In Vitro Fermentation of Beechwood Lignin–Carbohydrate Complexes Provides Evidence for Utilization by Gut Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Extraction of BW-LCC
2.2. Structural Characterization of BW-LCC
2.3. In Vitro Fermentation
2.4. Determination of SCFAs
2.5. Determination of Microbial Diversity
2.6. Metagenome Analysis
2.7. Data Statistics
3. Results and Discussions
3.1. Analysis of the Composition and Structure of Lignin in BW-LCC
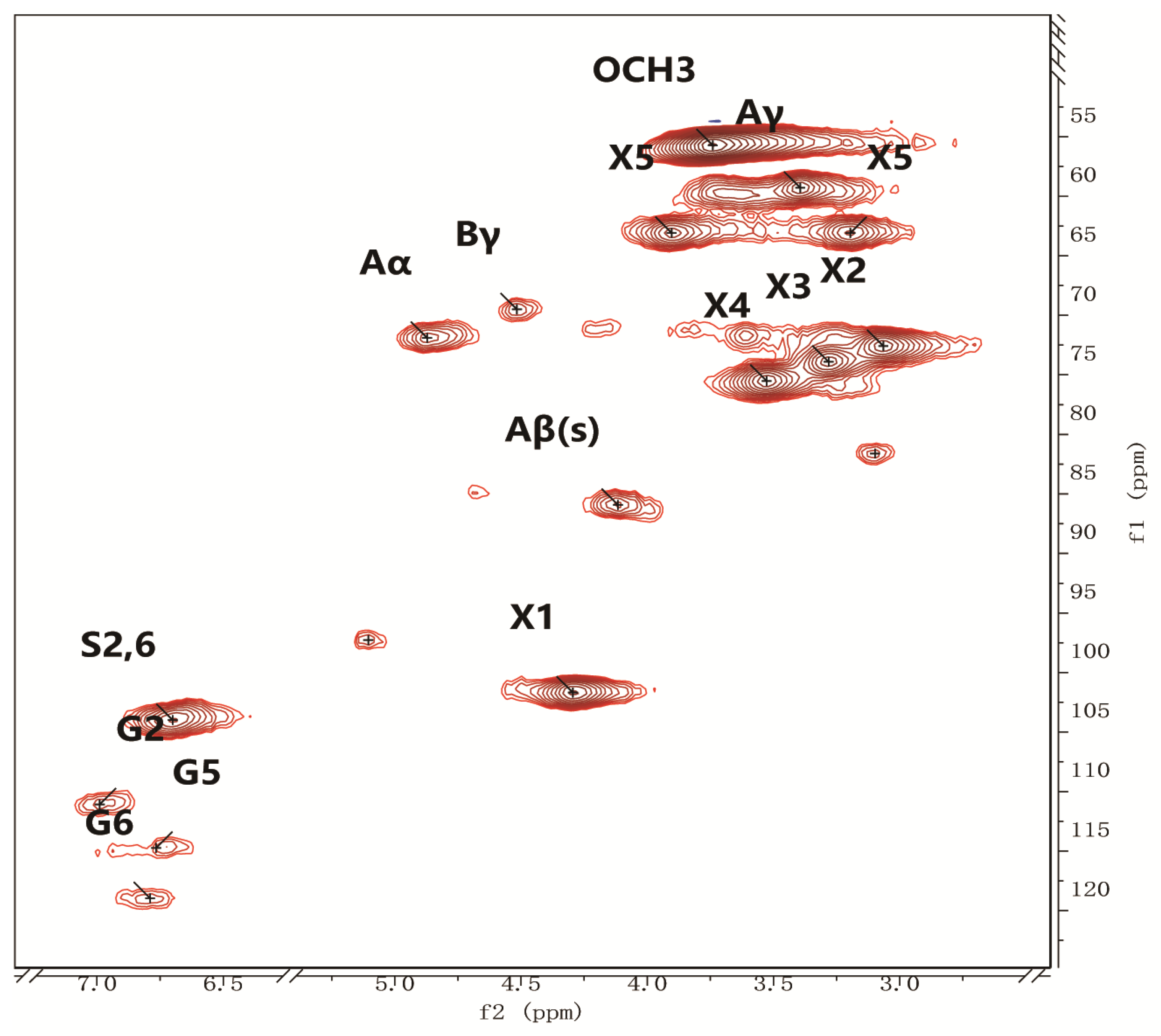
3.1.1. Structural Characterization of Lignin
