GC/MS-Based Analysis of Fatty Acids and Amino Acids in H460 Cells Treated with Short-Chain and Polyunsaturated Fatty Acids: A Highly Sensitive Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cultured Cells
2.3. Calibrators and Quality Control Samples
2.4. Method Validation
2.5. GC-MS Sample Preparation
2.6. GC-MS Analysis
2.7. Statistical Analysis
3. Results
3.1. Pretreatment Optimization
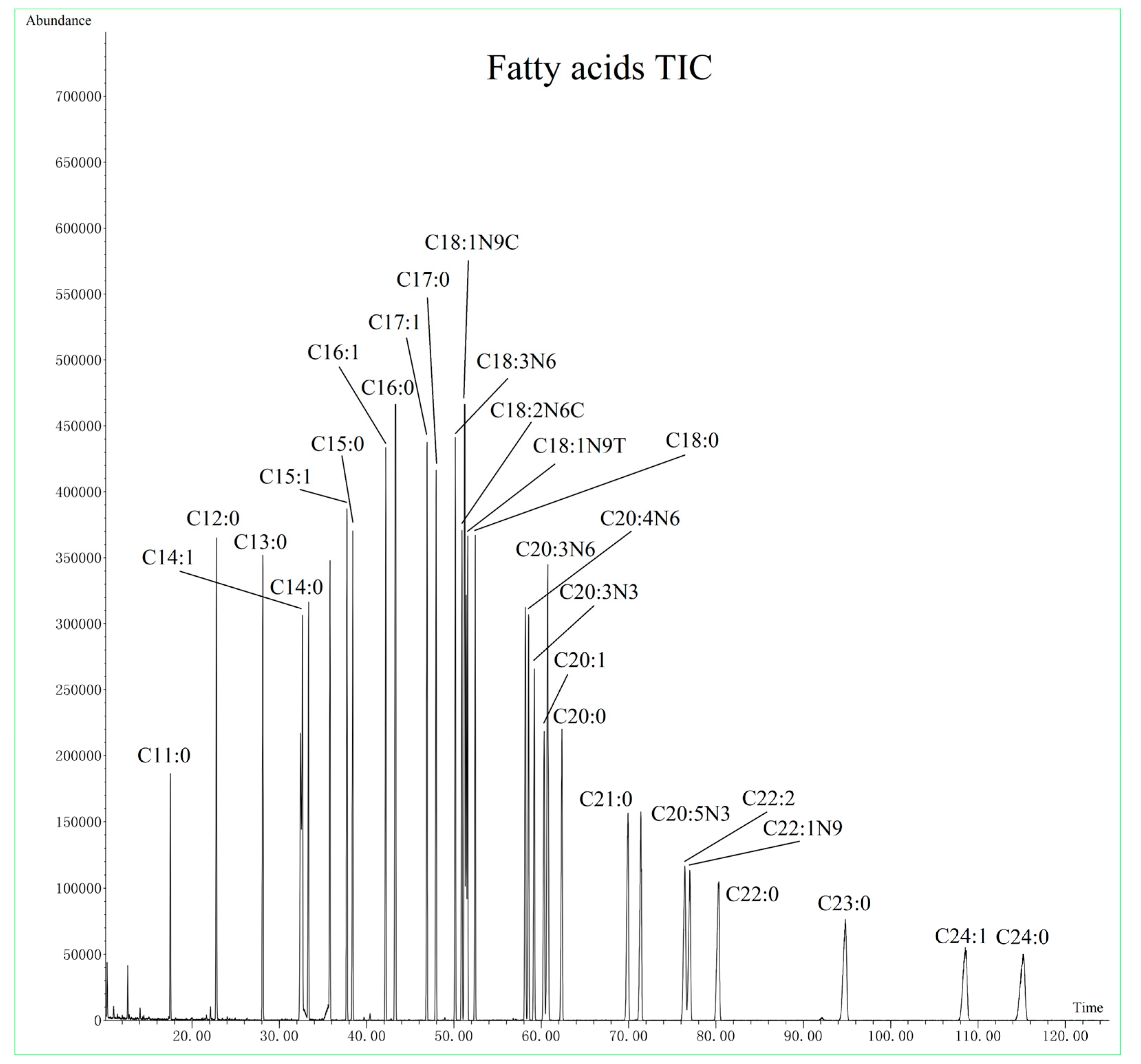
3.2. GC-MS Method Optimization
3.3. Methodology Validation
3.4. CCK8 Experiment
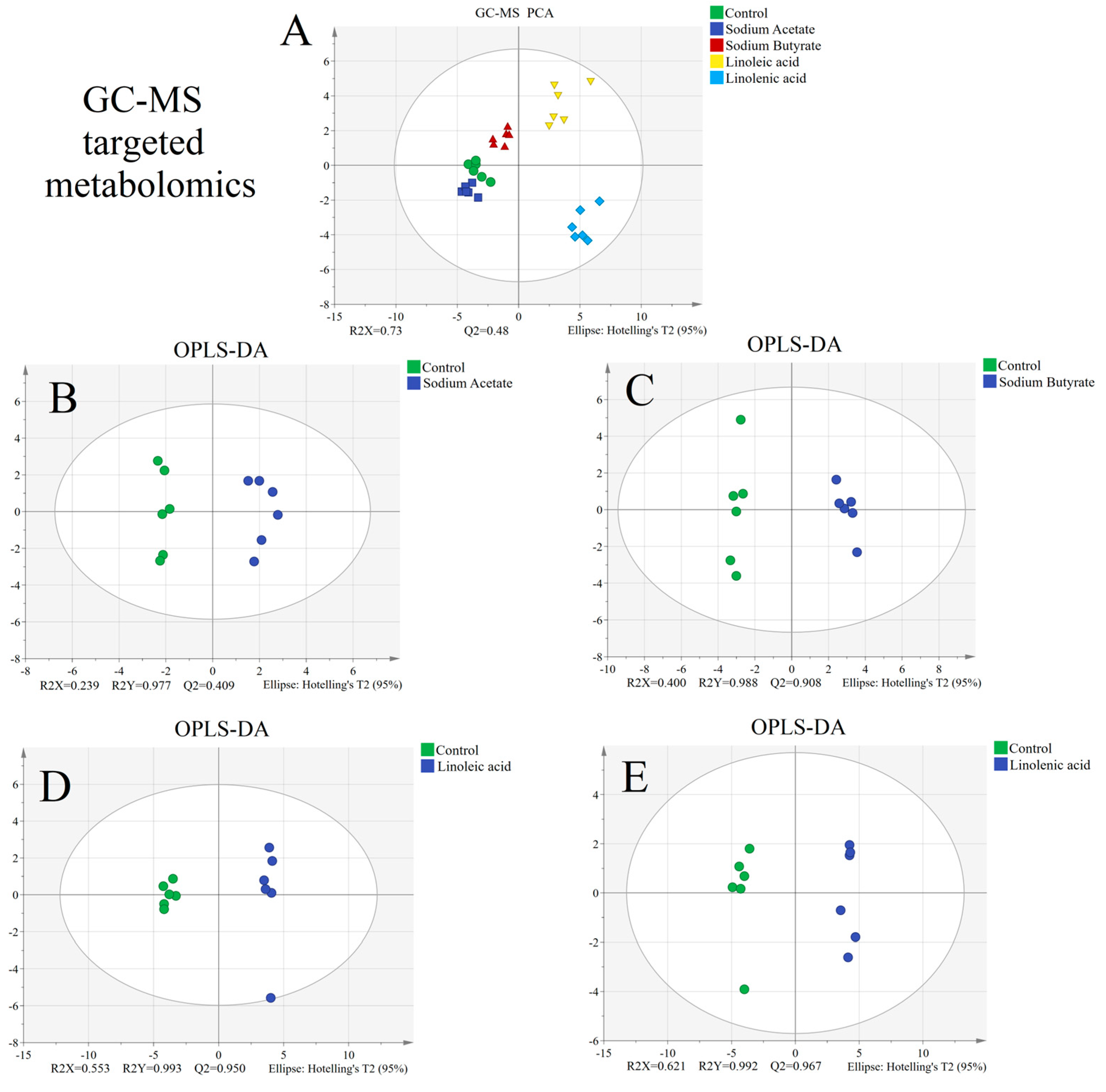
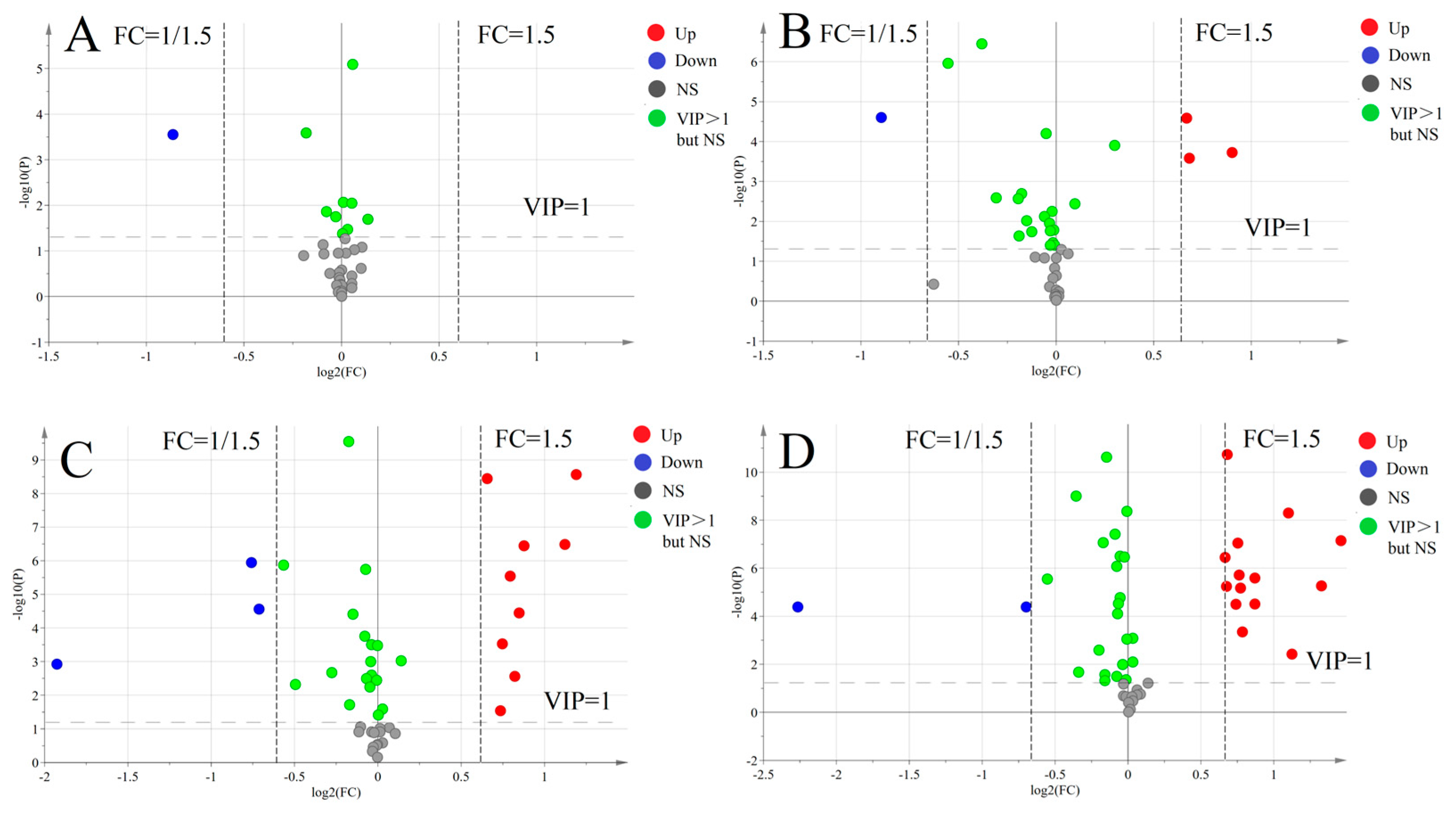
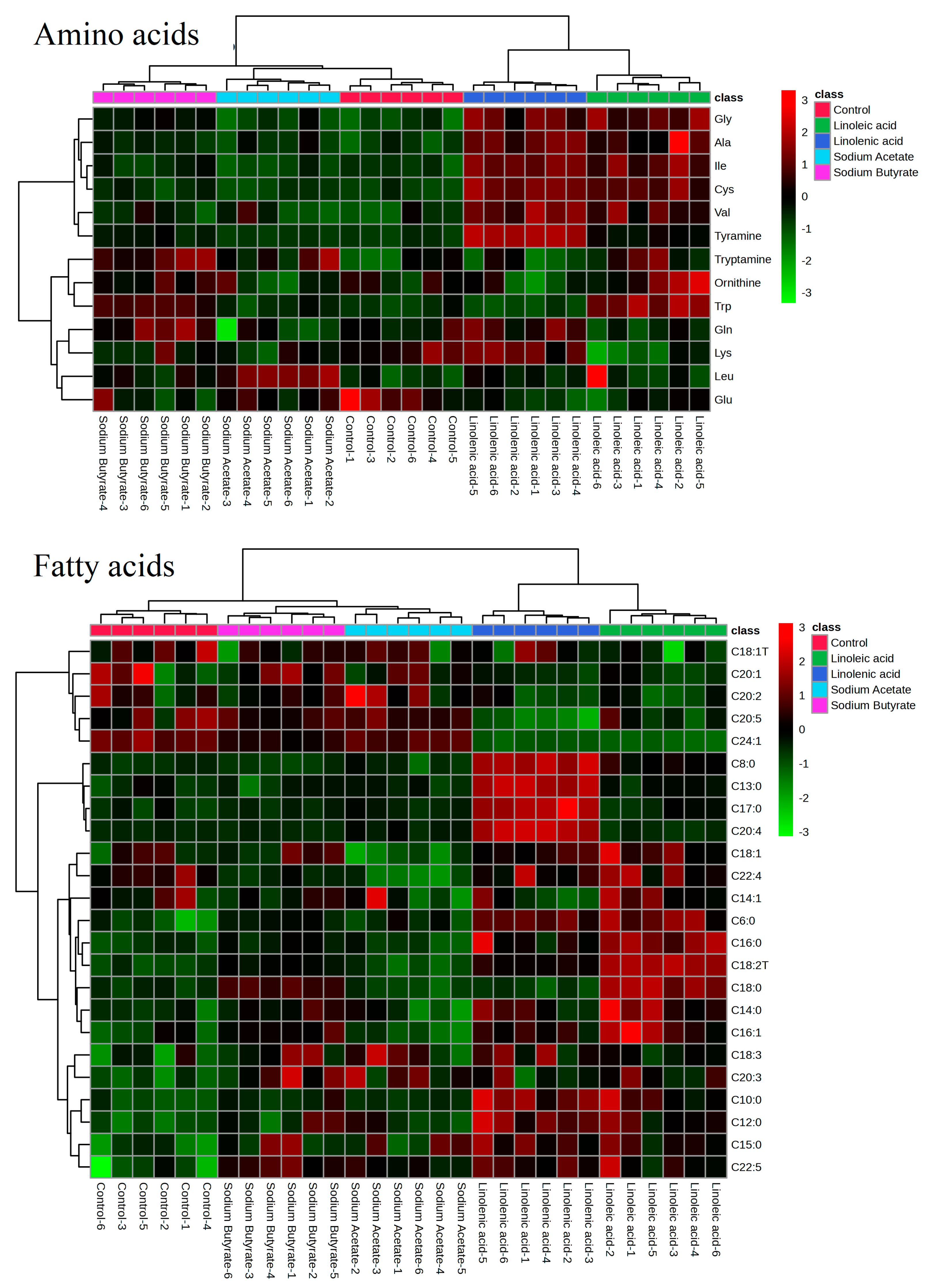
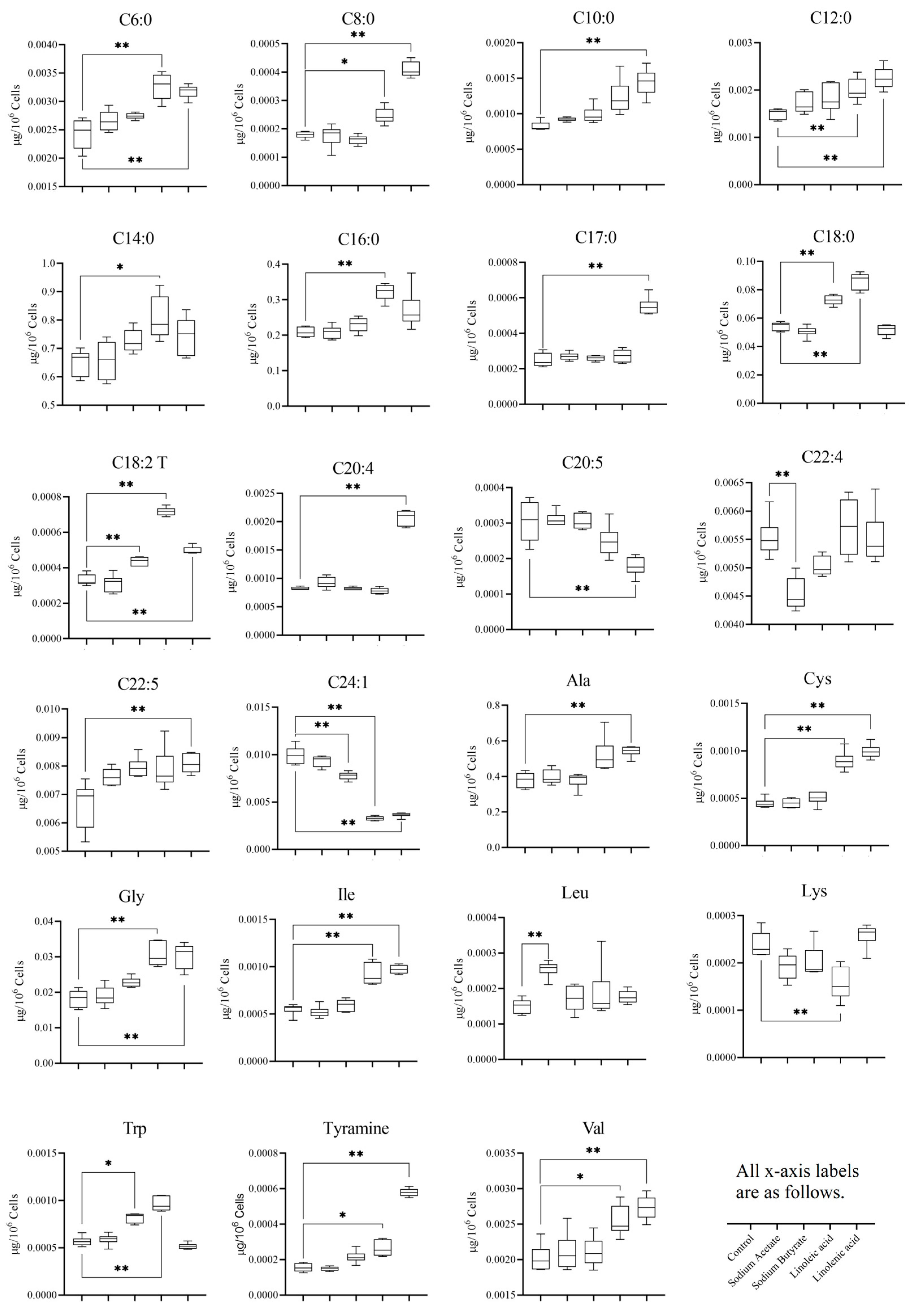
3.5. Statistical Analysis of Targeted Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ni, K.; Wang, D.; Xu, H.; Mei, F.; Wu, C.; Liu, Z.; Zhou, B. miR-21 promotes non-small cell lung cancer cells growth by regulating fatty acid metabolism. Cancer Cell Int. 2019, 19, 219. [Google Scholar] [CrossRef]
- Lemos, H.; Huang, L.; Prendergast, G.C.; Mellor, A.L. Immune control by amino acid catabolism during tumorigenesis and therapy. Nat. Rev. Cancer 2019, 19, 162–175. [Google Scholar] [CrossRef]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A.R. Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef]
