The Effect of Probiotic Bacteria on Composition and Metabolite Production of Faecal Microbiota Using In Vitro Batch Cultures
Abstract
1. Introduction
2. Methods
2.1. Preparation of Probiotic Strains
2.2. Faecal Sample Preparation
2.3. Batch Culture Fermentation
2.4. Preparation of Samples
2.4.1. Fluorescence In Situ Hybridisation with Flow Cytometry (Flow-FISH)
2.4.2. Gas Chromatography
2.4.3. Liquid Chromatography–Mass Spectroscopy
2.5. Statistical Analysis
3. Results
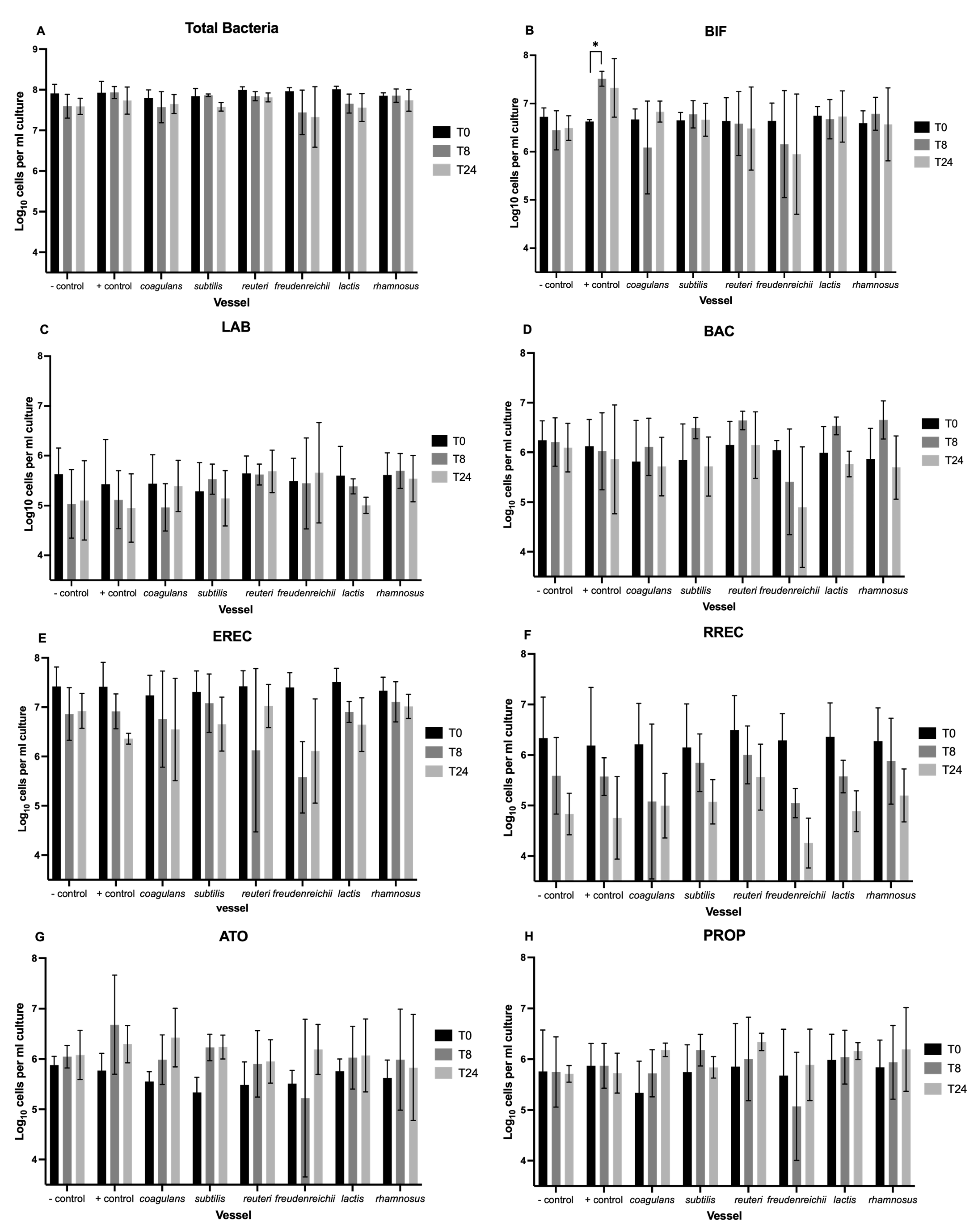
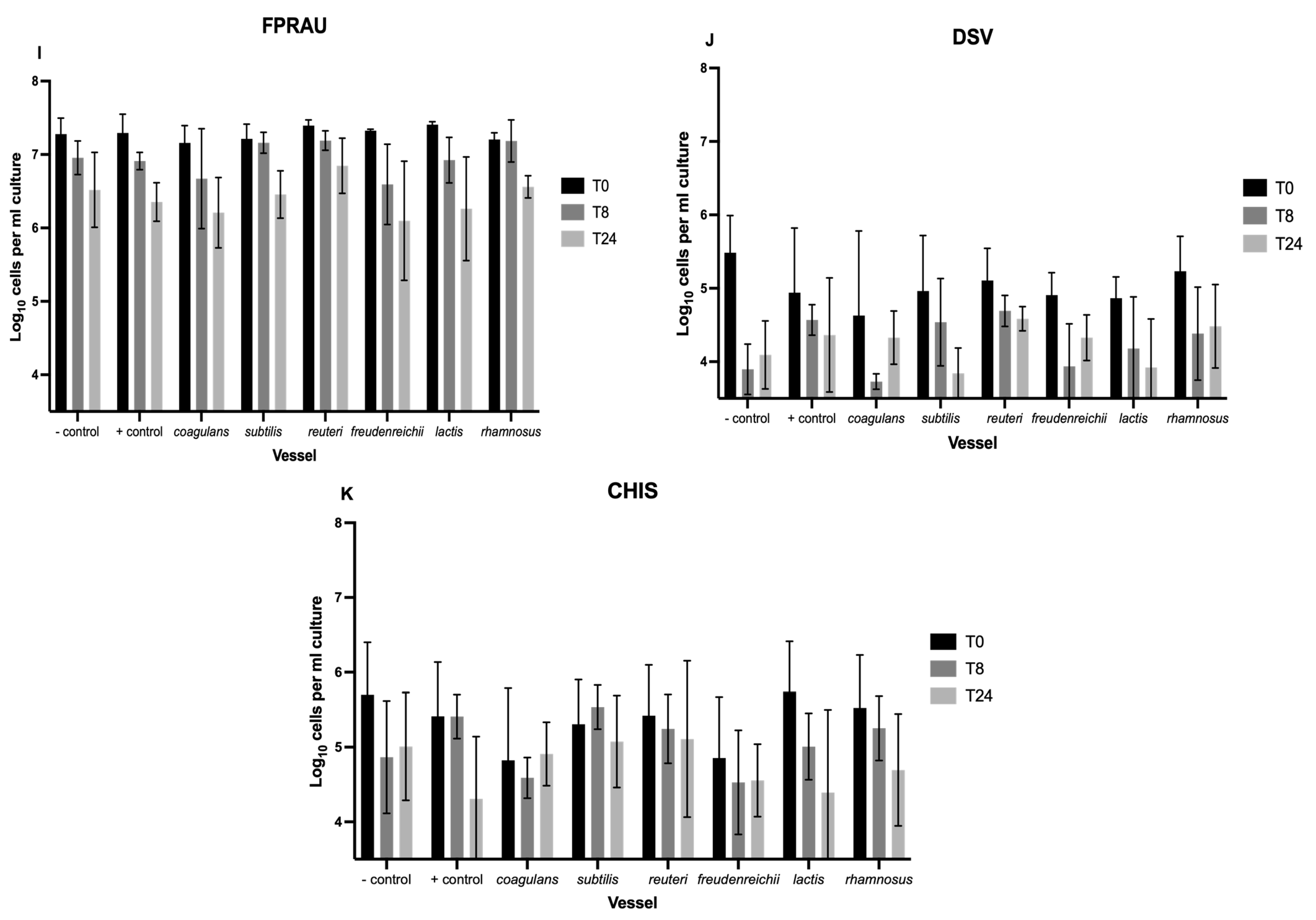
3.1. Enumeration of Bacteria with Flow-FISH
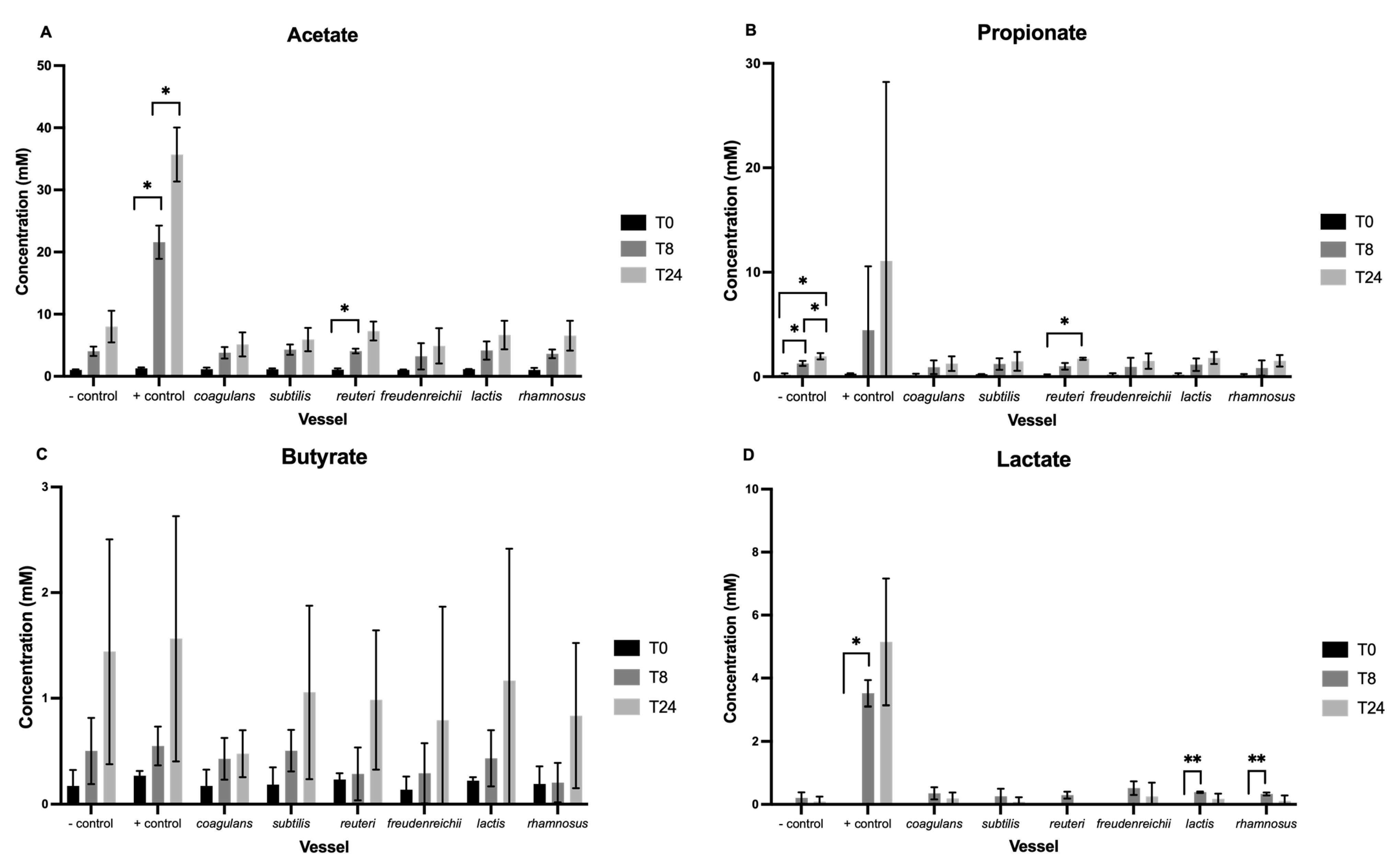
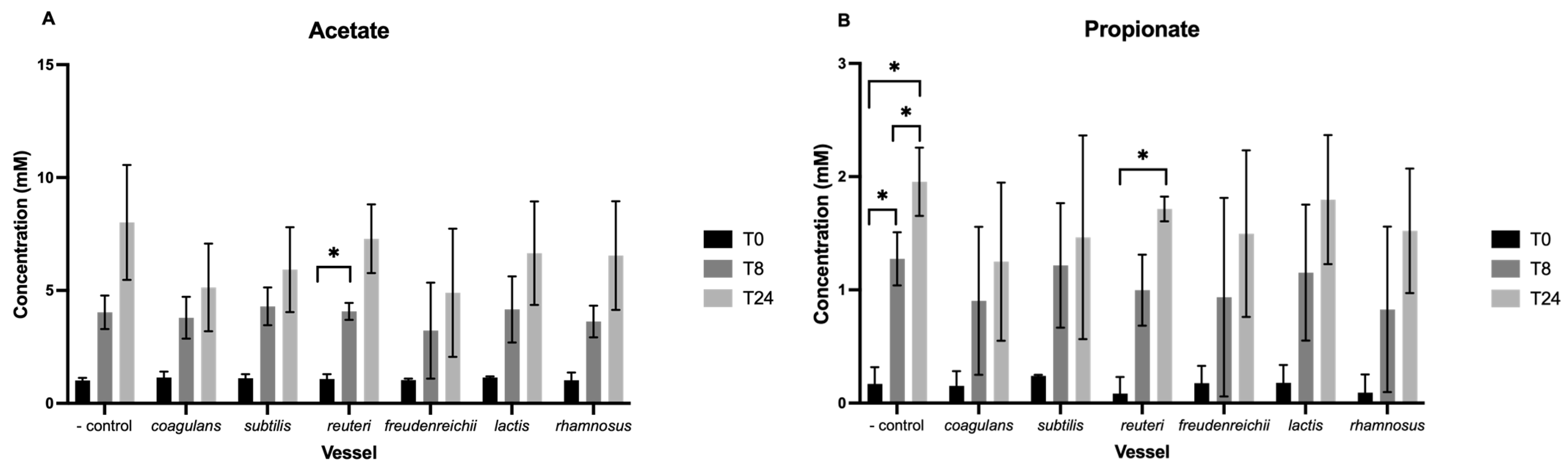
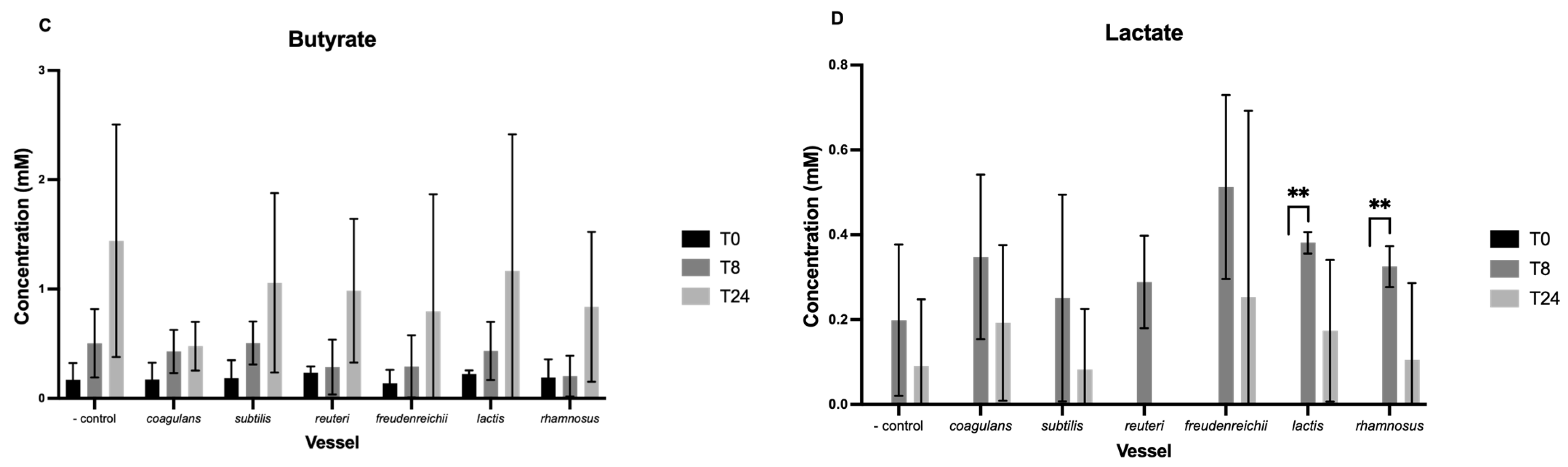
3.2. Short-Chain Fatty Acids
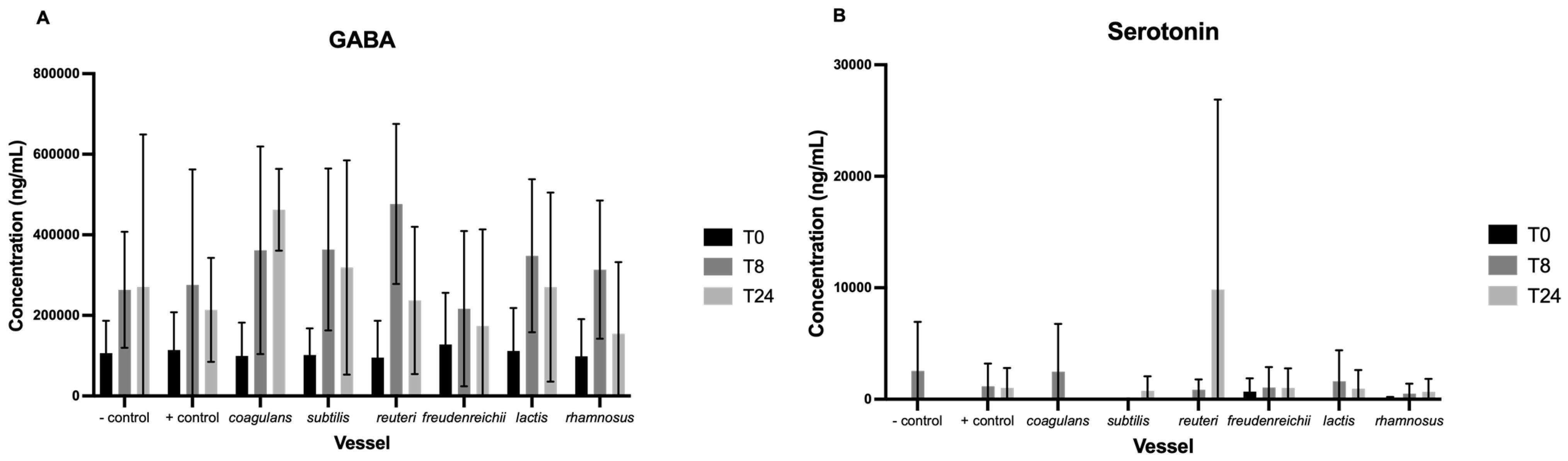
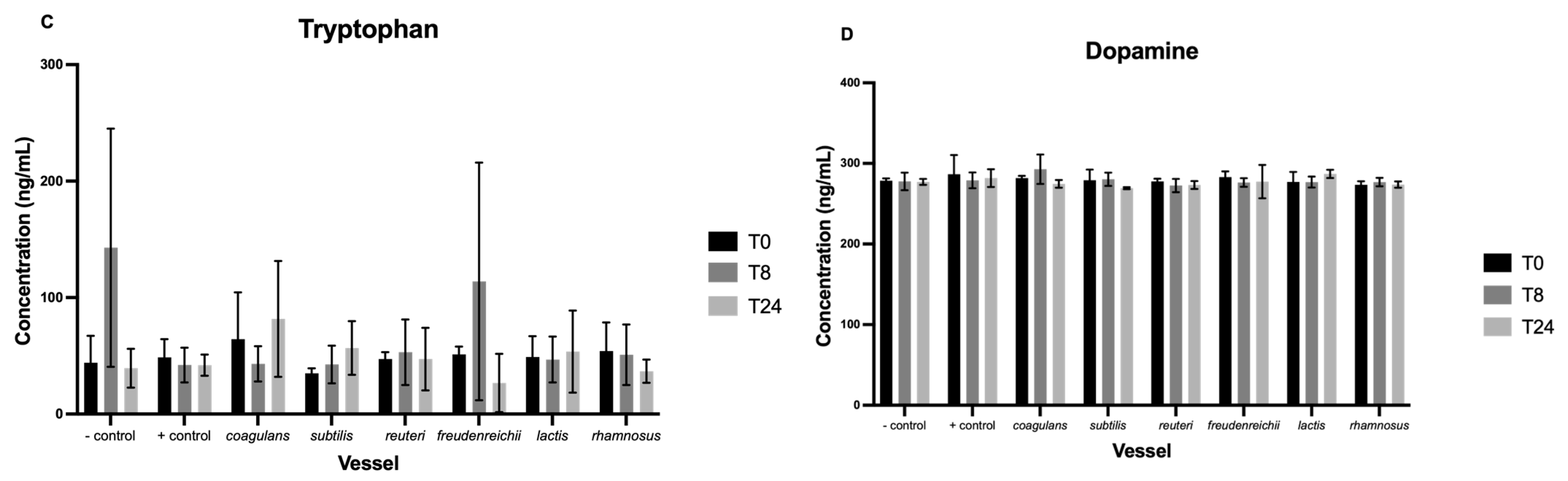
3.3. Neurotransmitters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nagpal, J.; Cryan, J.F. Microbiota-brain interactions: Moving toward mechanisms in model organisms. Neuron 2021, 109, 3930–3953. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.G.; Cryan, J.F.; Mayer, E.A. The microbiota-gut-brain axis: From motility to mood. Gastroenterology 2021, 160, 1486–1501. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Eastwood, J.; Walton, G.; Van Hemert, S.; Williams, C.; Lamport, D. The effect of probiotics on cognitive function across the human lifespan: A systematic review. Neurosci. Biobehav. Rev. 2021, 128, 311–327. [Google Scholar] [CrossRef]
