2018, 58, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, B.; Delattre, A.M.; Waltrick, A.P.F.; Araújo, G.; Suchecki, D.; Machado, R.B.; de Souza, L.E.R.; Zanata, S.M.; Zanoveli, J.M.; Ferraz, A.C. Fish-Oil Supplementation Decreases Indoleamine-2,3-Dioxygenase Expression and Increases Hippocampal Serotonin Levels in the LPS Depression Model. Behav. Brain Res. 2020, 390, 112675. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Liu, R.; Ma, Y.; Li, Y.; Chen, Z.; He, H.; Chen, J.; Tong, L.; Huang, C.; You, Q. Lipopolysaccharide-Induced Depression-like Behaviors Is Ameliorated by Sodium Butyrate via Inhibiting Neuroinflammation and Oxido-Nitrosative Stress. Pharmacology 2020, 105, 550–560. [Google Scholar] [CrossRef]
- Dang, R.; Zhou, X.; Tang, M.; Xu, P.; Gong, X.; Liu, Y.; Jiao, H.; Jiang, P. Fish Oil Supplementation Attenuates Neuroinflammation and Alleviates Depressive-like Behavior in Rats Submitted to Repeated Lipopolysaccharide. Eur. J. Nutr. 2018, 57, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Ren, H.; Huang, Z.; Peng, Y.; He, B.; Yao, X.; Yuan, T.-F.; Su, H. Fish Oil Prevents Lipopolysaccharide-Induced Depressive-Like Behavior by Inhibiting Neuroinflammation. Mol. Neurobiol. 2017, 54, 7327–7334. [Google Scholar] [CrossRef]
- Huang, N.; Hua, D.; Zhan, G.; Li, S.; Zhu, B.; Jiang, R.; Yang, L.; Bi, J.; Xu, H.; Hashimoto, K.; et al. Role of Actinobacteria and Coriobacteriia in the Antidepressant Effects of Ketamine in an Inflammation Model of Depression. Pharmacol. Biochem. Behav. 2019, 176, 93–100. [Google Scholar] [CrossRef]
- Wu, R.Y.; Määttänen, P.; Napper, S.; Scruten, E.; Li, B.; Koike, Y.; Johnson-Henry, K.C.; Pierro, A.; Rossi, L.; Botts, S.R.; et al. Non-Digestible Oligosaccharides Directly Regulate Host Kinome to Modulate Host Inflammatory Responses without Alterations in the Gut Microbiota. Microbiome 2017, 5, 135. [Google Scholar] [CrossRef]
- Bhatia, R.; Sharma, S.; Bhadada, S.K.; Bishnoi, M.; Kondepudi, K.K. Lactic Acid Bacterial Supplementation Ameliorated the Lipopolysaccharide-Induced Gut Inflammation and Dysbiosis in Mice. Front. Microbiol. 2022, 13, 930928. [Google Scholar] [CrossRef]
- Xu, B.; Yan, Y.; Yin, B.; Zhang, L.; Qin, W.; Niu, Y.; Tang, Y.; Zhou, S.; Yan, X.; Ma, L. Dietary Glycyl-Glutamine Supplementation Ameliorates Intestinal Integrity, Inflammatory Response, and Oxidative Status in Association with the Gut Microbiota in LPS-Challenged Piglets. Food Funct. 2021, 12, 3539–3551. [Google Scholar] [CrossRef] [PubMed]
- Suriguga, S.; Luangmonkong, T.; Mutsaers, H.A.M.; Groothuis, G.M.M.; Olinga, P. Host Microbiota Dictates the Proinflammatory Impact of LPS in the Murine Liver. Toxicol. Vitr. 2020, 67, 104920. [Google Scholar] [CrossRef] [PubMed]
- Caetano-Silva, M.E.; Rund, L.; Hutchinson, N.T.; Woods, J.A.; Steelman, A.J.; Johnson, R.W. Inhibition of Inflammatory Microglia by Dietary Fiber and Short-Chain Fatty Acids. Sci. Rep. 2023, 13, 2819. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Löf, M.; Chen, R.; Hultman, C.M.; Fang, F.; Sandin, S. Mediterranean Diet and Depression: A Population-Based Cohort Study. Int. J. Behav. Nutr. Phys. Act. 2021, 18, 153. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Shively, C.A.; Register, T.C.; Craft, S.; Yadav, H. Gut Microbiome-Mediterranean Diet Interactions in Improving Host Health. F1000Research 2019, 8, 699. [Google Scholar] [CrossRef] [PubMed]
