Peonidin-3-O-glucoside and Resveratrol Increase the Viability of Cultured Human hFOB Osteoblasts and Alter the Expression of Genes Associated with Apoptosis, Osteoblast Differentiation and Osteoclastogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture and Treatment of hFOB Osteoblasts
2.2. mRNAseq Library Preparation, Validation, Quantification and qPCR
2.3. Bioinformatics, Statistics and Database Annotation
2.4. Ingenuity® Pathway Analysis (IPA)
2.5. Data Availability and Gene Expression Omnibus (GEO) Deposition
3. Results
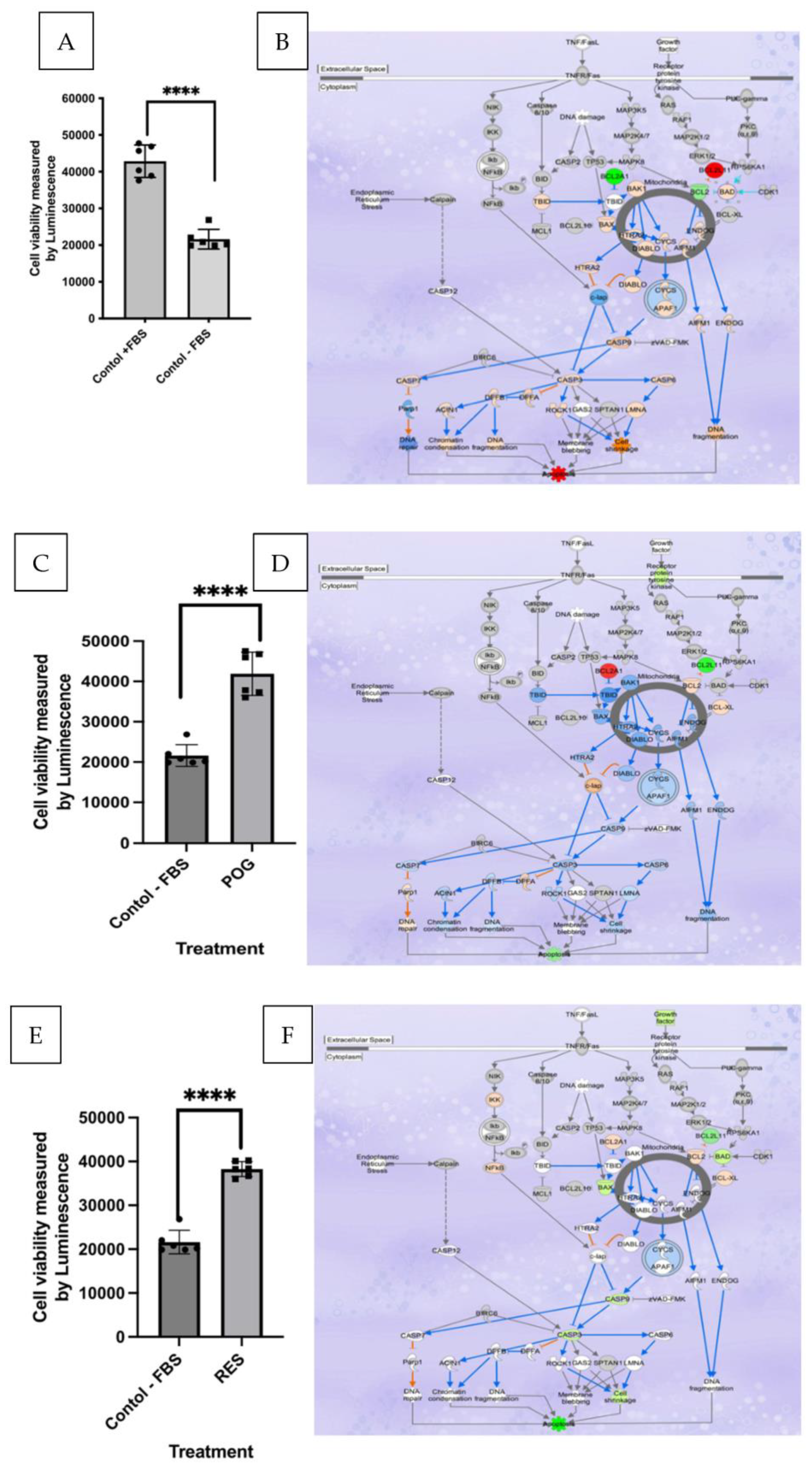
3.1. Serum Starvation Reduces Growth and Induces Apoptosis in Cultured hFOB Osteoblasts
3.2. Transcriptomic and IPA Analysis of Osteoblast Apoptosis
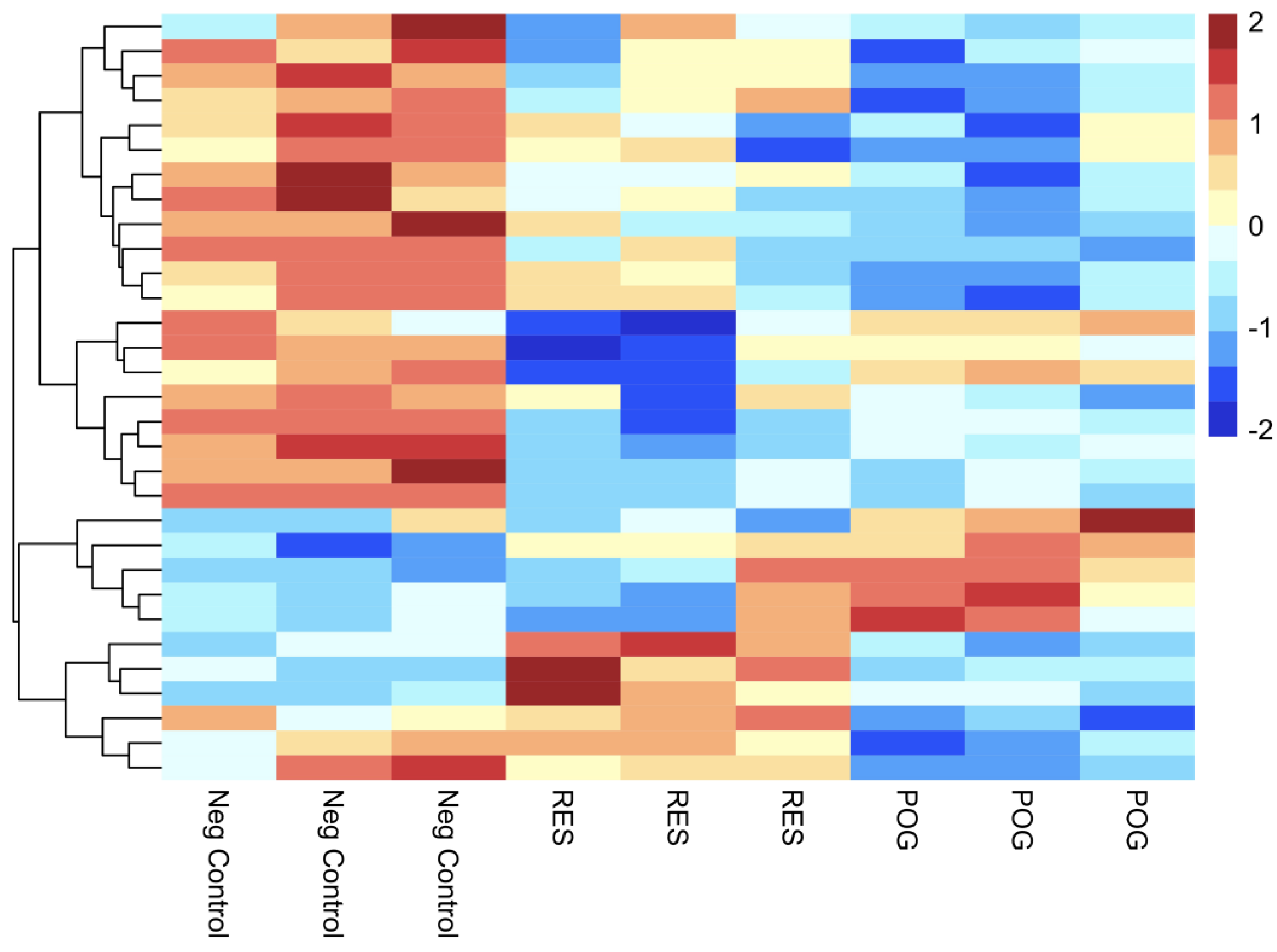
3.3. Transcriptomic Analysis of Treated Human hFOB Osteoblasts
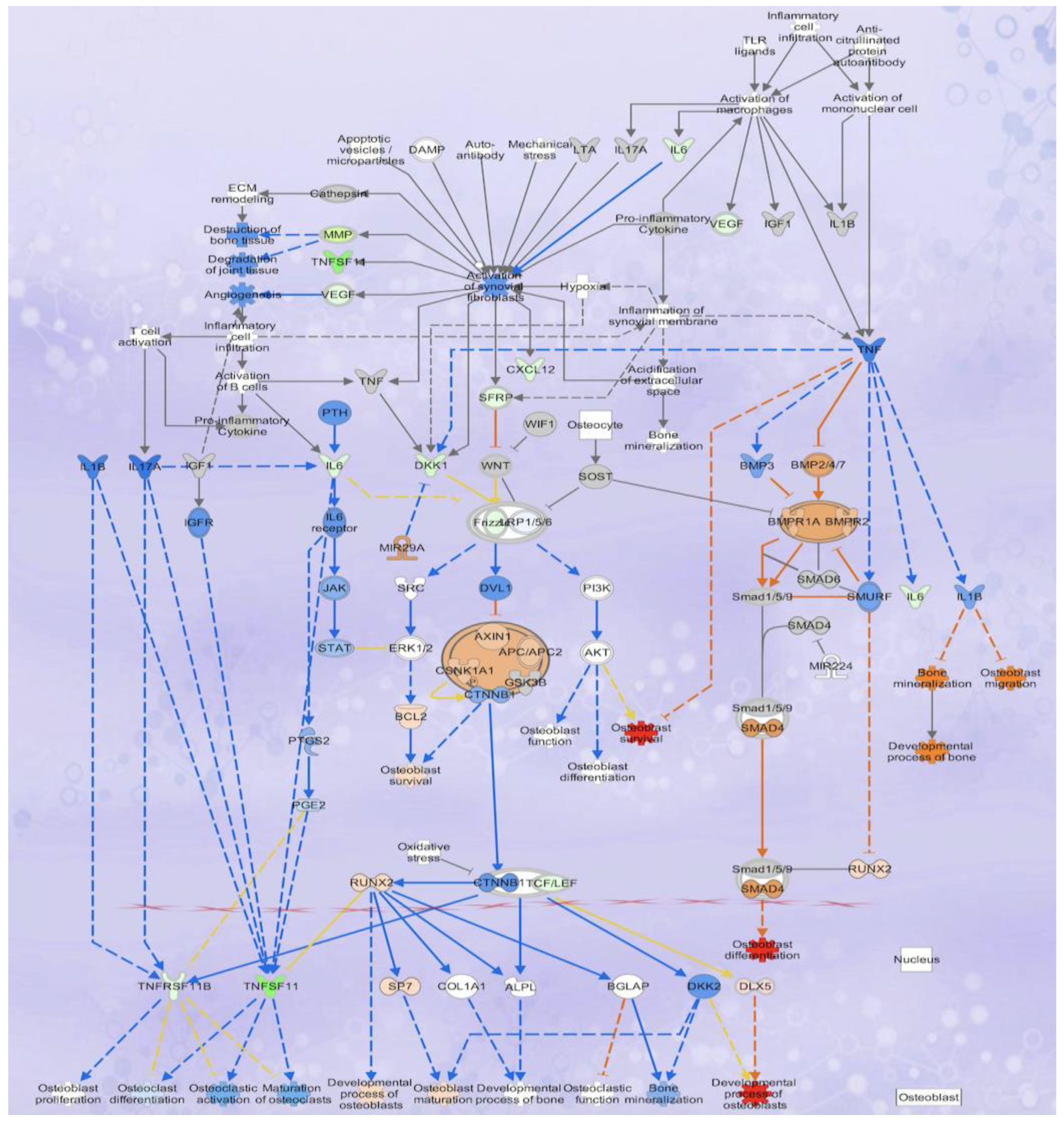
3.4. Analysis of Osteoblastogenesis, Rankl and HIF Signaling Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nazrun, A.S.; Tzar, M.N.; Mokhtar, S.A.; Mohamed, I.N. A systematic review of the outcomes of osteoporotic fracture patients after hospital discharge: Morbidity, subsequent fractures, and mortality. Ther. Clin. Risk Manag. 2014, 10, 937–948. [Google Scholar] [CrossRef]
- Salari, N.; Ghasemi, H.; Mohammadi, L.; Behzadi, M.H.; Rabieenia, E.; Shohaimi, S.; Mohammadi, M. The global prevalence of osteoporosis in the world: A comprehensive systematic review and meta-analysis. J. Orthop. Surg. Res. 2021, 16, 609. [Google Scholar] [CrossRef]
- Hadjidakis, D.J.; Androulakis, I.I. Bone remodeling. Ann. N. Y. Acad. Sci. 2006, 1092, 385–396. [Google Scholar] [CrossRef]
- Florencio-Silva, R.; Sasso, G.R.; Sasso-Cerri, E.; Simões, M.J.; Cerri, P.S. Biology of bone tissue: Structure, function, and factors that influence bone cells. Biomed. Res. Int. 2015, 2015, 421746. [Google Scholar] [CrossRef]
- Bolamperti, S.; Villa, I.; Rubinacci, A. Bone remodeling: An operational process ensuring survival and bone mechanical competence. Bone Res. 2022, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Burley, G.; Lin, S.; Shi, Y.C. Osteoporosis pathogenesis and treatment: Existing and emerging avenues. Cell. Mol. Biol. Lett. 2022, 27, 72. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Yang, H.; Zhang, Q.; Liu, C.; Zhao, J.; Zhang, L.; Chen, B. Natural products for treatment of osteoporosis: The effects and mechanisms on promoting osteoblast mediated bone formation. Life Sci. 2016, 147, 46–58. [Google Scholar] [CrossRef]