- Mörkl, S.; Butler, M.I.; Holl, A.; Cryan, J.F.; Dinan, T.G. Probiotics and the microbiota-gut-brain axis: Focus on psychiatry. Curr. Nutr. Rep. 2020, 9, 171–182. [Google Scholar] [CrossRef]
- Rieder, R.; Wisniewski, P.J.; Alderman, B.L.; Campbell, S.C. Microbes and mental health: A review. Brain Behav. Immun. 2017, 66, 9–17. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of action of probiotics. Adv. Nutr. 2019, 10 (Suppl. S1), S49–S66. [Google Scholar] [CrossRef] [PubMed]
- Pokusaeva, K.; Johnson, C.; Luk, B.; Uribe, G.; Fu, Y.; Oezguen, N.; Matsunami, R.K.; Lugo, M.; Major, A.; Mori-Akiyama, Y.; et al. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol. Motil. 2017, 29, e12904. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Tao, F.; Hu, Y.; Li, Z.; Zhang, Y.; Deng, B. Positive effects of a Clostridium butyricum- based compound probiotic on growth performance, immune responses, intestinal morphology, hypothalamic neurotransmitters, and colonic microbiota in weaned piglets. Food Funct. 2019, 10, 2926–2934. [Google Scholar] [CrossRef]
- Leblhuber, F.; Steiner, K.; Schuetz, B.; Fuchs, D.; Gostner, J.M. Probiotic supplementation in patients with Alzheimer’s dementia-an explorative intervention study. Curr. Alzheimer Res. 2018, 15, 1106–1113. [Google Scholar] [CrossRef]
