Activation of Liver X Receptors and Peroxisome Proliferator-Activated Receptors by Lipid Extracts of Brown Seaweeds: A Potential Application in Alzheimer’s Disease?
Abstract
1. Introduction
2. Materials and Methods
2.1. Seaweed Species
2.2. Preparation of Seaweed Extracts
2.3. Sterol Analysis
2.4. Lipomics Analysis
2.5. Arsenic and Cadmium Analysis
2.6. Cell Culture
2.7. Cell Transfection
2.8. LXR and PPAR Reporter Assays
2.9. Quantitative Real-Time PCR
2.10. Statistical Analysis
3. Results
3.1. Characteristics of the Tested Seaweeds and Seaweed Extracts
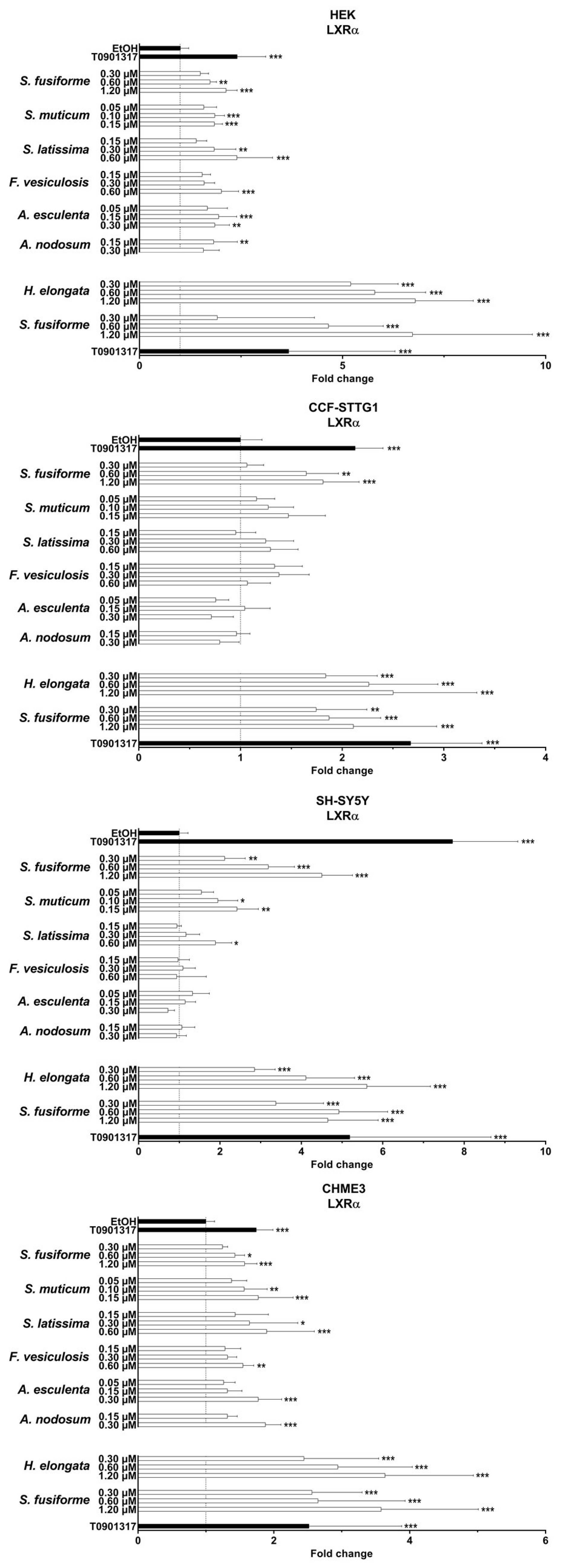
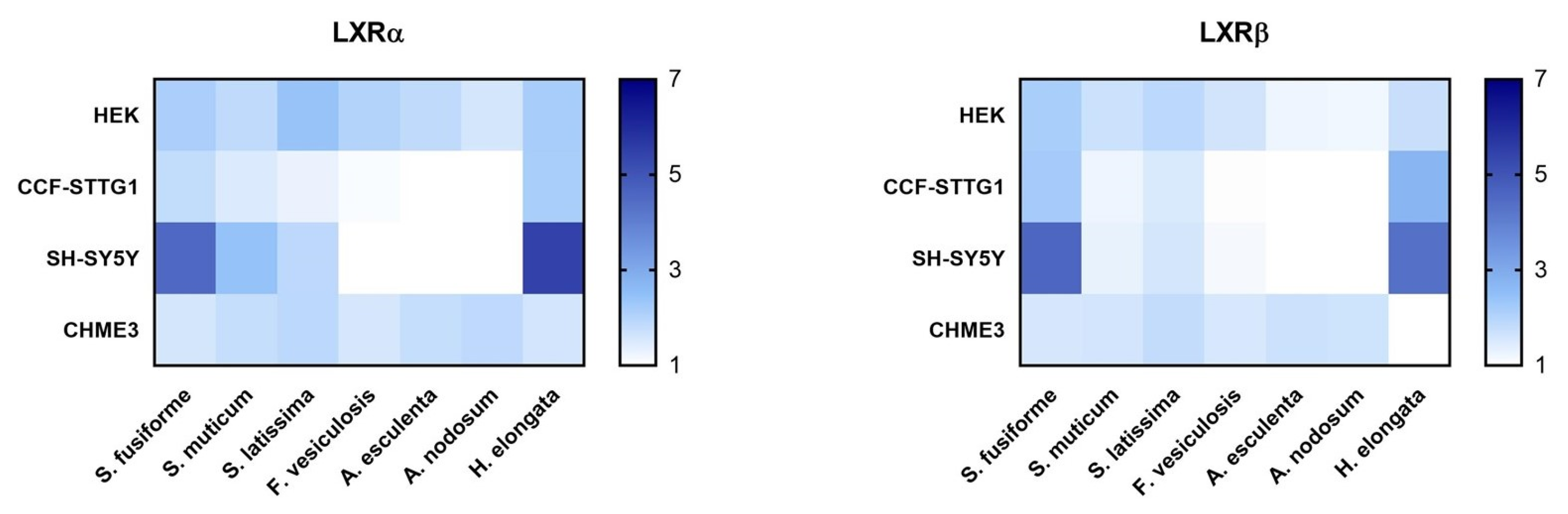
3.2. LXR Activating Capacity
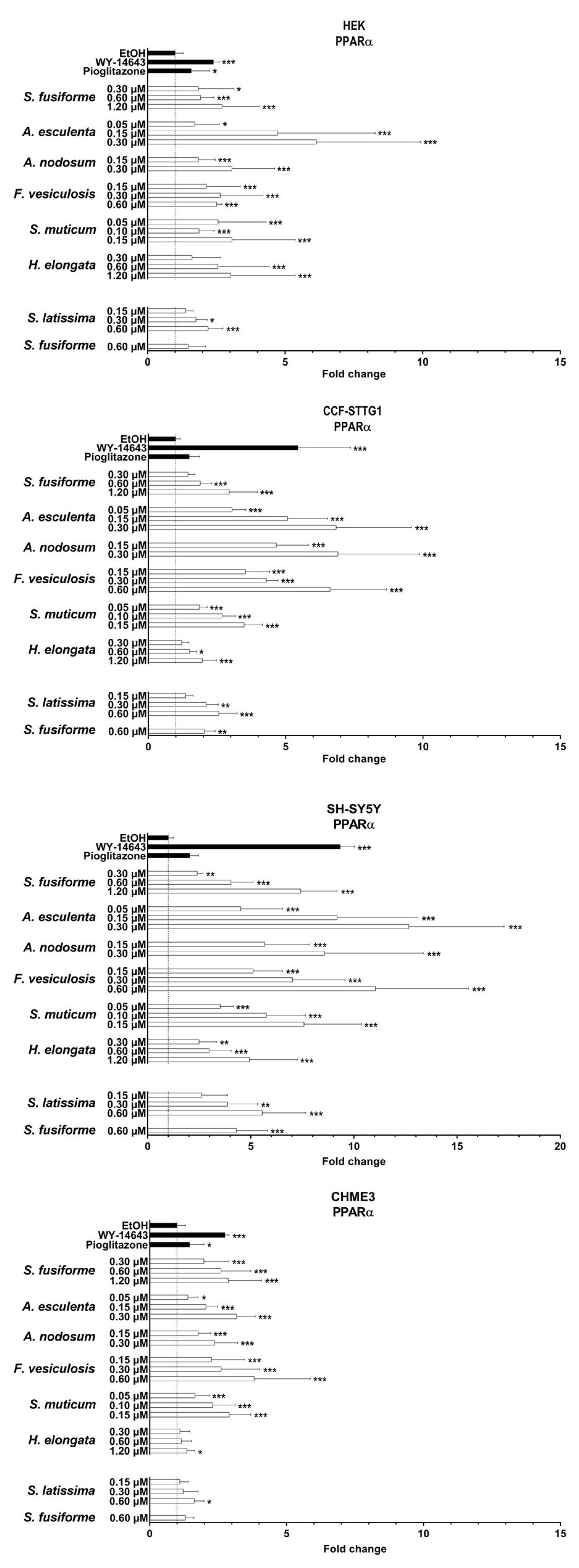
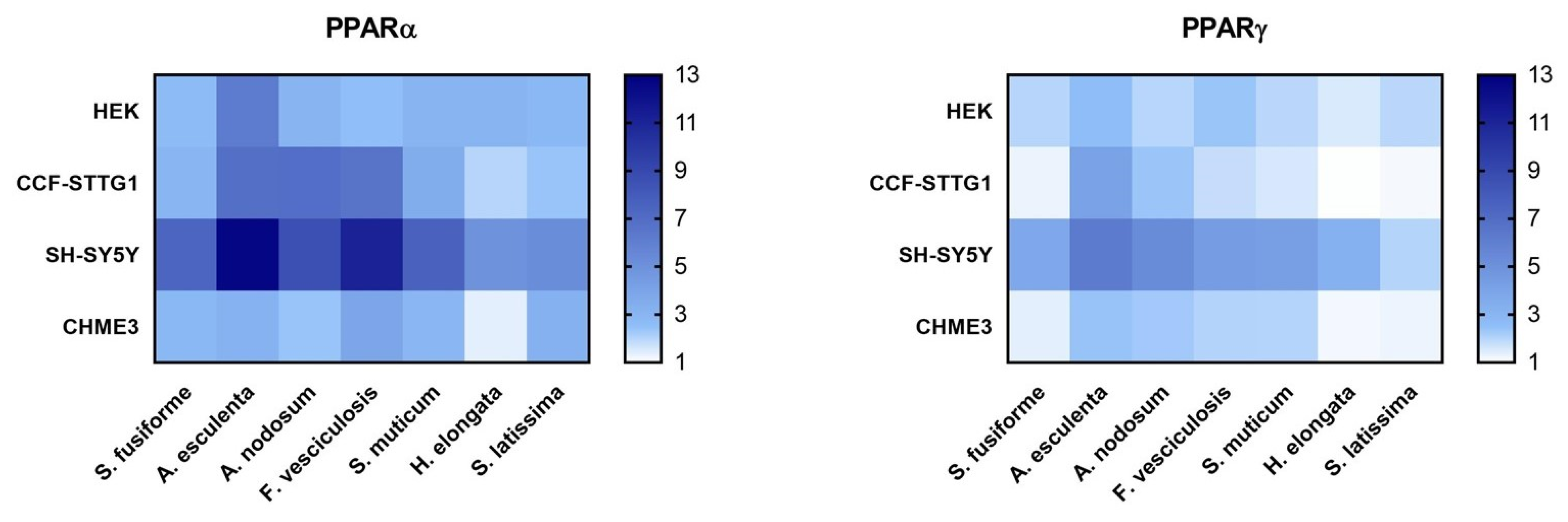
3.3. PPARα and PPARγ Activation by the Seaweed Extracts
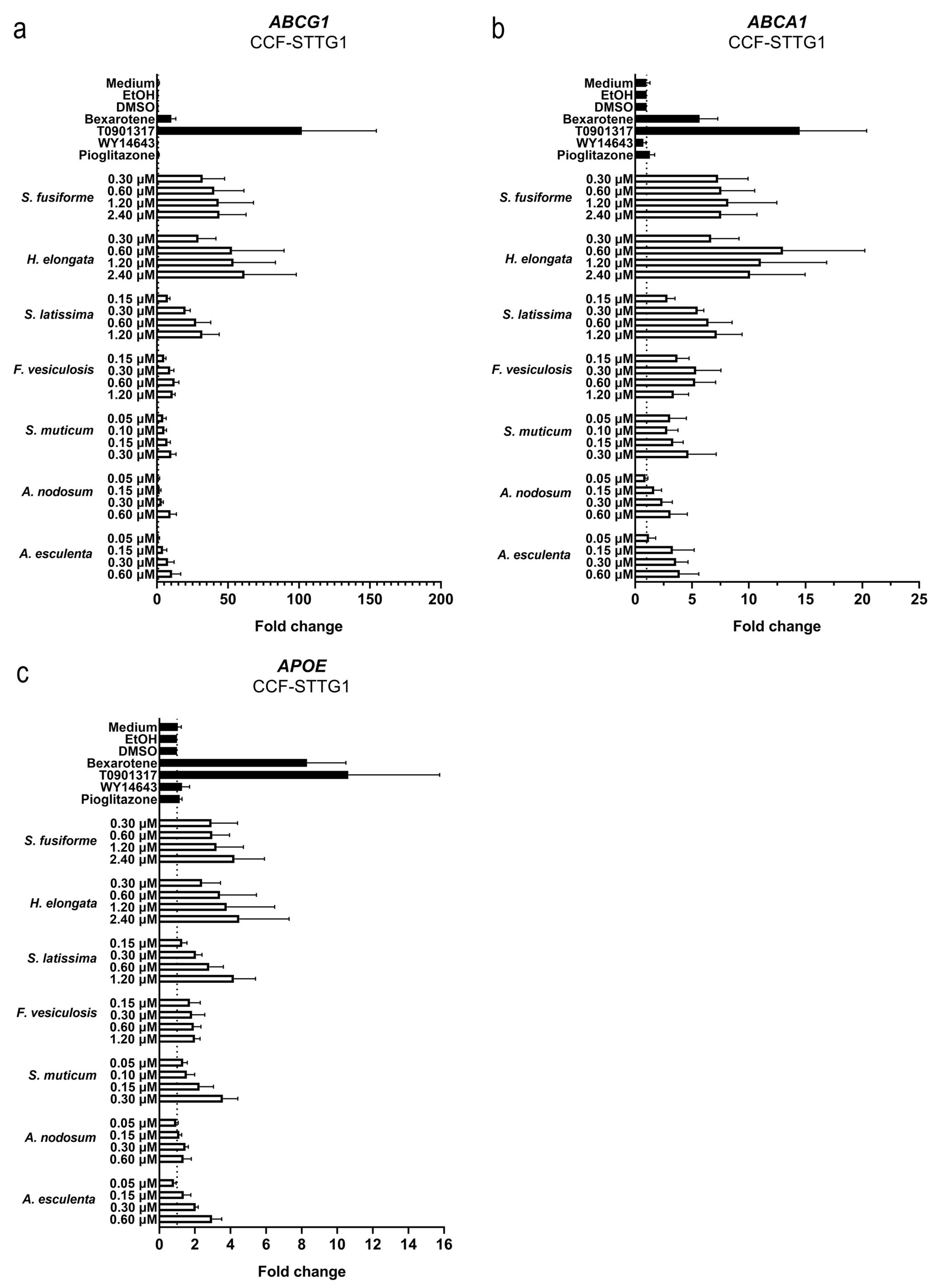
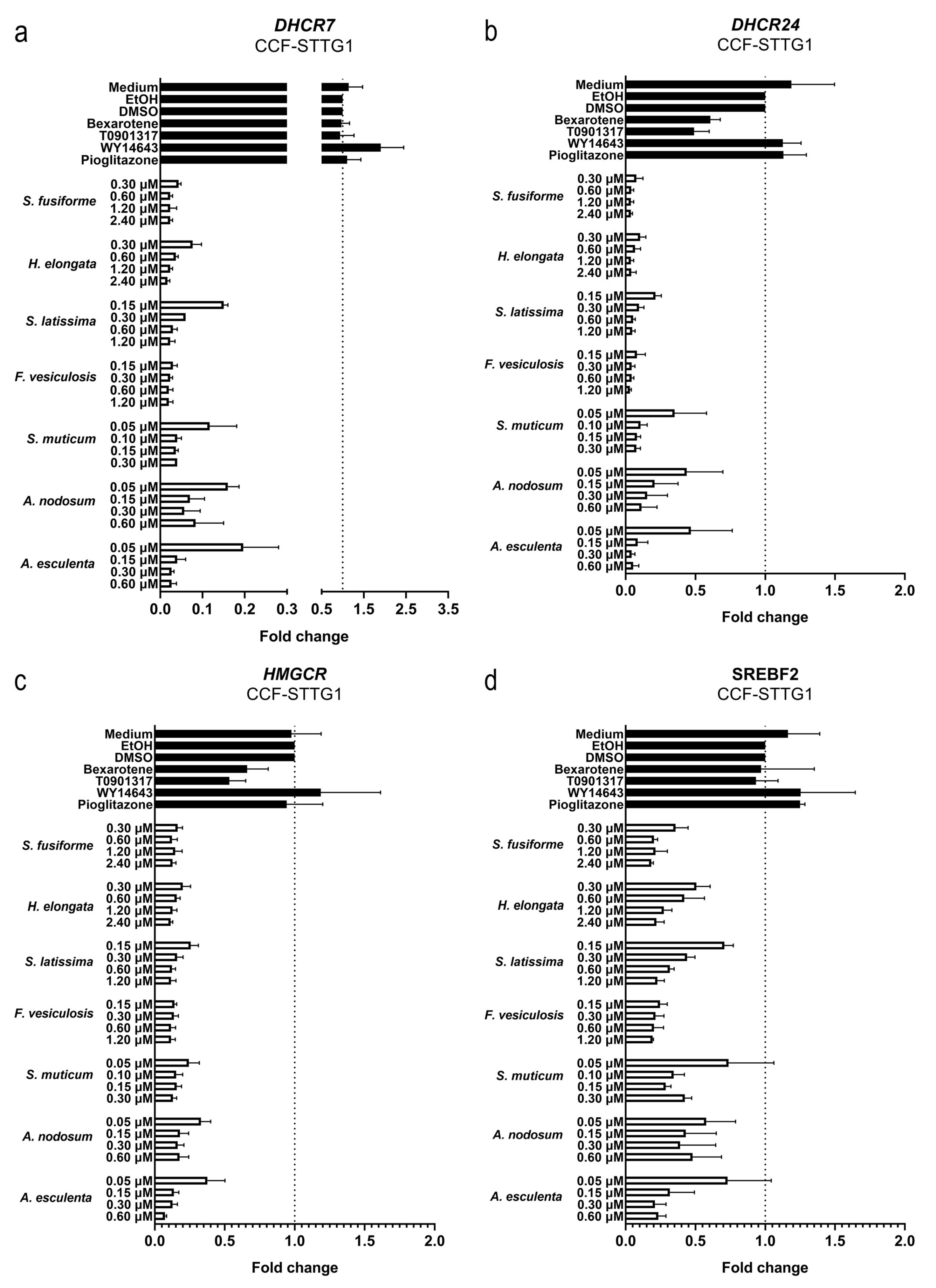
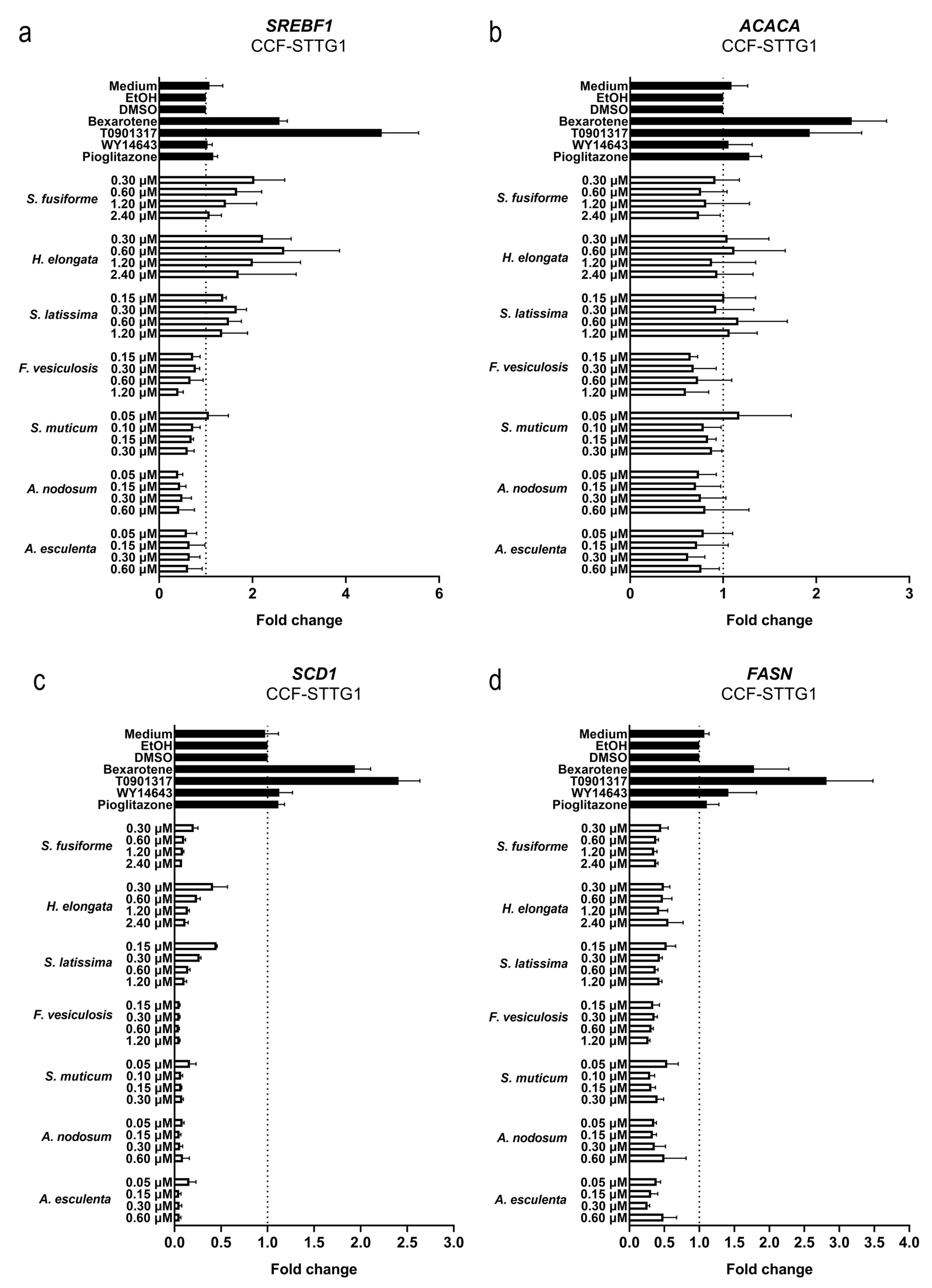
3.4. Effect of the Seaweed Extracts on the Expression of LXR and PPAR-Target Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Aβ | Amyloid-β |
| ABCA1 | ATP Binding Cassette Subfamily A Member 1 |
| ABCG1 | ATP Binding Cassette Subfamily G Member 1 |
| ACACA | Acetyl-CoA Carboxylase Alpha |
| ACTB | Actin Beta |
| AD | Alzheimer’s Disease |
| APOE | Apolipoprotein E |
| B2M | Beta-2-Microglobulin |
| cDNA | Complementary Deoxyribonucleic Acid |
| CNS | Central Nervous System |
| Ct | Cycle Threshold |
| DG | Diacylglycerol |
| DGDG | Digalactosyldiacylglycerol |
| DHCR7 | 7-Dehydrocholesterol Reductase |
| DHCR24 | 24-Dehydrocholesterol Reductase |
| DMSO | Dimethyl sulfoxide |
| DW | Dry Weight |
| EtOH | Ethyl Alcohol, Ethanol |
| FA | Fatty Acids |
| FASN | Fatty Acid Synthase |
| GFAP | Glial Fibrillary Acidic Protein |
| HMGCR | 3-Hydroxy-3-Methylglutaryl-CoA Reductase |
| HPRT1 | Hypoxanthine Phosphoribosyltransferase 1 |
| IA | Index of Atherogenicity |
| IT | Index of Thrombogenicity |
| LPA | Lysophosphatidic Acid |
| LPC | Lysophosphatidylcholine |
| LPE | Lysophosphatidylethanolamine |
| LPG | Lysophosphatidylglycerol |
| LPI | Lysophosphatidylinositol |
| LTP | Long-term Potentiation |
| LXR | Liver X Receptor |
| LXRE | Liver X Receptor Response Element |
| MGDG | Monogalactosyldiacylglycerol |
| MGMG | Monogalactosylmonoacylglycerol |
| MUFA | Monounsaturated Fatty Acid |
| NR1C1 | Nuclear Receptor Subfamily 1 Group C Member 1 |
| NR1C3 | Nuclear Receptor Subfamily 1 Group C Member 3 |
| NR1H2 | Nuclear Receptor Subfamily 1 Group H Member 2 |
| NR1H3 | Nuclear Receptor Subfamily 1 Group H Member 3 |
| PA | Phosphatidic Acid |
| PC | Phosphatidylcholine |
| PCR | Polymerase Chain Reaction |
| PE | Phosphatidylethanolamine |
| PG | Phosphatidylglycerol |
| PI | Phosphatidylinositol |
| PPAR | Peroxisome proliferator-Activated receptors |
| PPRE | Peroxisome Proliferator Response Element |
| PS | Phosphatidylserine |
| PUFA | Polyunsaturated Fatty Acid |
| qPCR | Quantitative Polymerase Chain Reaction |
| RC | Relative Content |
| RT | Retention Time |
| RXR | Retinoid X Receptor |
| SCAP | Sterol Regulatory Element-Binding Protein Cleavage-Activating Protein |
| SCD1 | Stearoyl-CoA Desaturase 1 |
| SD | Standard Deviation |
| SDHA | Succinate Dehydrogenase Complex Flavoprotein Subunit A |
| SFA | Saturated Fatty Acid |
| SQDG | Sulfoquinovosyl Diacylglycerol |
| SREBF1 | Sterol Regulatory Element Binding Transcription Factor 1 |
| SREBF2 | Sterol Regulatory Element Binding Transcription Factor 2 |
| SREBP | Sterol Regulatory Element-Binding Protein |
| TG | Triacylglycerol |
| UI | Unsaturation Index |
| UV | Ultraviolet |
| YWHAZ | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Zeta |
References
- Dixon, E.D.; Nardo, A.D.; Claudel, T.; Trauner, M. The Role of Lipid Sensing Nuclear Receptors (PPARs and LXR) and Metabolic Lipases in Obesity, Diabetes and NAFLD. Genes 2021, 12, 645. [Google Scholar] [CrossRef]