- Che, C.T.; Wong, M.; Lam, C. Natural products from Chinese medicines with potential benefits to bone health. Molecules 2016, 21, 239. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Sinclair, D.A. Therapeutic potential of resveratrol: The in vivo evidence. Nat. Rev. Drug Discov. 2006, 542, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Raut, N.; Lawal, T.O.; Wicks, S.M.; Patel, S.; Mahady, G.B. Epigenetic regulation of osteoblastogenesis by blackcurrant fruit extracts in vitro and in vivo. FASEB J. 2019, 33, 471. [Google Scholar] [CrossRef]
- Ren, Z.; Raut, N.A.; Lawal, T.O.; Patel, S.R.; Lee, S.M.; Mahady, G.B. Peonidin-3-O-glucoside and cyanidin increase osteoblast differentiation and reduce RANKL-induced bone resorption in transgenic medaka. Phytother. Res. 2021, 35, 6255–6269. [Google Scholar] [CrossRef] [PubMed]
- Imangali, N.; Phan, Q.T.; Mahady, G.B.; Winkler, C. The dietary anthocyanin delphinidin prevents bone resorption by inhibiting Rankl-induced differentiation of osteoclasts in a medaka (Oryzias latipes) model of osteoporosis. J. Fish Biol. 2020, 98, 1018–1030. [Google Scholar] [CrossRef]
- Raut, N.A.; Lawal, T.O.; Wicks, S.M.; Patel, S.; Mahady, G.B. Acai fruit extracts and anthocyanins alter osteoblast proliferation and the expression of HDAC1 and osterix/Sp7 in vitro and in vivo. FASEB J. 2019, 33, 515. [Google Scholar] [CrossRef]
- Kanabar, P.; Los, N.; Abrevia, Z.; Cline, M.; Patel, S.; Lawal, T.O.; Mahady, G.B. Transcriptomic analysis reveals that combinations of vitamins A, D2 and D3 have synergistic effects in HCT-116 colon cancer cells by altering the expression of genes involved in multiple canonical pathways including apoptosis, regulation of the epithelial mesenchymal transition and immunity. Funct. Foods Health Dis. 2021, 11, 154–178. [Google Scholar] [CrossRef]