- Alli, S.R.; Gorbovskaya, I.; Liu, J.C.W.; Kolla, N.J.; Brown, L.; Müller, D.J. The Gut Microbiome in Depression and Potential Benefit of Prebiotics, Probiotics and Synbiotics: A Systematic Review of Clinical Trials and Observational Studies. Int. J. Mol. Sci. 2022, 23, 4494. [Google Scholar] [CrossRef]
- Ma, J.; Sun, J.; Bai, H.; Ma, H.; Wang, K.; Wang, J.; Yu, X.; Pan, Y.; Yao, J. Influence of Flax Seeds on the Gut Microbiota of Elderly Patients with Constipation. J. Multidiscip. Healthc. 2022, 15, 2407–2418. [Google Scholar] [CrossRef] [PubMed]
- Power, K.A.; Lepp, D.; Zarepoor, L.; Monk, J.M.; Wu, W.; Tsao, R.; Liu, R. Dietary Flaxseed Modulates the Colonic Microenvironment in Healthy C57Bl/6 Male Mice Which May Alter Susceptibility to Gut-Associated Diseases. J. Nutr. Biochem. 2016, 28, 61–69. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, F.; Li, Q.; Odle, J.; Lin, X.; Zhu, H.; Pi, D.; Hou, Y.; Hong, Y.; Shi, H. Fish Oil Alleviates Activation of the Hypothalamic-Pituitary-Adrenal Axis Associated with Inhibition of TLR4 and NOD Signaling Pathways in Weaned Piglets after a Lipopolysaccharide Challenge. J. Nutr. 2013, 143, 1799–1807. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Shi, Z.; Kumaran Satyanarayanan, S.; He, C.; Li, P.; Wan, J.-B.; Su, H. Fish Oil Alleviates LPS-Induced Inflammation and Depressive-like Behavior in Mice via Restoration of Metabolic Impairments. Brain Behav. Immun. 2020, 90, 393–402. [Google Scholar] [CrossRef]
- Määttänen, P.; Lurz, E.; Botts, S.R.; Wu, R.Y.; Yeung, C.W.; Li, B.; Abiff, S.; Johnson-Henry, K.C.; Lepp, D.; Power, K.A. Ground Flaxseed Reverses Protection of a Reduced-Fat Diet against Citrobacter Rodentium-Induced Colitis. Am. J. Physiol.-Gastrointest. Liver Physiol. 2018, 315, G788–G798. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Sha, L.; Li, K.; Wang, Z.; Wang, T.; Li, Y.; Liu, P.; Dong, X.; Dong, Y.; Zhang, X.; et al. Dietary Flaxseed Oil Rich in Omega-3 Suppresses Severity of Type 2 Diabetes Mellitus via Anti-Inflammation and Modulating Gut Microbiota in Rats. Lipids Health Dis. 2020, 19, 20. [Google Scholar] [CrossRef] [PubMed]
- Taibi, A.; Ku, M.; Lin, Z.; Gargari, G.; Kubant, A.; Lepp, D.; Power, K.A.; Guglielmetti, S.; Thompson, L.U.; Comelli, E.M. Discriminatory and Cooperative Effects within the Mouse Gut Microbiota in Response to Flaxseed and Its Oil and Lignan Components. J. Nutr. Biochem. 2021, 98, 108818. [Google Scholar] [CrossRef] [PubMed]
- El Tanbouly, N.; El Sayed, A.M.; Ali, Z.Y.; Abdel Wahab, S.; El Gayed, S.H.; Ezzat, S.M.; El Senousy, A.S.; Choucry, M.A.; Abdel-Sattar, E. Antidepressant-like Effect of Selected Egyptian Cultivars of Flaxseed Oil on a Rodent Model of Postpartum Depression. Evid.-Based Complement. Altern. Med. 2017, 2017, 6405789. [Google Scholar] [CrossRef]
- Almousa, A.A.; Meurens, F.; Krol, E.S.; Alcorn, J. Linoorbitides and Enterolactone Mitigate Inflammation-Induced Oxidative Stress and Loss of Intestinal Epithelial Barrier Integrity. Int. Immunopharmacol. 2018, 64, 42–51. [Google Scholar] [CrossRef]