- Feng, Q.; Chen, W.-D.; Wang, Y.-D. Gut Microbiota: An Integral Moderator in Health and Disease. Front. Microbiol. 2018, 9, 151. [Google Scholar] [CrossRef]
- Hossain, F.; Al-Khami, A.A.; Wyczechowska, D.; Hernandez, C.; Zheng, L.; Reiss, K.; Valle, L.D.; Trillo-Tinoco, J.; Maj, T.; Zou, W.; et al. Inhibition of Fatty Acid Oxidation Modulates Immunosuppressive Functions of Myeloid-Derived Suppressor Cells and Enhances Cancer Therapies. Cancer Immunol. Res. 2015, 3, 1236–1247. [Google Scholar] [CrossRef]
- Li, M.; van Esch, B.C.A.M.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharmacol. 2018, 831, 52–59. [Google Scholar] [CrossRef]
- Baldi, S.; Menicatti, M.; Nannini, G.; Niccolai, E.; Russo, E.; Ricci, F.; Pallecchi, M.; Romano, F.; Pedone, M.; Poli, G.; et al. Free Fatty Acids Signature in Human Intestinal Disorders: Significant Association between Butyric Acid and Celiac Disease. Nutrients 2021, 13, 742. [Google Scholar] [CrossRef]
- Zuijdgeest-Van Leeuwen, S.D.; Van Der Heijden, M.S.; Rietveld, T.; Van Den Berg, J.W.O.; Tilanus, H.W.; Burgers, J.A.; Paul Wilson, J.H.; Dagnelie, P.C. Fatty acid composition of plasma lipids in patients with pancreatic, lung and oesophageal cancer in comparison with healthy subjects. Clin. Nutr. 2002, 21, 225–230. [Google Scholar] [CrossRef]