- Hong, C.; Tontonoz, P. Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr. Opin. Genet. Dev. 2008, 18, 461–467. [Google Scholar] [CrossRef]
- Monsalve, F.A.; Pyarasani, R.D.; Delgado-Lopez, F.; Moore-Carrasco, R. Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediat. Inflamm. 2013, 2013, 549627. [Google Scholar] [CrossRef]
- Perl, D.P. Neuropathology of Alzheimer’s disease. Mt. Sinai J. Med. 2010, 77, 32–42. [Google Scholar] [CrossRef]
- Swerdlow, R.H. Pathogenesis of Alzheimer’s disease. Clin. Interv. Aging 2007, 2, 347–359. [Google Scholar]
- Luchsinger, J.A.; Tang, M.X.; Shea, S.; Mayeux, R. Hyperinsulinemia and risk of Alzheimer disease. Neurology 2004, 63, 1187–1192. [Google Scholar] [CrossRef]
- Pierrot, N.; Ris, L.; Stancu, I.C.; Doshina, A.; Ribeiro, F.; Tyteca, D.; Baugé, E.; Lalloyer, F.; Malong, L.; Schakman, O.; et al. Sex-regulated gene dosage effect of PPARα on synaptic plasticity. Life Sci. Alliance 2019, 2, e201800262. [Google Scholar] [CrossRef]
- Vanmierlo, T.; Rutten, K.; Dederen, J.; Bloks, V.W.; van Vark-van der Zee, L.C.; Kuipers, F.; Kiliaan, A.; Blokland, A.; Sijbrands, E.J.G. Liver X receptor activation restores memory in aged AD mice without reducing amyloid. Neurobiol. Aging 2011, 32, 1262–1272. [Google Scholar] [CrossRef]
- Medrano-Jiménez, E.; Jiménez-Ferrer Carrillo, I.; Pedraza-Escalona, M.; Ramírez-Serrano, C.E.; Álvarez-Arellano, L.; Cortés-Mendoza, J.; Herrera-Ruiz, M.; Jiménez-Ferrer, E.; Zamilpa, A.; Tortoriello, J.; et al. Malva parviflora extract ameliorates the deleterious effects of a high fat diet on the cognitive deficit in a mouse model of Alzheimer’s disease by restoring microglial function via a PPAR-γ-dependent mechanism. J. Neuroinflamm. 2019, 16, 143. [Google Scholar] [CrossRef]
- Burns, M.P.; Vardanian, L.; Pajoohesh-Ganji, A.; Wang, L.; Cooper, M.; Harris, D.C.; Duff, K.; Rebeck, G.W. The effects of ABCA1 on cholesterol efflux and Abeta levels in vitro and in vivo. J. Neurochem. 2006, 98, 792–800. [Google Scholar] [CrossRef]
- Koldamova, R.P.; Lefterov, I.M.; Staufenbiel, M.; Wolfe, D.; Huang, S.; Glorioso, J.C.; Walter, M.; Roth, M.G.; Lazo, J.S. The liver X receptor ligand T0901317 decreases amyloid beta production in vitro and in a mouse model of Alzheimer’s disease. J. Biol. Chem. 2005, 280, 4079–4088. [Google Scholar] [CrossRef]
- Lefterov, I.; Bookout, A.; Wang, Z.; Staufenbiel, M.; Mangelsdorf, D.; Koldamova, R. Expression profiling in APP23 mouse brain: Inhibition of Abeta amyloidosis and inflammation in response to LXR agonist treatment. Mol. Neurodegener. 2007, 2, 20. [Google Scholar] [CrossRef]
- Riddell, D.R.; Zhou, H.; Comery, T.A.; Kouranova, E.; Lo, C.F.; Warwick, H.K.; Ring, R.H.; Kirksey, Y.; Aschmies, S.; Xu, J.; et al. The LXR agonist TO901317 selectively lowers hippocampal Abeta42 and improves memory in the Tg2576 mouse model of Alzheimer’s disease. Mol. Cell. Neurosci. 2007, 34, 621–628. [Google Scholar] [CrossRef]
- Carter, A.Y.; Letronne, F.; Fitz, N.F.; Mounier, A.; Wolfe, C.M.; Nam, K.N.; Reeves, V.L.; Kamboh, H.; Lefterov, I.; Koldamova, R. Liver X receptor agonist treatment significantly affects phenotype and transcriptome of APOE3 and APOE4 Abca1 haplo-deficient mice. PLoS ONE 2017, 12, e0172161. [Google Scholar] [CrossRef]
- Donkin, J.J.; Stukas, S.; Hirsch-Reinshagen, V.; Namjoshi, D.; Wilkinson, A.; May, S.; Chan, J.; Fan, J.; Collins, J.; Wellington, C.L. ATP-binding cassette transporter A1 mediates the beneficial effects of the liver X receptor agonist GW3965 on object recognition memory and amyloid burden in amyloid precursor protein/presenilin 1 mice. J. Biol. Chem. 2010, 285, 34144–34154. [Google Scholar] [CrossRef]
- Wesson, D.W.; Borkowski, A.H.; Landreth, G.E.; Nixon, R.A.; Levy, E.; Wilson, D.A. Sensory network dysfunction, behavioral impairments, and their reversibility in an Alzheimer’s β-amyloidosis mouse model. J. Neurosci. 2011, 31, 15962–15971. [Google Scholar] [CrossRef]