- Kanabar, P.; Los, N.; Patel, S.; Lawal, T.O.; Raut, N.; Abrevia, Z.; Cline, M.; Mahady, G.B. Combinations of vitamin A and D are synergistic in breast cancer cells and alter gene expression in the endoplasmic reticulum stress, unfolded protein and estrogen signaling canonical pathways. Funct. Foods Health Dis. 2023, 13, 135–155. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11–14. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An efficient, general-purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Fu, X.; Fu, N.; Guo, S.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Chen, W.; Li, Y.; Zeng, R. Estimating the accuracy of RNA-Seq and microarrays with proteomics. BMC Genom. 2009, 10, 161. [Google Scholar] [CrossRef]
- Bradford, J.R.; Hey, Y.; Yates, T.; Li, Y.; Pepper, S.D.; Miller, C.J. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genom. 2010, 11, 282. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J.; Tugendrich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Utting, J.C.; Robins, S.P.; Brandao-Burch, A.; Orriss, I.R.; Behar, J.; Arnett, T.R. Hypoxia inhibits the growth, differentiation, and bone-forming capacity of rat osteoblasts. Exp. Cell Res. 2006, 312, 1693–1702. [Google Scholar] [CrossRef]
- Wang, G.; Wang, J.; Sun, D.; Xin, J.; Wang, L.; Huang, D.; Wu, W.; Xian, C.J. Short-term hypoxia accelerates bone loss in ovariectomized rats by suppressing osteoblastogenesis but enhances osteoclastogenesis. Med. Sci. Monit. 2016, 22, 2962–2971. [Google Scholar] [CrossRef]
- Ahmad Hairi, H.; Jayusman, P.A.; Shuid, A.N. Revisiting resveratrol as an osteoprotective agent: Molecular evidence from in vivo and in vitro studies. Biomedicines 2023, 11, 1453. [Google Scholar] [CrossRef]
- Wallace, T.C. Dried plums, prunes and bone health: A comprehensive review. Nutrients 2017, 9, 401–415. [Google Scholar] [CrossRef]
- Bo, S.; Gambino, R.; Ponzo, V.; Cioffi, I.; Goitre, I.; Evangelista, A.; Ciccone, G.; Cassader, M.; Procopio, M. Effects of resveratrol on bone health in type 2 diabetic patients. A double-blind randomized-controlled trial. Nutr. Diabetes 2018, 8, 51. [Google Scholar] [CrossRef]
- Ornstrup, J.M.; Harsløf, T.; Kjær, T.N.; Langdahl, B.L.; Pedersen, S.B. Resveratrol increases bone mineral density and bone alkaline phosphatase in obese men: A randomized placebo-controlled trial. J. Clin. Endocrinol. Metab. 2014, 99, 4720–4729. [Google Scholar] [CrossRef] [PubMed]