- Liu, J.; Mazzone, P.J.; Cata, J.P.; Kurz, A.; Bauer, M.; Mascha, E.J.; Sessler, D.I. Serum Free Fatty Acid Biomarkers of Lung Cancer. Chest 2014, 146, 670–679. [Google Scholar] [CrossRef]
- Vriens, K.; Christen, S.; Parik, S.; Broekaert, D.; Yoshinaga, K.; Talebi, A.; Dehairs, J.; Escalona-Noguero, C.; Schmieder, R.; Cornfield, T.; et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 2019, 566, 403–406. [Google Scholar] [CrossRef]
- Schwarz, A.; Philippsen, R.; Schwarz, T. Induction of Regulatory T Cells and Correction of Cytokine Disbalance by Short-Chain Fatty Acids: Implications for Psoriasis Therapy. J. Investig. Dermatol. 2021, 141, 95–104.e2. [Google Scholar] [CrossRef]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Khani Ali Akbari, S.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef]
- Montecillo-Aguado, M.; Tirado-Rodriguez, B.; Antonio-Andres, G.; Morales-Martinez, M.; Tong, Z.; Yang, J.; Hammock, B.D.; Hernandez-Pando, R.; Huerta-Yepez, S. Omega-6 Polyunsaturated Fatty Acids Enhance Tumor Aggressiveness in Experimental Lung Cancer Model: Important Role of Oxylipins. Int. J. Mol. Sci. 2022, 23, 6179. [Google Scholar] [CrossRef]
- Nagarajan, S.R.; Butler, L.M.; Hoy, A.J. The diversity and breadth of cancer cell fatty acid metabolism. Cancer Metab. 2021, 9, 2. [Google Scholar] [CrossRef]