- Xu, X.; Xiao, X.; Yan, Y.; Zhang, T. Activation of liver X receptors prevents emotional and cognitive dysfunction by suppressing microglial M1-polarization and restoring synaptic plasticity in the hippocampus of mice. Brain Behav. Immun. 2021, 94, 111–124. [Google Scholar] [CrossRef]
- Jiang, Q.; Lee, C.D.; Mandrekar, S.; Wilkinson, B.; Cramer, P.; Zelcer, N.; Mann, K.; Lamb, B.; Willson, T.M. ApoE promotes the proteolytic degradation of Abeta. Neuron 2008, 58, 681–693. [Google Scholar] [CrossRef]
- Navas Guimaraes, M.E.; Lopez-Blanco, R.; Correa, J.; Fernandez-Villamarin, M.; Bistué, M.B.; Martino-Adami, P.; Morelli, L.; Kumar, V.; Wempe, M.F.; Cuello, A.C.; et al. Liver X Receptor Activation with an Intranasal Polymer Therapeutic Prevents Cognitive Decline without Altering Lipid Levels. ACS Nano 2021, 15, 4678–4687. [Google Scholar] [CrossRef]
- Grefhorst, A.; Elzinga, B.M.; Voshol, P.J.; Plo, T.; Kok, T.; Bloks, V.W.; van der Sluijs, F.H.; Havekes, L.M.; Romijn, J.A.; Verkade, H.J.; et al. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 2002, 277, 34182–34190. [Google Scholar] [CrossRef]
- Repa, J.J.; Liang, G.; Ou, J.; Bashmakov, Y.; Lobaccaro, J.M.; Shimomura, I.; Shan, B.; Brown, M.S.; Goldstein, J.L.; Mangelsdorf, D.J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000, 14, 2819–2830. [Google Scholar] [CrossRef]
- Schultz, J.R.; Tu, H.; Luk, A.; Repa, J.J.; Medina, J.C.; Li, L.; Schwendner, S.; Wang, S.; Thoolen, M.; Mangelsdorf, D.J.; et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000, 14, 2831–2838. [Google Scholar] [CrossRef]
- Vanmierlo, T.; Bogie, J.F.; Mailleux, J.; Vanmol, J.; Lütjohann, D.; Mulder, M.; Hendriks, J.J. Plant sterols: Friend or foe in CNS disorders? Prog. Lipid Res. 2015, 58, 26–39. [Google Scholar] [CrossRef]
- Bogie, J.; Hoeks, C.; Schepers, M.; Tiane, A.; Cuypers, A.; Leijten, F.; Chintapakorn, Y.; Suttiyut, T.; Pornpakakul, S.; Struik, D.; et al. Dietary Sargassum fusiforme improves memory and reduces amyloid plaque load in an Alzheimer’s disease mouse model. Sci. Rep. 2019, 9, 4908. [Google Scholar] [CrossRef]
- Zhan, N.; Wang, B.; Martens, N.; Liu, Y.; Zhao, S.; Voortman, G.; van Rooij, J.; Leijten, F.; Vanmierlo, T.; Kuipers, F.; et al. Identification of Side Chain Oxidized Sterols as Novel Liver X Receptor Agonists with Therapeutic Potential in the Treatment of Cardiovascular and Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 1290. [Google Scholar] [CrossRef]
- Martens, N.; Schepers, M.; Zhan, N.; Leijten, F.; Voortman, G.; Tiane, A.; Rombaut, B.; Poisquet, J.; Sande, N.V.; Kerksiek, A.; et al. 24(S)-Saringosterol Prevents Cognitive Decline in a Mouse Model for Alzheimer’s Disease. Mar. Drugs 2021, 19, 190. [Google Scholar] [CrossRef]
- Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar]
- Kliewer, S.A.; Sundseth, S.S.; Jones, S.A.; Brown, P.J.; Wisely, G.B.; Koble, C.S.; Devchand, P.; Wahli, W.; Willson, T.M.; Lenhard, J.M.; et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA 1997, 94, 4318–4323. [Google Scholar] [CrossRef]
- Dussault, I.; Forman, B.M. Prostaglandins and fatty acids regulate transcriptional signaling via the peroxisome proliferator activated receptor nuclear receptors. Prostaglandins Other Lipid Mediat. 2000, 62, 1–13. [Google Scholar] [CrossRef]
- Wang, L.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Blunder, M.; Liu, X.; Malainer, C.; Blazevic, T.; Schwaiger, S.; Rollinger, J.M.; Heiss, E.H.; et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): A review. Biochem. Pharmacol. 2014, 92, 73–89. [Google Scholar] [CrossRef]
- Wójtowicz, S.; Strosznajder, A.K.; Jeżyna, M.; Strosznajder, J.B. The Novel Role of PPAR Alpha in the Brain: Promising Target in Therapy of Alzheimer’s Disease and Other Neurodegenerative Disorders. Neurochem. Res. 2020, 45, 972–988. [Google Scholar] [CrossRef]
- Staels, B.; Dallongeville, J.; Auwerx, J.; Schoonjans, K.; Leitersdorf, E.; Fruchart, J.C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 1998, 98, 2088–2093. [Google Scholar] [CrossRef]