- Wang, Y.; Li, N.; Yang, J.-J.; Zhao, D.-M.; Chen, B.; Zhang, G.-Q.; Chen, S.; Cao, R.-F.; Yu, H.; Zhao, C.-Y.; et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol. Res. 2020, 157, 104784. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Jin, H.; Kwok, L.Y.; Sun, Z.; Liong, M.T.; Zhang, H. Probiotic consumption relieved human stress and anxiety symptoms possibly via modulating the neuroactive potential of the gut microbiota. Neurobiol. Stress 2021, 14, 100294. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Kaur, H.; Bose, C.; Mande, S.S. Tryptophan metabolism by gut microbiome and gut-brain-axis: An in silico analysis. Front. Neurosci. 2019, 13, 1365. [Google Scholar] [CrossRef]
- Cho, Y.R.; Chang, J.Y.; Chang, H.C. Production of gamma-Aminobutyric Acid (GABA) by Lactobacillus buchneri isolated from Kimchi and its neuroprotective effect on neuronal cells. J. Microbiol. Biotechnol. 2007, 17, 104–109. [Google Scholar]
- Barrett, E.; Ross, R.P.; O’Toole, P.W.; Fitzgerald, G.F.; Stanton, C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012, 113, 411–417. [Google Scholar] [CrossRef]
- Özoğul, F.; Kuley, E.; Özoğul, Y.; Özoğul, İ. The function of lactic acid bacteria on biogenic amines production by food-borne pathogens in arginine decarboxylase broth. Food Sci. Technol. Res. 2012, 18, 795–804. [Google Scholar] [CrossRef]
- Landete, J.M.; Ferrer, S.; Pardo, I. Biogenic amine production by lactic acid bacteria, acetic bacteria and yeast isolated from wine. Food Control 2007, 18, 1569–1574. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Botvinko, I.V.; Kudrin, V.S.; Oleskin, A.V. Detection of neurotransmitter amines in microorganisms with the use of high-performance liquid chromatography. Dokl. Biochem. 2000, 372, 115–117. [Google Scholar] [PubMed]