- Rauf, A.; Khalil, A.A.; Rahman, U.-U.; Khalid, A.; Naz, S.; Shariati, M.A.; Rebezov, M.; Urtecho, E.Z.; de Albuquerque, R.D.D.G.; Anwar, S.; et al. Recent advances in the therapeutic application of short-chain fatty acids (SCFAs): An updated review. Crit. Rev. Food Sci. Nutr. 2021, 62, 6034–6054. [Google Scholar] [CrossRef]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef]
- Zhou, T.; Yang, K.; Huang, J.; Fu, W.; Yan, C.; Wang, Y. Effect of Short-Chain Fatty Acids and Polyunsaturated Fatty Acids on Metabolites in H460 Lung Cancer Cells. Molecules 2023, 28, 2357. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Zhu, M.-J. A sensitive GC/MS detection method for analyzing microbial metabolites short chain fatty acids in fecal and serum samples. Talanta 2019, 196, 249–254. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, Z.; Li, M.; Wang, X. Study of the radical chemistry promoted by tributylborane. Res. Chem. Intermed. 2012, 38, 1893–1907. [Google Scholar] [CrossRef]
- Qiao, J.-Q.; Xu, D.; Lian, H.-Z.; Ge, X. Analysis of related substances in synthetical arbutin and its intermediates by HPLC–UV and LC–ESI–MS. Res. Chem. Intermed. 2013, 41, 691–703. [Google Scholar] [CrossRef]