- Heneka, M.T.; Landreth, G.E. PPARs in the brain. Biochim. Biophys. Acta 2007, 1771, 1031–1045. [Google Scholar] [CrossRef]
- Pedersen, W.A.; McMillan, P.J.; Kulstad, J.J.; Leverenz, J.B.; Craft, S.; Haynatzki, G.R. Rosiglitazone attenuates learning and memory deficits in Tg2576 Alzheimer mice. Exp. Neurol. 2006, 199, 265–273. [Google Scholar] [CrossRef]
- De Felice, F.G.; Lourenco, M.V.; Ferreira, S.T. How does brain insulin resistance develop in Alzheimer’s disease? Alzheimer’s Dement. 2014, 10 (Suppl. 1), S26–S32. [Google Scholar] [CrossRef]
- Santos, M.J.; Quintanilla, R.A.; Toro, A.; Grandy, R.; Dinamarca, M.C.; Godoy, J.A.; Inestrosa, N.C. Peroxisomal proliferation protects from beta-amyloid neurodegeneration. J. Biol. Chem. 2005, 280, 41057–41068. [Google Scholar] [CrossRef]
- Esmaeili, M.A.; Yadav, S.; Gupta, R.K.; Waggoner, G.R.; Deloach, A.; Calingasan, N.Y.; Beal, M.F.; Kiaei, M. Preferential PPAR-α activation reduces neuroinflammation, and blocks neurodegeneration in vivo. Hum. Mol. Genet. 2016, 25, 317–327. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular Actions of PPARα in Lipid Metabolism and Inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef]
- Inestrosa, N.C.; Carvajal, F.J.; Zolezzi, J.M.; Tapia-Rojas, C.; Serrano, F.; Karmelic, D.; Toledo, E.M.; Toro, A.; Toro, J.; Santos, M.J. Peroxisome proliferators reduce spatial memory impairment, synaptic failure, and neurodegeneration in brains of a double transgenic mice model of Alzheimer’s disease. J. Alzheimer’s Dis. 2013, 33, 941–959. [Google Scholar] [CrossRef]
- Kummer, M.P.; Heneka, M.T. PPARs in Alzheimer’s Disease. PPAR Res. 2008, 2008, 403896. [Google Scholar] [CrossRef]
- Yan, Q.; Zhang, J.; Liu, H.; Babu-Khan, S.; Vassar, R.; Biere, A.L.; Citron, M.; Landreth, G. Anti-inflammatory drug therapy alters beta-amyloid processing and deposition in an animal model of Alzheimer’s disease. J. Neurosci. 2003, 23, 7504–7509. [Google Scholar] [CrossRef]
- Sastre, M.; Dewachter, I.; Landreth, G.E.; Willson, T.M.; Klockgether, T.; van Leuven, F.; Heneka, M.T. Nonsteroidal anti-inflammatory drugs and peroxisome proliferator-activated receptor-gamma agonists modulate immunostimulated processing of amyloid precursor protein through regulation of beta-secretase. J. Neurosci. 2003, 23, 9796–9804. [Google Scholar] [CrossRef]
- Luna-Medina, R.; Cortes-Canteli, M.; Alonso, M.; Santos, A.; Martínez, A.; Perez-Castillo, A. Regulation of inflammatory response in neural cells in vitro by thiadiazolidinones derivatives through peroxisome proliferator-activated receptor gamma activation. J. Biol. Chem. 2005, 280, 21453–21462. [Google Scholar] [CrossRef]
- d’Angelo, M.; Castelli, V.; Catanesi, M.; Antonosante, A.; Dominguez-Benot, R.; Ippoliti, R.; Benedetti, E.; Cimini, A. PPARγ and Cognitive Performance. Int. J. Mol. Sci. 2019, 20, 5068. [Google Scholar] [CrossRef]
- Luo, R.; Su, L.Y.; Li, G.; Yang, J.; Liu, Q.; Yang, L.X.; Zhang, D.F.; Zhou, H.; Xu, M.; Fan, Y.; et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 2020, 16, 52–69. [Google Scholar] [CrossRef]
- Chandra, S.; Roy, A.; Jana, M.; Pahan, K. Cinnamic acid activates PPARα to stimulate Lysosomal biogenesis and lower Amyloid plaque pathology in an Alzheimer’s disease mouse model. Neurobiol. Dis. 2019, 124, 379–395. [Google Scholar] [CrossRef]
- Chandra, S.; Pahan, K. Gemfibrozil, a Lipid-Lowering Drug, Lowers Amyloid Plaque Pathology and Enhances Memory in a Mouse Model of Alzheimer’s Disease via Peroxisome Proliferator-Activated Receptor α. J. Alzheimer’s Dis. Rep. 2019, 3, 149–168. [Google Scholar] [CrossRef]
- Escribano, L.; Simón, A.M.; Gimeno, E.; Cuadrado-Tejedor, M.; Lopez de Maturana, R.; García-Osta, A.; Ricobaraza, A.; Pérez-Mediavilla, A.; Del Río, J.; Frechilla, D. Rosiglitazone rescues memory impairment in Alzheimer’s transgenic mice: Mechanisms involving a reduced amyloid and tau pathology. Neuropsychopharmacology 2010, 35, 1593–1604. [Google Scholar] [CrossRef]