- Welch, A.; MacGregor, A.; Jennings, A.; Fairweather-Tait, S.; Spector, T.; Cassidy, A. Habitual flavonoid intakes are positively associated with bone mineral density in women. J. Bone Miner. Res. 2012, 27, 1872–1878. [Google Scholar] [CrossRef]
- Wong, R.H.; Thaung Zaw, J.J.; Xian, C.J.; Howe, P.R. Regular supplementation with resveratrol improves bone mineral density in postmenopausal women: A randomized, placebo-controlled trial. J. Bone Miner. Res. 2020, 35, 2121–2131. [Google Scholar] [CrossRef] [PubMed]
- Raut, N.; Wicks, S.M.; Lawal, T.O.; Mahady, G.B. Epigenetic regulation of bone remodelling by natural compounds. Pharmacol. Res. 2019, 147, 104350. [Google Scholar] [CrossRef]
- Tseng, P.C.; Hou, S.M.; Chen, R.J.; Peng, H.W.; Hsieh, C.F.; Kuo, M.L.; Yen, M.L. Resveratrol promotes osteogenesis of human mesenchymal stem cells by upregulating RUNX2 gene expression via the SIRT1/FOXO3A axis. J. Bone Miner. Res. 2011, 26, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, K.; Ikeda, K.; Kawai, Y.; Yamori, Y. Resveratrol stimulates the proliferation and differentiation of osteoblastic MC3T3-E1 cells. Biochem. Biophys. Res. Commun. 1998, 253, 859–863. [Google Scholar] [CrossRef]
- Moriwaki, S.; Suzuki, K.; Muramatsu, M.; Nomura, A.; Inoue, F. Delphinidin, one of the major anthocyanidins, prevents bone loss through the inhibition of excessive osteoclastogenesis in osteoporosis model mice. PLoS ONE 2014, 9, 97177. [Google Scholar] [CrossRef]
- Park, K.H.; Gu, D.R.; So, H.S.; Kim, K.; Lee, S.H. Dual Role of cyanidin-3-glucoside on the differentiation of bone cells. J. Dent. Res. 2015, 94, 1676–1683. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Vogler, M. BCL2A1: The underdog in the BCL2 family. Cell Death Differ. 2012, 19, 67–74. [Google Scholar] [CrossRef]
- Sionov, R.V.; Vlahopoulos, S.A.; Granot, Z. Regulation of Bim in health and disease. Oncotarget 2015, 6, 23058–23134. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Shimamura, T.; Perera, S.; Carlson, N.E.; Cai, D.; Shapiro, G.I.; Wong, K.K.; Letai, A. Proapoptotic BH3-Only BCL-2 family protein BIM connects death signaling from epidermal growth factor receptor inhibition to the mitochondrion. Cancer Res. 2007, 67, 11867–11875. [Google Scholar] [CrossRef]