- Girvin, G.T.; Stevenson, J.W. Cell free “choline acetylase” from Lactobacillus plantarum. Can. J. Biochem. Physiol. 1954, 32, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Poveda, C.; Jenkins, P.E.; Walton, G.E. An in vitro approach to studying the microbial community and impact of pre and probiotics under anorexia nervosa related dietary restrictions. Nutrients 2021, 13, 4447. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short- chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [PubMed]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F., III; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Tzounis, X.; Vulevic, J.; Kuhnle, G.G.; George, T.; Leonczak, J.; Gibson, G.R.; Kwik-Uribe, C.; Spencer, J.P.E. Flavanol monomer-induced changes to the human faecal microflora. Br. J. Nutr. 2008, 99, 782–792. [Google Scholar] [CrossRef]
- Otaru, N.; Ye, K.; Mujezinovic, D.; Berchtold, L.; Constancias, F.; Cornejo, F.A.; Kryzstek, A.; de Wouters, T.; Braegger, C.; Lacroix, C.; et al. GABA production by human intestinal Bacteroides spp.: Prevalence, regulation, and role in acid stress tolerance. Front. Microbiol. 2021, 12, 656895. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Fanti, V.; Rodriguez-Sobstel, C.; Gibson, G.; Wijeyesekera, A.; Karatzas, K.A.; Chakrabarti, B. Gamma aminobutyric acid production by commercially available probiotic strains. J. Appl. Microbiol. 2022, 134, lxac066. [Google Scholar] [CrossRef]