- Denner, L.A.; Rodriguez-Rivera, J.; Haidacher, S.J.; Jahrling, J.B.; Carmical, J.R.; Hernandez, C.M.; Zhao, Y.; Sadygov, R.G.; Starkey, J.M.; Spratt, H.; et al. Cognitive enhancement with rosiglitazone links the hippocampal PPARγ and ERK MAPK signaling pathways. J. Neurosci. 2012, 32, 16725–16735a. [Google Scholar] [CrossRef]
- Sato, T.; Hanyu, H.; Hirao, K.; Kanetaka, H.; Sakurai, H.; Iwamoto, T. Efficacy of PPAR-γ agonist pioglitazone in mild Alzheimer disease. Neurobiol. Aging 2011, 32, 1626–1633. [Google Scholar] [CrossRef]
- Hanyu, H.; Sato, T.; Kiuchi, A.; Sakurai, H.; Iwamoto, T. Pioglitazone improved cognition in a pilot study on patients with Alzheimer’s disease and mild cognitive impairment with diabetes mellitus. J. Am. Geriatr. Soc. 2009, 57, 177–179. [Google Scholar] [CrossRef]
- Risner, M.E.; Saunders, A.M.; Altman, J.F.; Ormandy, G.C.; Craft, S.; Foley, I.M.; Zvartau-Hind, M.E.; Hosford, D.A.; Roses, A.D. Efficacy of rosiglitazone in a genetically defined population with mild-to-moderate Alzheimer’s disease. Pharmacogenom. J. 2006, 6, 246–254. [Google Scholar] [CrossRef]
- Watson, G.S.; Cholerton, B.A.; Reger, M.A.; Baker, L.D.; Plymate, S.R.; Asthana, S.; Fishel, M.A.; Kulstad, J.J.; Green, P.S.; Cook, D.G.; et al. Preserved cognition in patients with early Alzheimer disease and amnestic mild cognitive impairment during treatment with rosiglitazone: A preliminary study. Am. J. Geriatr. Psychiatry 2005, 13, 950–958. [Google Scholar] [CrossRef]
- Sagheddu, C.; Melis, M.; Muntoni, A.L.; Pistis, M. Repurposing Peroxisome Proliferator-Activated Receptor Agonists in Neurological and Psychiatric Disorders. Pharmaceuticals 2021, 14, 1025. [Google Scholar] [CrossRef]
- Lütjohann, D.; Brzezinka, A.; Barth, E.; Abramowski, D.; Staufenbiel, M.; von Bergmann, K.; Beyreuther, K.; Multhaup, G.; Bayer, T.A. Profile of cholesterol-related sterols in aged amyloid precursor protein transgenic mouse brain. J. Lipid Res. 2002, 43, 1078–1085. [Google Scholar] [CrossRef]
- Loera-Valencia, R.; Goikolea, J.; Parrado-Fernandez, C.; Merino-Serrais, P.; Maioli, S. Alterations in cholesterol metabolism as a risk factor for developing Alzheimer’s disease: Potential novel targets for treatment. J. Steroid Biochem. Mol. Biol. 2019, 190, 104–114. [Google Scholar] [CrossRef]
- Yin, M.; Chen, M.; Matsuoka, R.; Song, X.; Xi, Y.; Zhang, L.; Wang, X. UHPLC-Q-Exactive Orbitrap MS/MS based untargeted lipidomics reveals fatty acids and lipids profiles in different parts of capelin (Mallotus villosus). J. Food Compos. Anal. 2023, 116, 105096. [Google Scholar] [CrossRef]
- Ma, W.-P.; Yin, S.-N.; Chen, J.-P.; Geng, X.-C.; Liu, M.-F.; Li, H.-H.; Liu, M.; Liu, H.-B. Stimulating the hematopoietic effect of simulated digestive product of fucoidan from Sargassum fusiforme on cyclophosphamide-induced hematopoietic damage in mice and its protective mechanisms based on serum lipidomics. Mar. Drugs 2022, 20, 201. [Google Scholar] [CrossRef]
- Zwarts, I.; van Zutphen, T.; Kruit, J.K.; Liu, W.; Oosterveer, M.H.; Verkade, H.J.; Uhlenhaut, N.H.; Jonker, J.W. Identification of the fructose transporter GLUT5 (SLC2A5) as a novel target of nuclear receptor LXR. Sci. Rep. 2019, 9, 9299. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Dixon, W.J. Analysis of extreme values. Ann. Math. Stat. 1950, 21, 488–506. [Google Scholar] [CrossRef]
- Dixon, W.J. Ratios involving extreme values. Ann. Math. Stat. 1951, 22, 68–78. [Google Scholar] [CrossRef]
- Yan, Y.; Niu, Z.; Wang, B.; Zhao, S.; Sun, C.; Wu, Y.; Li, Y.; Ying, H.; Liu, H. Saringosterol from sargassum fusiforme modulates cholesterol metabolism and alleviates atherosclerosis in ApoE-deficient mice. Mar. Drugs 2021, 19, 485. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Akay, L.A.; Davila-Velderrain, J.; von Maydell, D.; Mathys, H.; Davidson, S.M.; Effenberger, A.; Chen, C.Y.; Maner-Smith, K.; Hajjar, I.; et al. APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes. Nature 2022, 611, 769–779. [Google Scholar] [CrossRef]