- Corsini, E.; Dell’Agli, M.; Facchi, A.; De Fabiani, E.; Lucchi, L.; Boraso, M.S.; Marinovich, M.; Galli, C.L. Enterodiol and Enterolactone Modulate the Immune Response by Acting on Nuclear Factor-ΚB (NF-ΚB) Signaling. J. Agric. Food Chem. 2010, 58, 6678–6684. [Google Scholar] [CrossRef] [PubMed]
- Rom, S.; Zuluaga-Ramirez, V.; Reichenbach, N.L.; Erickson, M.A.; Winfield, M.; Gajghate, S.; Christofidou-Solomidou, M.; Jordan-Sciutto, K.L.; Persidsky, Y. Secoisolariciresinol Diglucoside Is a Blood-Brain Barrier Protective and Anti-Inflammatory Agent: Implications for Neuroinflammation. J. Neuroinflamm. 2018, 15, 25. [Google Scholar] [CrossRef]
- Kolmogorova, D.; Murray, E.; Ismail, N. Monitoring Pathogen-induced Sickness in Mice and Rats. Curr. Protoc. Mouse Biol. 2017, 7, 65–76. [Google Scholar] [CrossRef]
- Deacon, R.M.J. Assessing Nest Building in Mice. Nat. Protoc. 2006, 1, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Gaskill, B.N.; Pritchett-Corning, K.R. Nest Building as an Indicator of Illness in Laboratory Mice. Appl. Anim. Behav. Sci. 2016, 180, 140–146. [Google Scholar] [CrossRef]
- Liverani, E.; Rico, M.C.; Yaratha, L.; Tsygankov, A.Y.; Kilpatrick, L.E.; Kunapuli, S.P. LPS-Induced Systemic Inflammation Is More Severe in P2Y12 Null Mice. J. Leukoc. Biol. 2014, 95, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Franklin, K.B.J. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates; Academic Press: Cambridge, MA, USA, 2019; ISBN 0128161582. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation Evaluation and Phylogenetic Diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley Statsref Stat. Ref. Online 2014, 1–15. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.; McDonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic Placement of Exact Amplicon Sequences Improves Associations with Clinical Information. Msystems 2018, 3, e00021-18. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Douglas, G.M. Gavin Douglas. Available online: https://github.com/picrust/picrust2/wiki/Frequently-Asked-Questions#how-can-i-run-categorize_by_functionpy-like-in-picrust1 (accessed on 9 July 2022).
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Kleigrewe, K.; Haack, M.; Baudin, M.; Ménabréaz, T.; Crovadore, J.; Masri, M.; Beyrer, M.; Andlauer, W.; Lefort, F.; Dawid, C. Dietary Modulation of the Human Gut Microbiota and Metabolome with Flaxseed Preparations. Int. J. Mol. Sci. 2022, 23, 10473. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Z.; Deng, Q.; Huang, Q.; Wang, X.; Huang, F. Beneficial Effects of Flaxseed Polysaccharides on Metabolic Syndrome via Gut Microbiota in High-Fat Diet Fed Mice. Food Res. Int. 2020, 131, 108994. [Google Scholar] [CrossRef]
- Badger, R.; Aho, K.; Serve, K. Short-Term Exposure to Synthetic Flaxseed Lignan LGM2605 Alters Gut Microbiota in Mice. Microbiologyopen 2021, 10, e1185. [Google Scholar] [CrossRef]