- Grimaldi, R.; Cela, D.; Swann, J.R.; Vulevic, J.; Gibson, G.R.; Tzortzis, G.; Costabile, A. In vitro fermentation of B-GOS: Impact on faecal bacterial populations and metabolic activity in autistic and non-autistic children. FEMS Microbiol. Ecol. 2017, 93, fiw233. [Google Scholar] [CrossRef]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with rRNA- targeted oligonucleotide probes for flow cytometric identification ofmicroorganisms. Cytom. J. Int. Soc. Anal. Cytol. 1993, 14, 136–143. [Google Scholar]
- Daims, H.; Brühl, A.; Amann, R.; Schleifer, K.H.; Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Langendijk, P.S.; Schut, F.; Jansen, G.J.; Raangs, G.C.; Kamphuis, G.R.; Wilkinson, M.H.; Welling, G.W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus- specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 1995, 61, 3069–3075. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Elfferich, P.M.; Schut, F.; Welling, G.W. A 16S rRNA-targeted Probe for Detection of Lactobacilli and Enterococci in Faecal Samples by FluorescentIn SituHybridization. Microb. Ecol. Health Dis. 1999, 11, 3–12. [Google Scholar] [CrossRef]
- Manz, W.; Amann, R.; Ludwig, W.; Vancanneyt, M.; Schleifer, K.H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga- flavobacter-bacteroides in the natural environment. Microbiology 1996, 142, 1097–1106. [Google Scholar] [CrossRef]