- Varma, V.R.; Büşra Lüleci, H.; Oommen, A.M.; Varma, S.; Blackshear, C.T.; Griswold, M.E.; An, Y.; Roberts, J.A.; O’Brien, R.; Pletnikova, O.; et al. Abnormal brain cholesterol homeostasis in Alzheimer’s disease-a targeted metabolomic and transcriptomic study. NPJ Aging Mech. Dis. 2021, 7, 11. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Q. Cholesterol metabolism and homeostasis in the brain. Protein Cell 2015, 6, 254–264. [Google Scholar] [CrossRef]
- Sodero, A.O. 24S-hydroxycholesterol: Cellular effects and variations in brain diseases. J. Neurochem. 2021, 157, 899–918. [Google Scholar] [CrossRef]
- Mitchell, R.W.; Hatch, G.M. Fatty acid transport into the brain: Of fatty acid fables and lipid tails. Prostaglandins Leukot. Essent. Fat. Acids 2011, 85, 293–302. [Google Scholar] [CrossRef]
- Kim, J.; Yoon, H.; Basak, J.; Kim, J. Apolipoprotein E in synaptic plasticity and Alzheimer’s disease: Potential cellular and molecular mechanisms. Mol. Cells 2014, 37, 767–776. [Google Scholar] [CrossRef]
- Koldamova, R.; Fitz, N.F.; Lefterov, I. ATP-binding cassette transporter A1: From metabolism to neurodegeneration. Neurobiol. Dis. 2014, 72, 13–21. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Das, S.; Hyman, B.T. APOE and Alzheimer’s disease: Advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021, 20, 68–80. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Sun, L.P.; Kwon, H.J.; Brown, M.S.; Goldstein, J.L. Direct binding of cholesterol to the purified membrane region of SCAP: Mechanism for a sterol-sensing domain. Mol. Cell 2004, 15, 259–268. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Ikeda, Y.; Kwon, H.J.; Brown, M.S.; Goldstein, J.L. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc. Natl. Acad. Sci. USA 2007, 104, 6511–6518. [Google Scholar] [CrossRef]
- Roy, A.; Jana, M.; Corbett, G.T.; Ramaswamy, S.; Kordower, J.H.; Gonzalez, F.J.; Pahan, K. Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor α. Cell Rep. 2013, 4, 724–737. [Google Scholar] [CrossRef]
- Xu, H.E.; Lambert, M.H.; Montana, V.G.; Parks, D.J.; Blanchard, S.G.; Brown, P.J.; Sternbach, D.D.; Lehmann, J.M.; Wisely, G.B.; Willson, T.M.; et al. Molecular recognition of fatty acids by peroxisome proliferator–activated receptors. Mol. Cell 1999, 3, 397–403. [Google Scholar] [CrossRef]
- Ricote, M.; Li, A.C.; Willson, T.M.; Kelly, C.J.; Glass, C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature 1998, 391, 79–82. [Google Scholar] [CrossRef]
- Tugal, D.; Liao, X.; Jain, M.K. Transcriptional control of macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1135–1144. [Google Scholar] [CrossRef]
- Thomas, D.G.; Doran, A.C.; Fotakis, P.; Westerterp, M.; Antonson, P.; Jiang, H.; Jiang, X.-C.; Gustafsson, J.-Å.; Tabas, I.; Tall, A.R. LXR suppresses inflammatory gene expression and neutrophil migration through cis-repression and cholesterol efflux. Cell Rep. 2018, 25, 3774–3785.e4. [Google Scholar] [CrossRef]
- Jesse, S.; Steinacker, P.; Cepek, L.; von Arnim, C.A.F.; Tumani, H.; Lehnert, S.; Kretzschmar, H.A.; Baier, M.; Otto, M. Glial fibrillary acidic protein and protein S-100B: Different concentration pattern of glial proteins in cerebrospinal fluid of patients with Alzheimer’s disease and Creutzfeldt-Jakob disease. J. Alzheimer’s Dis. 2009, 17, 541–551. [Google Scholar] [CrossRef]
- Elahi, F.M.; Casaletto, K.B.; La Joie, R.; Walters, S.M.; Harvey, D.; Wolf, A.; Edwards, L.; Rivera-Contreras, W.; Karydas, A.; Cobigo, Y.; et al. Plasma biomarkers of astrocytic and neuronal dysfunction in early-and late-onset Alzheimer’s disease. Alzheimer’s Dement. 2019, 16, 681–695. [Google Scholar] [CrossRef]