- Zhu, Y.-Q.; Yin, Q.-H.; Yang, J.; Yang, C.-F.; Sun, X.-D. Quality assessment of Panax notoginseng flowers based on fingerprinting using high-performance liquid chromatography–PDA. Res. Chem. Intermed. 2013, 40, 1641–1653. [Google Scholar] [CrossRef]
- Cha, D.; Liu, M.; Zeng, Z.; Cheng, D.; Zhan, G. Analysis of fatty acids in lung tissues using gas chromatography-mass spectrometry preceded by derivatization-solid-phase microextraction with a novel fiber. Anal. Chim. Acta 2006, 572, 47–54. [Google Scholar] [CrossRef]
- Ni, J.; Xu, L.; Li, W.; Zheng, C.; Wu, L. Targeted metabolomics for serum amino acids and acylcarnitines in patients with lung cancer. Exp. Ther. Med. 2019, 18, 188–198. [Google Scholar] [CrossRef]
- Shi, F.; Wang, C.; Wang, L.; Song, X.; Yang, H.; Fu, Q.; Zhao, W. Preparative isolation and purification of steroidal glycoalkaloid from the ripe berries of Solanum nigrum L. by preparative HPLC-MS and UHPLC-TOF-MS/MS and its anti-non-small cell lung tumors effects in vitro and in vivo. J. Sep. Sci. 2019, 42, 2471–2481. [Google Scholar] [CrossRef]
- Mattarozzi, M.; Riboni, N.; Maffini, M.; Scarpella, S.; Bianchi, F.; Careri, M. Reversed-phase and weak anion-exchange mixed-mode stationary phase for fast separation of medium-, long- and very long chain free fatty acids by ultra-high-performance liquid chromatography-high resolution mass spectrometry. J. Chromatogr. A 2021, 1648, 462209. [Google Scholar] [CrossRef]