- Franks, A.H.; Harmsen, H.J.; Raangs, G.C.; Jansen, G.J.; Schut, F.; Welling, G.W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group- specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 1998, 64, 3336–3345. [Google Scholar] [CrossRef]
- Walker, A.W.; Duncan, S.H.; McWilliam Leitch, E.C.; Child, M.W.; Flint, H.J. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 2005, 71, 3692–3700. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Wildeboer-Veloo, A.C.; Grijpstra, J.; Knol, J.; Degener, J.E.; Welling, G.W. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 2000, 66, 4523–4527. [Google Scholar] [CrossRef]
- Hold, G.L.; Schwiertz, A.; Aminov, R.I.; Blaut, M.; Flint, H.J. Oligonucleotide Probes That Detect Quantitatively Significant Groups of Butyrate-Producing Bacteria in Human Feces. Appl. Environ. Microbiol. 2003, 69, 4320–4324. [Google Scholar] [CrossRef] [PubMed]
- Devereux, R.; Kane, M.D.; Winfrey, J.; Stahl, D.A. Genus-and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 1992, 15, 601–609. [Google Scholar] [CrossRef]
- Richardson, A.J.; Calder, A.G.; Stewart, C.S.; Smith, A. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 1989, 9, 5–8. [Google Scholar] [CrossRef]
- Liu, Y.; Gibson, G.R.; Walton, G.E. An in vitro approach to study effects of prebiotics and probiotics on the faecal microbiota and selected immune parameters relevant to the elderly. PLoS ONE 2016, 11, e0162604. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.r-project.org/ (accessed on 2 April 2023).