- Oeckl, P.; Halbgebauer, S.; Anderl-Straub, S.; Steinacker, P.; Huss, A.M.; Neugebauer, H.; von Arnim, C.A.F.; Diehl-Schmid, J.; Grimmer, T.; Kornhuber, J.; et al. Glial fibrillary acidic protein in serum is increased in Alzheimer’s disease and correlates with cognitive impairment. J. Alzheimer’s Dis. 2019, 67, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Liberato, M.V.; Nascimento, A.S.; Ayers, S.D.; Lin, J.Z.; Cvoro, A.; Silveira, R.L.; Martínez, L.; Souza, P.C.; Saidemberg, D.; Deng, T.; et al. Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) γ activators and pan-PPAR partial agonists. PLoS ONE 2012, 7, e36297. [Google Scholar] [CrossRef] [PubMed]
- Göttlicher, M.; Widmark, E.; Li, Q.; Gustafsson, J.A. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 1992, 89, 4653–4657. [Google Scholar] [CrossRef] [PubMed]
- Venn-Watson, S.; Lumpkin, R.; Dennis, E.A. Efficacy of dietary odd-chain saturated fatty acid pentadecanoic acid parallels broad associated health benefits in humans: Could it be essential? Sci. Rep. 2020, 10, 8161. [Google Scholar] [CrossRef]
- Coccia, E.; Varricchio, E.; Vito, P.; Turchini, G.M.; Francis, D.S.; Paolucci, M. Fatty acid-specific alterations in leptin, PPARα, and CPT-1 gene expression in the rainbow trout. Lipids 2014, 49, 1033–1046. [Google Scholar] [CrossRef]
- Ren, B.; Thelen, A.P.; Peters, J.M.; Gonzalez, F.J.; Jump, D.B. Polyunsaturated Fatty Acid Suppression of Hepatic Fatty Acid Synthase and S14 Gene Expression Does Not Require Peroxisome Proliferator-activated Receptor α*. J. Biol. Chem. 1997, 272, 26827–26832. [Google Scholar] [CrossRef]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef]
- Yu, K.; Bayona, W.; Kallen, C.B.; Harding, H.P.; Ravera, C.P.; McMahon, G.; Brown, M.; Lazar, M.A. Differential Activation of Peroxisome Proliferator-activated Receptors by Eicosanoids (∗). J. Biol. Chem. 1995, 270, 23975–23983. [Google Scholar] [CrossRef]
- Hontecillas, R.; Wannemeulher, M.J.; Zimmerman, D.R.; Hutto, D.L.; Wilson, J.H.; Ahn, D.U.; Bassaganya-Riera, J. Nutritional regulation of porcine bacterial-induced colitis by conjugated linoleic acid. J. Nutr. 2002, 132, 2019–2027. [Google Scholar] [CrossRef]
- de Almeida, M.M.; Luquetti, S.C.; Sabarense, C.M.; Corrêa, J.O.; dos Reis, L.G.; Conceição, E.P.; Lisboa, P.C.; de Moura, E.G.; Gameiro, J.; da Gama, M.A.; et al. Butter naturally enriched in cis-9, trans-11 CLA prevents hyperinsulinemia and increases both serum HDL cholesterol and triacylglycerol levels in rats. Lipids Health Dis. 2014, 13, 200. [Google Scholar] [CrossRef]
- Berger, J.; Moller, D.E. The mechanisms of action of PPARs. Annu. Rev. Med. 2002, 53, 409–435. [Google Scholar] [CrossRef]
- Yu, Y.; Correll, P.H.; Vanden Heuvel, J.P. Conjugated linoleic acid decreases production of pro-inflammatory products in macrophages: Evidence for a PPAR gamma-dependent mechanism. Biochim. Biophys. Acta 2002, 1581, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Marion-Letellier, R.; Butler, M.; Déchelotte, P.; Playford, R.J.; Ghosh, S. Comparison of cytokine modulation by natural peroxisome proliferator-activated receptor gamma ligands with synthetic ligands in intestinal-like Caco-2 cells and human dendritic cells--potential for dietary modulation of peroxisome proliferator-activated receptor gamma in intestinal inflammation. Am. J. Clin. Nutr. 2008, 87, 939–948. [Google Scholar] [PubMed]
- Salakhutdinov, N.F.; Laev, S.S. Triglyceride-lowering agents. Bioorganic Med. Chem. 2014, 22, 3551–3564. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.-H.; Kaya, T.; Waxman, D.J.; Vajda, S. Exploring the Binding Site Structure of the PPARγ Ligand-Binding Domain by Computational Solvent Mapping. Biochemistry 2005, 44, 1193–1209. [Google Scholar] [CrossRef]
- Altinoz, M.A.; Ozpinar, A. PPAR-δ and erucic acid in multiple sclerosis and Alzheimer’s Disease. Likely benefits in terms of immunity and metabolism. Int. Immunopharmacol. 2019, 69, 245–256. [Google Scholar] [CrossRef]
- Altinoz, M.A.; Ozpinar, A.; Ozpinar, A.; Hacker, E. Erucic acid, a nutritional PPARδ-ligand may influence Huntington’s disease pathogenesis. Metab. Brain Dis. 2020, 35, 1–9. [Google Scholar] [CrossRef]



| Gene | Gene Name | Primer Sequence |
|---|---|---|