- Fürstenberger, G.; Krieg, P.; Müller-Decker, K.; Habenicht, A.J.R. What are cyclooxygenases and lipoxygenases doing in the driver’s seat of carcinogenesis? Int. J. Cancer 2006, 119, 2247–2254. [Google Scholar] [CrossRef] [PubMed]
- Luu, H.N.; Cai, H.; Murff, H.J.; Xiang, Y.-B.; Cai, Q.; Li, H.; Gao, J.; Yang, G.; Lan, Q.; Gao, Y.-T.; et al. A prospective study of dietary polyunsaturated fatty acids intake and lung cancer risk. Int. J. Cancer 2018, 143, 2225–2237. [Google Scholar] [CrossRef]
- Davidson, S.M.; Papagiannakopoulos, T.; Olenchock, B.A.; Heyman, J.E.; Keibler, M.A.; Luengo, A.; Bauer, M.R.; Jha, A.K.; O’Brien, J.P.; Pierce, K.A.; et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016, 23, 517–528. [Google Scholar] [CrossRef]
- Schiliro, C.; Firestein, B.L. Mechanisms of Metabolic Reprogramming in Cancer Cells Supporting Enhanced Growth and Proliferation. Cells 2021, 10, 1056. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Criado-Navarro, I.; Mena-Bravo, A.; Calderón-Santiago, M.; Priego-Capote, F. Profiling analysis of phospholipid fatty acids in serum as a complement to the comprehensive fatty acids method. J. Chromatogr. A 2020, 1619, 460965. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Yang, K.; Ma, Y.; Huang, J.; Fu, W.; Yan, C.; Li, X.; Wang, Y. GC/MS-Based Analysis of Fatty Acids and Amino Acids in H460 Cells Treated with Short-Chain and Polyunsaturated Fatty Acids: A Highly Sensitive Approach. Nutrients 2023, 15, 2342. https://doi.org/10.3390/nu15102342
Zhou T, Yang K, Ma Y, Huang J, Fu W, Yan C, Li X, Wang Y. GC/MS-Based Analysis of Fatty Acids and Amino Acids in H460 Cells Treated with Short-Chain and Polyunsaturated Fatty Acids: A Highly Sensitive Approach. Nutrients. 2023; 15(10):2342. https://doi.org/10.3390/nu15102342
Chicago/Turabian StyleZhou, Tianxiao, Kaige Yang, Yinjie Ma, Jin Huang, Wenchang Fu, Chao Yan, Xinyan Li, and Yan Wang. 2023. "GC/MS-Based Analysis of Fatty Acids and Amino Acids in H460 Cells Treated with Short-Chain and Polyunsaturated Fatty Acids: A Highly Sensitive Approach" Nutrients 15, no. 10: 2342. https://doi.org/10.3390/nu15102342
APA StyleZhou, T., Yang, K., Ma, Y., Huang, J., Fu, W., Yan, C., Li, X., & Wang, Y. (2023). GC/MS-Based Analysis of Fatty Acids and Amino Acids in H460 Cells Treated with Short-Chain and Polyunsaturated Fatty Acids: A Highly Sensitive Approach. Nutrients, 15(10), 2342. https://doi.org/10.3390/nu15102342