- Meyer, D.; Stasse-Wolthuis, M. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur. J. Clin. Nutr. 2009, 63, 1277–1289. [Google Scholar] [CrossRef]
- van der Beek, C.M.; Canfora, E.E.; Kip, A.M.; Gorissen, S.H.; Damink, S.W.O.; van Eijk, H.M.; Holst, J.J.; Blaak, E.E.; Dejong, C.H.C.; Lanaerts, K. The prebiotic inulin improves substrate metabolism and promotes short-chain fatty acid production in overweight to obese men. Metabolism 2018, 87, 25–35. [Google Scholar] [CrossRef]
- Villegas, J.M.; Brown, L.; de Giori, G.S.; Hebert, E.M. Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. LWT—Food Sci. Technol. 2016, 67, 22–26. [Google Scholar] [CrossRef]
- Li, H.; Cao, Y. Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 2010, 39, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.J.; Reay, A.J.; Whitwood, A.C.; Fairlamb, I.J. A mild and selective Pd-mediated methodology for the synthesis of highly fluorescent 2-arylated tryptophans and tryptophan- containing peptides: A catalytic role for Pd 0 nanoparticles? Chem. Commun. 2014, 50, 3052–3054. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Hyland, N.P.; Cavanaugh, C.R.; Hornby, P.J. Emerging effects of tryptophan pathway metabolites and intestinal microbiota on metabolism and intestinal function. Amino Acids 2022, 54, 57–70. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.B.; Powley, T.L. Vagal innervation of intestines: Afferent pathways mapped with new en bloc horseradish peroxidase adaptation. Cell Tissue Res. 2007, 329, 221–230. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Kwon, S.; Yoo, I.K.; Lee, W.G.; Chang, H.N.; Chang, Y.K. High-rate continuous production of lactic acid by Lactobacillus rhamnosus in a two-stage membrane cell-recycle bioreactor. Biotechnol. Bioeng. 2001, 73, 25–34. [Google Scholar] [CrossRef]
- Song, A.A.L.; In, L.L.; Lim, S.H.E.; Rahim, R.A. A review on Lactococcus lactis: From food to factory. Microb. Cell Factories 2017, 16, 1–15. [Google Scholar]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015, 74, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Xiao, W.; Li, J.; Gao, X.; Yang, H.; Su, J.; Weng, R.; Gao, Y.; Ni, W.; Gu, Y. Involvement of the gut-brain axis in vascular depression via tryptophan metabolism: A benefit of short chain fatty acids. Exp. Neurol. 2022, 358, 114225. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Duncan, S.H.; Sheridan, P.O.; Walker, A.W.; Flint, H.J. Microbial lactate utilisation and the stability of the gut microbiome. Gut Microbiome 2022, 3, e3. [Google Scholar] [CrossRef]
- Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; Del Rio, B.; Alvarez, M.A. GABA-producing Lactococcus lactis strains isolated from camel’s milk as starters for the production of GABA- enriched cheese. Foods 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Tette, F.M.; Kwofie, S.K.; Wilson, M.D. Therapeutic anti-depressant potential of microbial GABA produced by Lactobacillus rhamnosus strains for GABAergic signaling restoration and inhibition of addiction-induced HPA axis hyperactivity. Curr. Issues Mol. Biol. 2022, 44, 1434–1451. [Google Scholar] [CrossRef]
- Bouhnik, Y.; Raskine, L.; Simoneau, G.; Vicaut, E.; Neut, C.; Flourié, B.; Brouns, F.; Bornet, F.R. The capacity of nondigestible carbohydrates to stimulate fecal bifidobacteria in healthy humans: A double-blind, randomized, placebo-controlled, parallel-group, dose-response relation study. Am. J. Clin. Nutr. 2004, 80, 1658–1664. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; Lange, K.; Havinga, R.; van Dijk, T.H.; Gerding, A.; van Eunen, K.; Müller, M.; Groen, A.K.; Hooiveld, G.J.; Bakker, B.M.; et al. Gut-derived short-chain fatty acids are vividly assimilated into host carbohydrates and lipids. Am. J. Physiol.—Gastrointest. Liver Physiol. 2013, 305, G900–G910. [Google Scholar] [CrossRef] [PubMed]