| ABCA1 | ATP Binding Cassette Subfamily A Member 1 | F: TCTCTGTTCGGCTGAGCTAC |
| R: TGCAGAGGGCATGGCTTTAT | ||
| ABCG1 | ATP Binding Cassette Subfamily G Member 1 | F: GGTCGCTCCATCATTTGCAC |
| R: GCAGACTTTTCCCCGGTACA | ||
| ACACA | Acetyl-CoA Carboxylase Alpha | F: GGGTCAAGTCCTTCCTGCTC |
| R: GGACTGTCGAGTCACCTTAAGTA | ||
| ACTB | Actin Beta | F: CTCCCTGGAGAAGAGCTACG |
| R: GAAGGAAGGCTGGAAGAGTG | ||
| APOE | Apolipoprotein E | F: ACCCAGGAACTGAGGGC |
| R: CTCCTTGGACAGCCGTG | ||
| B2M | Beta-2-Microglobulin | F: CTCCGTGGCCTTAGCTGTG |
| R: TTTGGAGTACGCTGGATAGCCT | ||
| DHCR7 | 7-Dehydrocholesterol Reductase | F: TGGGCCAAGACTCCACCTAT |
| R: ACGTGTACAGAAGCACCTGG | ||
| DHCR24 | 24-Dehydrocholesterol Reductase | F: GTCTCACTACGTGTCGGGAA |
| R: CTCCACACGGACAATCTGTTTC | ||
| FASN | Fatty Acid Synthase | F: CACAGACGAGAGCACCTTTGA |
| R: CAGGTCTATGAGGCCTATCTGG | ||
| GFAP | Glial Fibrillary Acidic Protein | F: GGCCCGCCACTTGCA |
| R: GGGAATGGTGATCCGGTTCT | ||
| HMGCR | 3-Hydroxy-3-Methylglutaryl-CoA Reductase | F: GCAGGACCCCTTTGCTTAGA |
| R: GCACCTCCACCAAGACCTAT | ||
| HPRT1 | Hypoxanthine Phosphoribosyltransferase 1 | F: TGACACTGGCAAAACAATGCA |
| R: GGTCCTTTTCACCAGCAAGCT | ||
| SCD1 | Stearoyl-CoA Desaturase 1 | F: GCTGTCAAAGAGAAGGGGAGT |
| R: AGCCAGGTTTGTAGTACCTCCT | ||
| SDHA | Succinate Dehydrogenase Complex Flavoprotein Subunit A | F: TGGGAACAAGAGGGCATCTG |
| R: CCACCACTGCATCAAATTCATG | ||
| SREBF1 | Sterol Regulatory Element Binding Transcription Factor 1 | F: ACAGCCATGAAGACAGACGG |
| R: CAAGATGGTTCCGCCACTCA | ||
| SREBF2 | Sterol Regulatory Element Binding Transcription Factor 2 | F: GATCACGCCAACATTCAGCA |
| R: GACTTGAGGCTGAAGGACTTGAA | ||
| YWHAZ | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase Activation Protein Zeta | F: ACTTTTGGTACATTGTGGCTTCAA |
| R: CCGCCAGGACAAACCAGTAT |
| Seaweed Species | Crude Seaweed | Extract | ||
|---|---|---|---|---|
| Saringosterol (μg/mg DW) | Fucosterol (μg/mg DW) | Saringosterol (mM) | Fucosterol (mM) | |
| Alaria esculenta | 0.008 | 0.130 | 0.2 | 9.9 |
| Ascophyllum nodosum | 0.002 | 0.495 | 0.2 | 12.2 |
| Fucus vesiculosus | 0.034 | 0.407 | 0.7 | 13.0 |
| Himanthalia elongata | 0.018 | 0.771 | 1.8 | 7.3 |
| Saccharina latissima | 0.002 | 0.037 | 0.7 | 6.1 |
| Sargassum fusiforme | 0.026 | 0.209 | 1.1 | 7.0 |
| Sargassum muticum | 0.032 | 0.325 | 0.1 | 12.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martens, N.; Zhan, N.; Voortman, G.; Leijten, F.P.J.; van Rheenen, C.; van Leerdam, S.; Geng, X.; Huybrechts, M.; Liu, H.; Jonker, J.W.; et al. Activation of Liver X Receptors and Peroxisome Proliferator-Activated Receptors by Lipid Extracts of Brown Seaweeds: A Potential Application in Alzheimer’s Disease? Nutrients 2023, 15, 3004. https://doi.org/10.3390/nu15133004
Martens N, Zhan N, Voortman G, Leijten FPJ, van Rheenen C, van Leerdam S, Geng X, Huybrechts M, Liu H, Jonker JW, et al. Activation of Liver X Receptors and Peroxisome Proliferator-Activated Receptors by Lipid Extracts of Brown Seaweeds: A Potential Application in Alzheimer’s Disease? Nutrients. 2023; 15(13):3004. https://doi.org/10.3390/nu15133004
Chicago/Turabian StyleMartens, Nikita, Na Zhan, Gardi Voortman, Frank P. J. Leijten, Connor van Rheenen, Suzanne van Leerdam, Xicheng Geng, Michiel Huybrechts, Hongbing Liu, Johan W. Jonker, and et al. 2023. "Activation of Liver X Receptors and Peroxisome Proliferator-Activated Receptors by Lipid Extracts of Brown Seaweeds: A Potential Application in Alzheimer’s Disease?" Nutrients 15, no. 13: 3004. https://doi.org/10.3390/nu15133004
APA StyleMartens, N., Zhan, N., Voortman, G., Leijten, F. P. J., van Rheenen, C., van Leerdam, S., Geng, X., Huybrechts, M., Liu, H., Jonker, J. W., Kuipers, F., Lütjohann, D., Vanmierlo, T., & Mulder, M. T. (2023). Activation of Liver X Receptors and Peroxisome Proliferator-Activated Receptors by Lipid Extracts of Brown Seaweeds: A Potential Application in Alzheimer’s Disease? Nutrients, 15(13), 3004. https://doi.org/10.3390/nu15133004