- He, N.; Zhu, X.W.; He, W.; Zhao, S.W.; Zhao, W.Y.; Zhu, C.L. Resveratrol inhibits the hydrogen dioxide-induced apoptosis via Sirt 1 activation in osteoblast cells. Biosci. Biotechnol. Biochem. 2015, 79, 1779–1786. [Google Scholar] [CrossRef]
- Tang, X.Y.; Zhu, H.; Cao, C. Effects of resveratrol on proliferation and differentiation of osteoblasts induced by high glucose through SIRT1/FOXO1 pathway. J. Oral Sci. Res. 2020, 36, 79–84. [Google Scholar] [CrossRef]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Kawane, T.; Komori, H.; Liu, W.; Moriishi, T.; Miyazaki, T.; Mori, M.; Matsuo, Y.; Takada, Y.; Izumi, S.; Jiang, Q.; et al. Dlx5 and mef2 regulate a novel runx2 enhancer for osteoblast-specific expression. J. Bone Miner. Res. 2014, 29, 1960–1969. [Google Scholar] [CrossRef]
- Heo, J.S.; Lee, S.G.; Kim, H.O. Distal-less homeobox 5 is a master regulator of the osteogenesis of human mesenchymal stem cells. Int. J. Mol. Med. 2017, 40, 1486–1494. [Google Scholar] [CrossRef]
- Samee, N.; Geoffroy, V.; Marty, C.; Schiltz, C.; Vieux-Rochas, M.; Levi, G.; de Vernejoul, M.C. Dlx5, a positive regulator of osteoblastogenesis, is essential for osteoblast-osteoclast coupling. Am. J. Pathol. 2008, 173, 773–780. [Google Scholar] [CrossRef]
- Shakibaei, M.; Shayan, P.; Busch, F.; Aldinger, C.; Buhrmann, C.; Lueders, C.; Mobasheri, A. Resveratrol mediated modulation of Sirt-1/Runx2 promotes osteogenic differentiation of mesenchymal stem cells: Potential role of Runx2 deacetylation. PLoS ONE 2012, 7, e35712. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, E.F. Cellular mechanisms of bone remodeling. Rev. Endocr. Metab. Disord. 2010, 11, 219–227. [Google Scholar] [CrossRef]
- Hikita, A.; Yana, I.; Wakeyama, H. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappaB ligand. J. Biol. Chem. 2006, 281, 36846–36855. [Google Scholar] [CrossRef]
- Tobeiha, M.; Moghadasian, M.H.; Amin, N.; Jafarnejad, S. RANKL/RANK/OPG Pathway: A mechanism involved in exercise-induced bone remodeling. Biomed. Res. Int. 2020, 2020, 6910312. [Google Scholar] [CrossRef]
- Shakibaei, M.; Buhrmann, C.; Mobasheri, A. Resveratrol-mediated SIRT-1 interactions with p300 modulate receptor activator of NF-kappaB ligand (RANKL) activation of NF-kappaB signaling and inhibit osteoclastogenesis in bone-derived cells. J. Biol. Chem. 2011, 286, 11492–11505. [Google Scholar] [CrossRef]