- Poveda, C.; Pereira, D.I.; Lewis, M.C.; Walton, G.E. The impact of low-level iron supplements on the faecal microbiota of irritable bowel syndrome and healthy donors using in vitro batch cultures. Nutrients 2020, 12, 3819. [Google Scholar] [CrossRef]
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl. Environ. Microbiol. 1988, 54, 2750–2755. [Google Scholar] [CrossRef]
| Probe | Sequence | Target Species |
|---|---|---|
| Non-Eub | ACTCCTAGGGAGGCAGA | Control probe for EUB338 [32] |
| Eub338I+ | GCTGCCTCCCGTAGGAGT | Most bacteria [33] |
| Eub338II+ | GCAGCCACCCGTAGGTGT | Planctomycetales [33] |
| Eub338III+ | GCTGCCACCCGTAGGTGT | Verrucomicrobialesm [33] |
| Bif164 | CATCCGGCATTACCACCC | Bifidobacterium spp. [34] |
| Lab158 | GGTATTAGCAYCTGTTTGGA | Lactobacillus and Enterococcus [35] |
| Bac303 | CCAATGTGGGGGACCTT | Bacteroidaceae, Prevotellaceae [36] |
| Erec482 | GCTTCTTAGTCARGTACCG | Most of the Clostridium coccoides-Eubacterium rectale group [37] |
| Rrec584 | TCAGACTTGCCGYACCGC | Roseburia [38] |
| Ato291 | GGTCGGTCTCTCAACCC | Atopobium cluster [39] |
| Prop853 | ATTGCGTTAACTCCGGCAC | Clostridium cluster IX [38] |
| Fprau655 | CGCCTACCTCTGCACTAC | Faecalibacterium prausnitzii and relatives [40] |
| DSV687 | TACGGATTTCACTCCT | Desulfovibrio genus [41] |
| Chis150 | TTATGCGGTATTAATCTYCCTTT | Most of the Clostridium histolyticum group [37] |
| Compound | Retention Time (Min) | Retention Time Window (Min) | Precursor Ion (m/z) | Product Ion (m/z) | Fragment Or (V) | Collision Energy (V) | Classification |
|---|---|---|---|---|---|---|---|
| GABA | 1.90 | 3 | 104 | 87 | 50 | 4 | Organic acid |
| 3 | 104 | 45 | 50 | 20 | |||
| Norepinephrine | 2.50 | 3 | 152 | 107 | 116 | 16 | Catecholamine |
| 152 | 77 | 116 | 30 | ||||
| Epinephrine | 4.60 | 3 | 184 | 166 | 70 | 8 | Catecholamine |
| 184 | 107 | 70 | 24 | ||||
| Dopamine | 7.00 | 3 | 154 | 137 | 75 | 8 | Catecholamine |
| 154 | 91 | 75 | 28 | ||||
| Serotonin | 9.70 | 3 | 177 | 160 | 45 | 4 | Amino acid derivative |
| 177 | 115 | 45 | 30 | ||||
| Kynurenic acid | 9.77 | 3 | 190 | 144 | 100 | 16 | Organic acid |
| 190 | 172 | 100 | 4 | ||||
| Tryptophan | 10.20 | 3 | 205 | 188 | 78 | 4 | Amino acid |
| 205 | 146 | 78 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eastwood, J.; van Hemert, S.; Poveda, C.; Elmore, S.; Williams, C.; Lamport, D.; Walton, G. The Effect of Probiotic Bacteria on Composition and Metabolite Production of Faecal Microbiota Using In Vitro Batch Cultures. Nutrients 2023, 15, 2563. https://doi.org/10.3390/nu15112563
Eastwood J, van Hemert S, Poveda C, Elmore S, Williams C, Lamport D, Walton G. The Effect of Probiotic Bacteria on Composition and Metabolite Production of Faecal Microbiota Using In Vitro Batch Cultures. Nutrients. 2023; 15(11):2563. https://doi.org/10.3390/nu15112563
Chicago/Turabian StyleEastwood, Jessica, Saskia van Hemert, Carlos Poveda, Stephen Elmore, Claire Williams, Daniel Lamport, and Gemma Walton. 2023. "The Effect of Probiotic Bacteria on Composition and Metabolite Production of Faecal Microbiota Using In Vitro Batch Cultures" Nutrients 15, no. 11: 2563. https://doi.org/10.3390/nu15112563
APA StyleEastwood, J., van Hemert, S., Poveda, C., Elmore, S., Williams, C., Lamport, D., & Walton, G. (2023). The Effect of Probiotic Bacteria on Composition and Metabolite Production of Faecal Microbiota Using In Vitro Batch Cultures. Nutrients, 15(11), 2563. https://doi.org/10.3390/nu15112563