- Biesmans, S.; Meert, T.F.; Bouwknecht, J.A.; Acton, P.D.; Davoodi, N.; De Haes, P.; Kuijlaars, J.; Langlois, X.; Matthews, L.J.R.; Ver Donck, L.; et al. Systemic Immune Activation Leads to Neuroinflammation and Sickness Behavior in Mice. Mediat. Inflamm. 2013, 2013, 271359. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Wu, X.; Block, M.L.; Liu, Y.; Breese, G.R.; Hong, J.; Knapp, D.J.; Crews, F.T. Systemic LPS Causes Chronic Neuroinflammation and Progressive Neurodegeneration. Glia 2007, 55, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Mittli, D.; Tukacs, V.; Ravasz, L.; Csősz, É.; Kozma, T.; Kardos, J.; Juhász, G.; Kékesi, K.A. LPS-Induced Acute Neuroinflammation, Involving Interleukin-1 Beta Signaling, Leads to Proteomic, Cellular, and Network-Level Changes in the Prefrontal Cortex of Mice. Brain Behav. Immun. Health 2023, 28, 100594. [Google Scholar] [CrossRef] [PubMed]
- Goossens, E.; Li, J.; Callens, C.; Van Rysselberghe, N.; Kettunen, H.; Vuorenmaa, J.; Garcia Gonzalez, N.; Libert, C.; Ducatelle, R.; Van Immerseel, F. Acute Endotoxemia-Induced Respiratory and Intestinal Dysbiosis. Int. J. Mol. Sci. 2022, 23, 11602. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Hasani, A.; Ebrahimzadeh, S.; Hemmati, F.; Khabbaz, A.; Hasani, A.; Gholizadeh, P. The Role of Akkermansia Muciniphila in Obesity, Diabetes and Atherosclerosis. J. Med. Microbiol. 2021, 70, 001435. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Yoo, B.B.; Runia, T.F.; Debelius, J.W.; Singh, S.; Nelson, C.A.; Kanner, R.; Bencosme, Y.; Lee, Y.K.; Hauser, S.L.; et al. Gut Bacteria from Multiple Sclerosis Patients Modulate Human T Cells and Exacerbate Symptoms in Mouse Models. Proc. Natl. Acad. Sci. USA 2017, 114, 10713–10718. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, A.; Bazzaz, S.; Da Cruz, A.G.; Khorshidian, N.; Saadat, Y.R.; Sabahi, S.; Ozma, M.A.; Lahouty, M.; Aslani, R.; Mortazavian, A.M. A Critical Review on Akkermansia Muciniphila: Functional Mechanisms, Technological Challenges, and Safety Issues. Probiot. Antimicrob. Proteins 2023. [Google Scholar] [CrossRef]
- Cheng, L.-H.; Liu, Y.-W.; Wu, C.-C.; Wang, S.; Tsai, Y.-C. Psychobiotics in Mental Health, Neurodegenerative and Neurodevelopmental Disorders. J. Food Drug Anal. 2019, 27, 632–648. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Fujita, Y.; Ren, Q.; Ma, M.; Dong, C.; Hashimoto, K. Bifidobacterium in the Gut Microbiota Confer Resilience to Chronic Social Defeat Stress in Mice. Sci. Rep. 2017, 7, 45942. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Kazemi Sefat, N.A.; Mohammadi, M.M.; Hadjati, J.; Talebi, S.; Ajami, M.; Daneshvar, H. Sodium Butyrate as a Histone Deacetylase Inhibitor Affects Toll-Like Receptor 4 Expression in Colorectal Cancer Cell Lines. Immunol. Investig. 2019, 48, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Qian, Y.; Mo, C.; Ai, P.; Yang, X.; Xiao, Q. Sodium Butyrate Ameliorates Gut Dysfunction and Motor Deficits in a Mouse Model of Parkinson’s Disease by Regulating Gut Microbiota. Front. Aging Neurosci. 2023, 15, 1099018. [Google Scholar] [CrossRef]
- Matt, S.M.; Allen, J.M.; Lawson, M.A.; Mailing, L.J.; Woods, J.A.; Johnson, R.W. Butyrate and Dietary Soluble Fiber Improve Neuroinflammation Associated with Aging in Mice. Front. Immunol. 2018, 9, 1832. [Google Scholar] [CrossRef] [PubMed]