- Ameen, O.; Yassien, R.I.; Naguib, Y.M. Activation of FoxO1/SIRT1/RANKL/OPG pathway may underlie the therapeutic effects of resveratrol on aging-dependent male osteoporosis. BMC Musculoskelet. Disord. 2020, 21, 375. [Google Scholar] [CrossRef]
- Basu, S.; Michaelsson, K.; Olofsson, H.; Johansson, S.; Melhus, H. Association between oxidative stress and bone mineral density. Biochem. Biophys. Res. Commun. 2001, 288, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Jagger, C.J.; Lean, J.M.; Davies, J.T.; Chambers, T.J. Tumor necrosis factor-a mediates osteopenia caused by depletion of antioxidants. Endocrinology 2004, 146, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Altindag, O.; Erel, O.; Soran, N.; Celik, H.; Selek, S. Total oxidative/anti-oxidative status and relation to bone mineral density in osteoporosis. Rheumatol. Int. 2008, 28, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Usategui-Martín, R.; Rigual, R.; Ruiz-Mambrilla, M.; Fernández-Gómez, J.-M.; Dueñas, A.; Pérez-Castrillón, J.L. Molecular mechanisms involved in hypoxia-induced alterations in bone remodeling. Int. J. Mol. Sci. 2022, 23, 3233. [Google Scholar] [CrossRef]
- Corrado, C.; Fontana, S. Hypoxia and HIF Signaling: One Axis with Divergent Effects. Int. J. Mol. Sci. 2020, 21, 5611. [Google Scholar] [CrossRef]
- Komori, T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 2019, 20, 1694. [Google Scholar] [CrossRef]
- Ontiveros, C.; Irwin, R.; Wiseman, R.W.; McCabe, L.R. Hypoxia suppresses runx2 independent of modeled microgravity. J. Cell. Physiol. 2004, 200, 169–176. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, Z.; Guo, L.; Liu, R.; Li, R.; Chu, X.; Yang, J.; Luo, J.; Chen, F.; Deng, M. Hypoxia mediates Runt-related transcription factor 2 expression via induction of vascular endothelial growth factor in periodontal ligament stem cells. Mol. Cells 2019, 42, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wu, P.; Yu, F.; Luo, G.; Qing, L.; Tang, J. HIF-1α Regulates bone homeostasis and angiogenesis, participating in the occurrence of bone metabolic diseases. Cells 2022, 11, 3552. [Google Scholar] [CrossRef] [PubMed]
- Horwood, N.J.; Elliott, J.; Martin, T.J.; Gillespie, M.T. IL-12 alone and in synergy with IL-18 inhibits osteoclast formation in vitro. J. Immunol. 2001, 166, 4915–4921. [Google Scholar] [CrossRef]
- Amarasekara, D.S.; Yun, H.; Kim, S.; Lee, N.; Kim, H.; Rho, J. Regulation of osteoclast differentiation by cytokine networks. Immune Netw. 2018, 18, e8. [Google Scholar] [CrossRef]
- Kitaura, H.; Tatamiya, M.; Nagata, N.; Fujimura, Y.; Eguchi, T.; Yoshida, N.; Nakayama, K. IL-18 induces apoptosis of adherent bone marrow cells in TNF-α mediated osteoclast formation in synergy with IL-12. Immunol. Lett. 2006, 107, 22–31. [Google Scholar] [CrossRef]
- Raggatt, L.J.; Qin, L.; Tamasi, J.; Bevelock, L.; Feyen, J.H.; Partridge, N.C. Interleukin-18 is regulated by parathyroid hormone and is required for its bone anabolic actions. J. Biol. Chem. 2008, 283, 6790–6798. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.M.; Nishioka, K.; Yudoh, K. Interleukin (IL) 18 stimulates osteoclast formation through synovial T cells in rheumatoid arthritis: Comparison with IL1 beta and tumour necrosis factor alpha. Ann. Rheum. Dis. 2004, 63, 1379–1386. [Google Scholar] [CrossRef]
- Lorenzo, J.A. The role of Interleukin-6 in bone. J. Endocr. Soc. 2020, 4, bvaa112. [Google Scholar] [CrossRef] [PubMed]




| A | B | ||
| Geneid | Gene name | Geneid | Gene name |
| ENSG00000120915 | EPHX2 | ENSG00000282608 | ADORA3 |
| ENSG00000267677 | RP11-27G24.1 | ENSG00000213761 | MT1P1 |
| ENSG00000273313 | RBAKDN | ENSG00000283045 | RP11-764D10.2 |
| ENSG00000259215 | RP11-253M7.4 | ENSG00000227517 | LINC01483 |
| ENSG00000251600 | RP11-673E1.1 | ENSG00000169442 | CD52 |
| ENSG00000268750 | CTD-2583A14.10 | ENSG00000274421 | RP11-386J22.3 |
| ENSG00000159496 | RGL4 | ENSG00000280639 | LINC02204 |
| ENSG00000110328 | GALNT18 | ENSG00000267938 | EIF1P6 |
| ENSG00000260331 | RP11-111J6.2 | ENSG00000198844 | ARHGEF15 |
| ENSG00000233087 | WTH3DI | ENSG00000261783 | RP11-252K23.2 |
| C | D | ||
| Geneid | Gene name | Geneid | Gene name |
| ENSG00000196415 | PRTN3 | ENSG00000159239 | C2orf81 |
| ENSG00000120915 | EPHX2 | ENSG00000118017 | A4GNT |
| ENSG00000227076 | RP11-4C20.4 | ENSG00000241112 | RPL29P14 |
| ENSG00000183484 | GPR132 | ENSG00000109424 | UCP1 |