| Ingredients | BD (g/Kg) | BD + 10% FS (g/Kg) | BD + 4% FO (g/Kg) |
|---|---|---|---|
| Casein | 200 | 173 | 200 |
| L-Cystine | 3 | 3 | 3 |
| Corn starch | 397.49 | 387.22 | 397.48 |
| Maltodextrin | 132 | 132 | 132 |
| Sucrose | 100 | 100 | 100 |
| Corn oil | 70 | 30.5 | 30.3 |
| Cellulose | 50 | 19.4 | 50 |
| Mineral mix (AIN-93G-MX) | 35 | 35 | 35 |
| Vitamin mix (AIN-93-VX) | 10 | 10 | 10 |
| Choline bitartrate | 2.5 | 2.5 | 2.5 |
| TBHQ a | 0.014 | 0.014 | 0.014 |
| Ground flaxseed | 0 | 100 | 0 |
| Flaxseed oil | 0 | 0 | 39.7 |
| Caloric density (Kcal/g) | 3.8 | 3.7 | 3.8 |
| Site | Target | SAL | LPS | ||||
|---|---|---|---|---|---|---|---|
| BD | FS | FO | BD | FS | FO | ||
| HIP | IL-1β | 0.31 ± 0.053 c | 0.49 ± 0.067 bc | 0.33 ± 0.041 c | 10.59 ± 1.8 a | 9.13 ± 1.1 ab | 17.79 ± 2.6 a |
| TNF-α | 14.14 ± 3.69 c | 28.79 ± 6.43 bc | 23.65 ± 3.53 bc | 69.78 ± 15.86 a | 60.85 ± 14.13 ab | 75.35 ± 10.43 a | |
| IL-10 | 0.71 ± 0.15 b | 1.11 ± 0.26 b | 0.97 ± 0.18 b | 4.63 ± 0.67 a | 3.41 ± 0.59 a | 3.16 ± 0.47 a | |
| TLR-4 | 9.19 ± 0.55 bc | 8.67 ± 0.8 c | 7.73 ± 0.57 c | 13.15 ± 1.29 a | 12.77 ± 0.88 a | 11.92 ± 0.8 ab | |
| mPFC | IL-1β | 0.94 ± 0.11 b | 0.9 ± 0.13 b | 0.84 ± 0.14 b | 22.15 ± 3.61 a | 14.53 ± 3.47 a | 17.78 ± 2.38 a |
| TNF-α | 15.95 ± 5.11 b | 16.72 ± 5.8 b | 13.71 ± 5.12 b | 56.42 ± 10.53 a | 40.22 ± 8.75 ab | 25.3 ± 5.43 ab | |
| IL-10 | 1.16 ± 0.17 b | 1.22 ± 0.3 b | 1.41 ± 0.3 b | 2.9 ± 0.36 a | 1.96 ± 0.3 ab | 2.92 ± 0.4 a | |
| TLR-4 | 17.65 ± 1.98 | 14.54 ± 1.69 | 20.12 ± 2.3 | 20.03 ± 2.04 | 16.43 ± 1.61 | 17.21 ± 1.25 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Livingston, D.B.H.; Sweet, A.; Rodrigue, A.; Kishore, L.; Loftus, J.; Ghali, F.; Mahmoodianfard, S.; Celton, C.; Hosseinian, F.; Power, K.A. Dietary Flaxseed and Flaxseed Oil Differentially Modulate Aspects of the Microbiota Gut–Brain Axis Following an Acute Lipopolysaccharide Challenge in Male C57Bl/6 Mice. Nutrients 2023, 15, 3542. https://doi.org/10.3390/nu15163542
Livingston DBH, Sweet A, Rodrigue A, Kishore L, Loftus J, Ghali F, Mahmoodianfard S, Celton C, Hosseinian F, Power KA. Dietary Flaxseed and Flaxseed Oil Differentially Modulate Aspects of the Microbiota Gut–Brain Axis Following an Acute Lipopolysaccharide Challenge in Male C57Bl/6 Mice. Nutrients. 2023; 15(16):3542. https://doi.org/10.3390/nu15163542
Chicago/Turabian StyleLivingston, Dawson B. H., Allison Sweet, Alexane Rodrigue, Lalit Kishore, Julia Loftus, Farida Ghali, Salma Mahmoodianfard, Colleen Celton, Farah Hosseinian, and Krista A. Power. 2023. "Dietary Flaxseed and Flaxseed Oil Differentially Modulate Aspects of the Microbiota Gut–Brain Axis Following an Acute Lipopolysaccharide Challenge in Male C57Bl/6 Mice" Nutrients 15, no. 16: 3542. https://doi.org/10.3390/nu15163542
APA StyleLivingston, D. B. H., Sweet, A., Rodrigue, A., Kishore, L., Loftus, J., Ghali, F., Mahmoodianfard, S., Celton, C., Hosseinian, F., & Power, K. A. (2023). Dietary Flaxseed and Flaxseed Oil Differentially Modulate Aspects of the Microbiota Gut–Brain Axis Following an Acute Lipopolysaccharide Challenge in Male C57Bl/6 Mice. Nutrients, 15(16), 3542. https://doi.org/10.3390/nu15163542