| ENSG00000173930 | SLCO4C1 | ENSG00000261832 | RP11-435I10.4 |
| ENSG00000260947 | RP11-384P7.7 | ENSG00000144868 | TMEM108 |
| ENSG00000149633 | KIAA1755 | ENSG00000236670 | KRT18P5 |
| ENSG00000137843 | PAK6 | ENSG00000235651 | AC064850.4 |
| ENSG00000144488 | ESPNL | ENSG00000205502 | C2CD4B |
| ENSG00000183807 | FAM162B | ENSG00000198535 | C2CD4A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ostos Mendoza, K.C.; Garay Buenrostro, K.D.; Kanabar, P.N.; Maienschein-Cline, M.; Los, N.S.; Arbieva, Z.; Raut, N.A.; Lawal, T.O.; López, A.M.; Cabada-Aguirre, P.; et al. Peonidin-3-O-glucoside and Resveratrol Increase the Viability of Cultured Human hFOB Osteoblasts and Alter the Expression of Genes Associated with Apoptosis, Osteoblast Differentiation and Osteoclastogenesis. Nutrients 2023, 15, 3233. https://doi.org/10.3390/nu15143233
Ostos Mendoza KC, Garay Buenrostro KD, Kanabar PN, Maienschein-Cline M, Los NS, Arbieva Z, Raut NA, Lawal TO, López AM, Cabada-Aguirre P, et al. Peonidin-3-O-glucoside and Resveratrol Increase the Viability of Cultured Human hFOB Osteoblasts and Alter the Expression of Genes Associated with Apoptosis, Osteoblast Differentiation and Osteoclastogenesis. Nutrients. 2023; 15(14):3233. https://doi.org/10.3390/nu15143233
Chicago/Turabian StyleOstos Mendoza, Keila C., Karen D. Garay Buenrostro, Pinal N. Kanabar, Mark Maienschein-Cline, Nina S. Los, Zarema Arbieva, Nishikant A. Raut, Temitope O. Lawal, Alice M. López, Paulina Cabada-Aguirre, and et al. 2023. "Peonidin-3-O-glucoside and Resveratrol Increase the Viability of Cultured Human hFOB Osteoblasts and Alter the Expression of Genes Associated with Apoptosis, Osteoblast Differentiation and Osteoclastogenesis" Nutrients 15, no. 14: 3233. https://doi.org/10.3390/nu15143233
APA StyleOstos Mendoza, K. C., Garay Buenrostro, K. D., Kanabar, P. N., Maienschein-Cline, M., Los, N. S., Arbieva, Z., Raut, N. A., Lawal, T. O., López, A. M., Cabada-Aguirre, P., Luna-Vital, D. A., & Mahady, G. B. (2023). Peonidin-3-O-glucoside and Resveratrol Increase the Viability of Cultured Human hFOB Osteoblasts and Alter the Expression of Genes Associated with Apoptosis, Osteoblast Differentiation and Osteoclastogenesis. Nutrients, 15(14), 3233. https://doi.org/10.3390/nu15143233