3.1.2. Quantification of Lignin Linkages
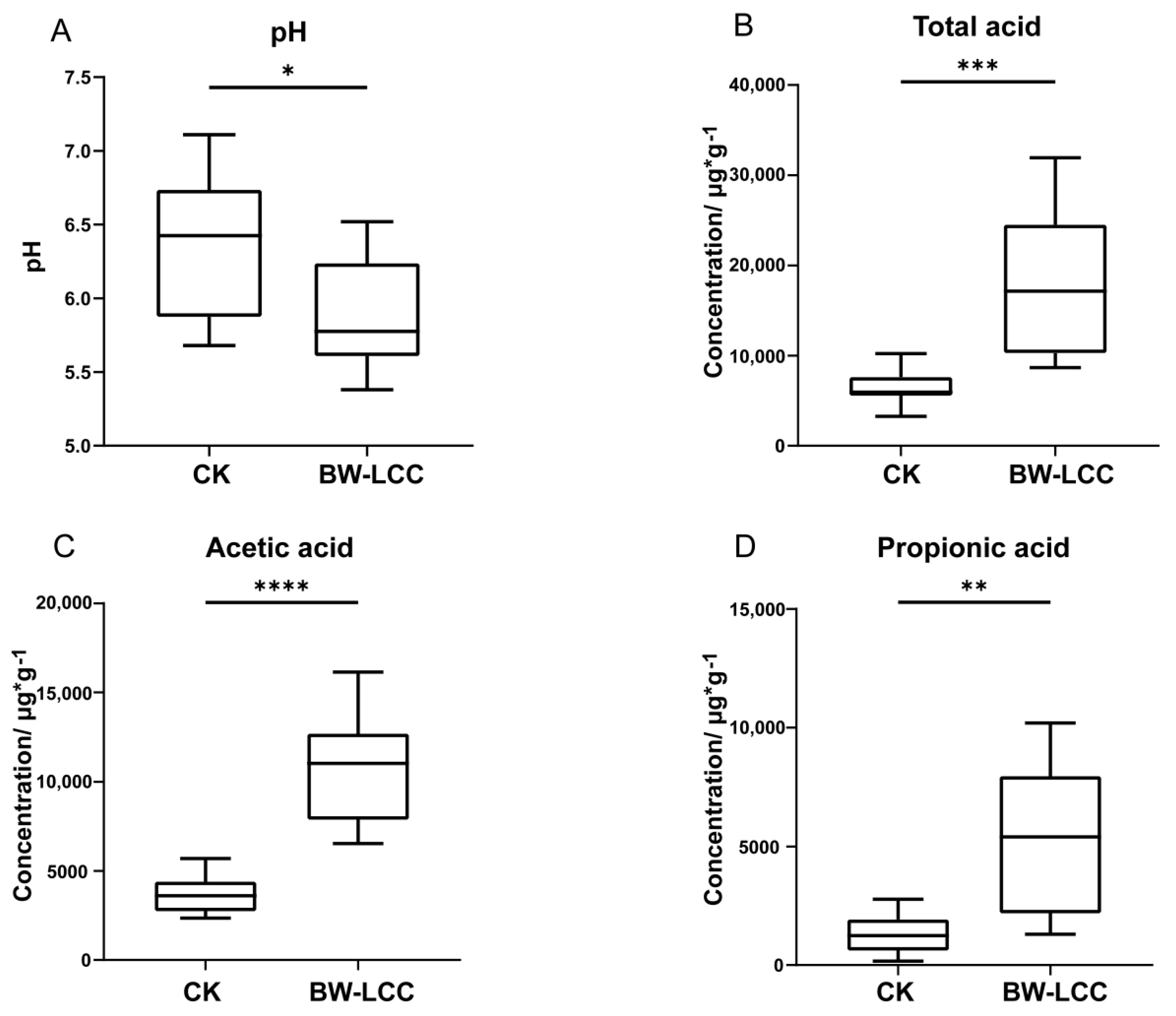
3.2. pH and SCFAs Analysis
3.3. The Impact of BW-LCC on Gut Microbiota Communities
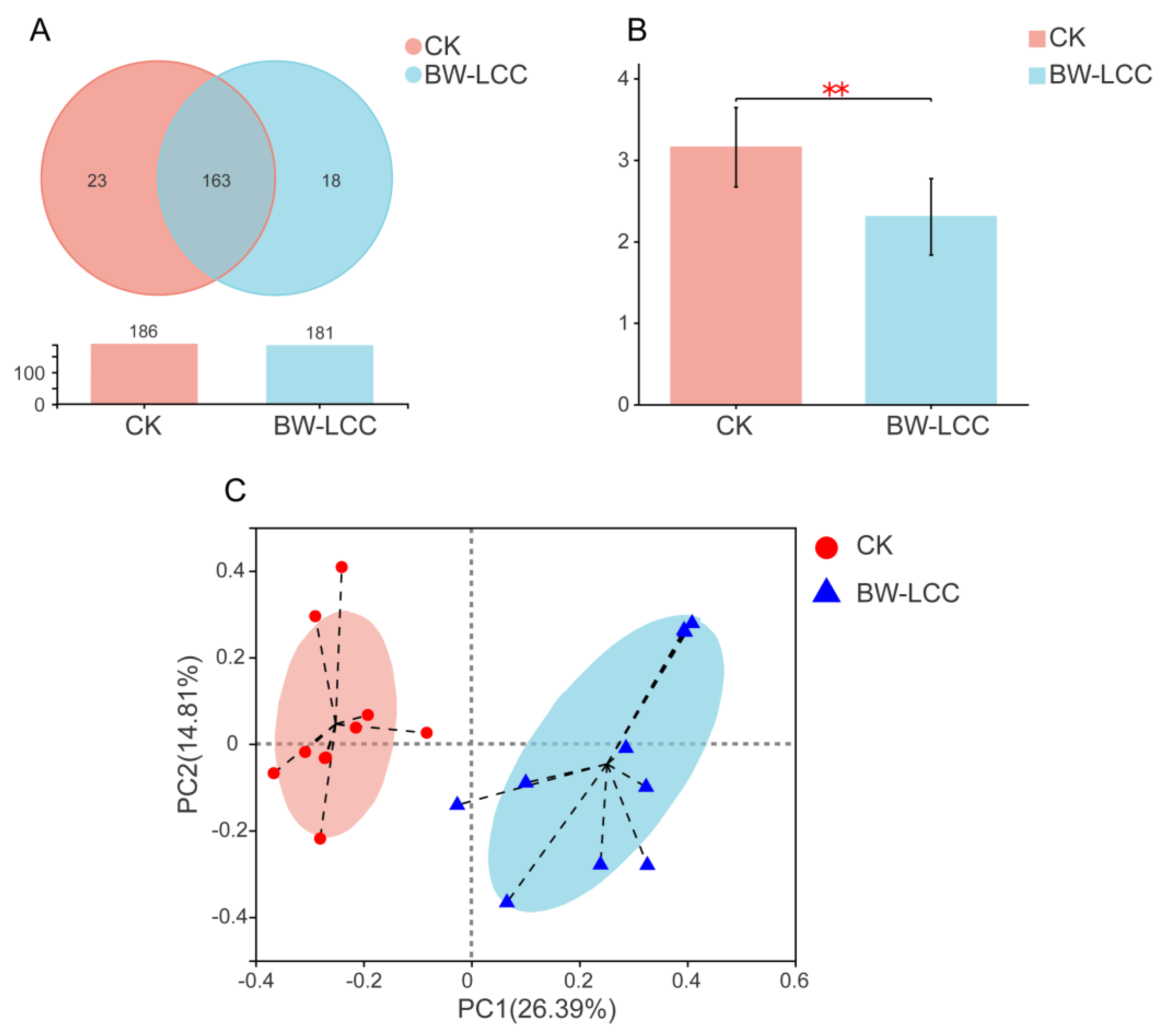
3.3.1. Changes in Intestinal Microbial Diversity
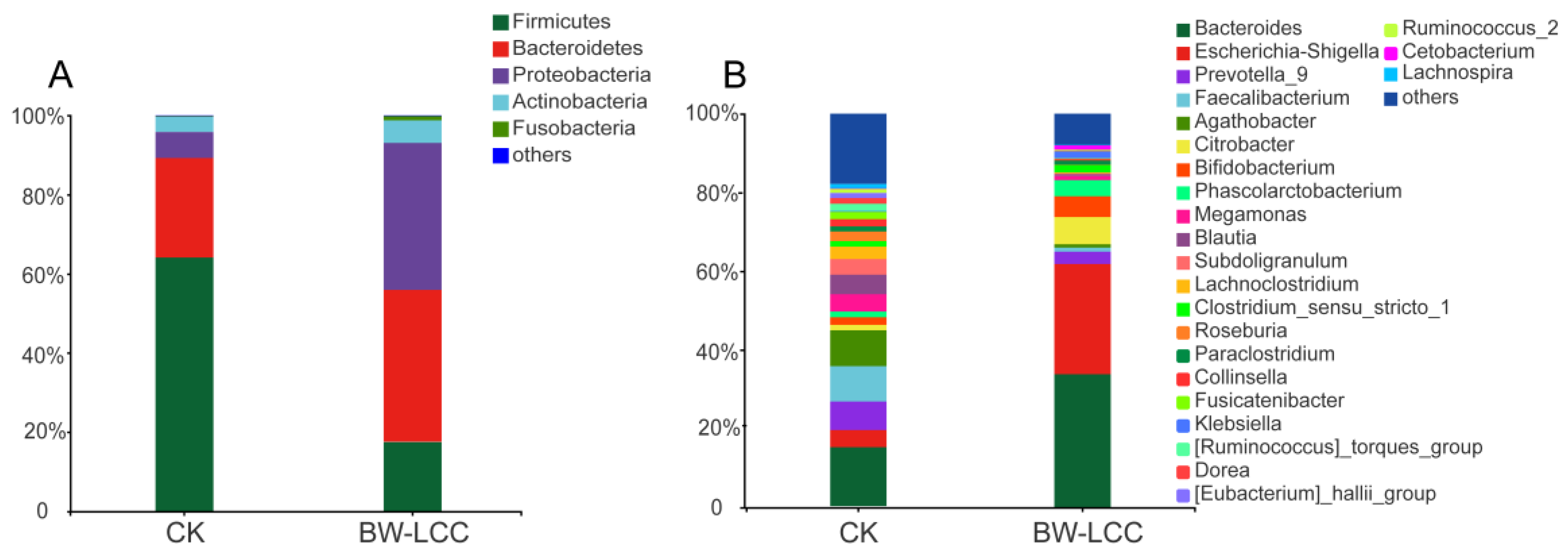
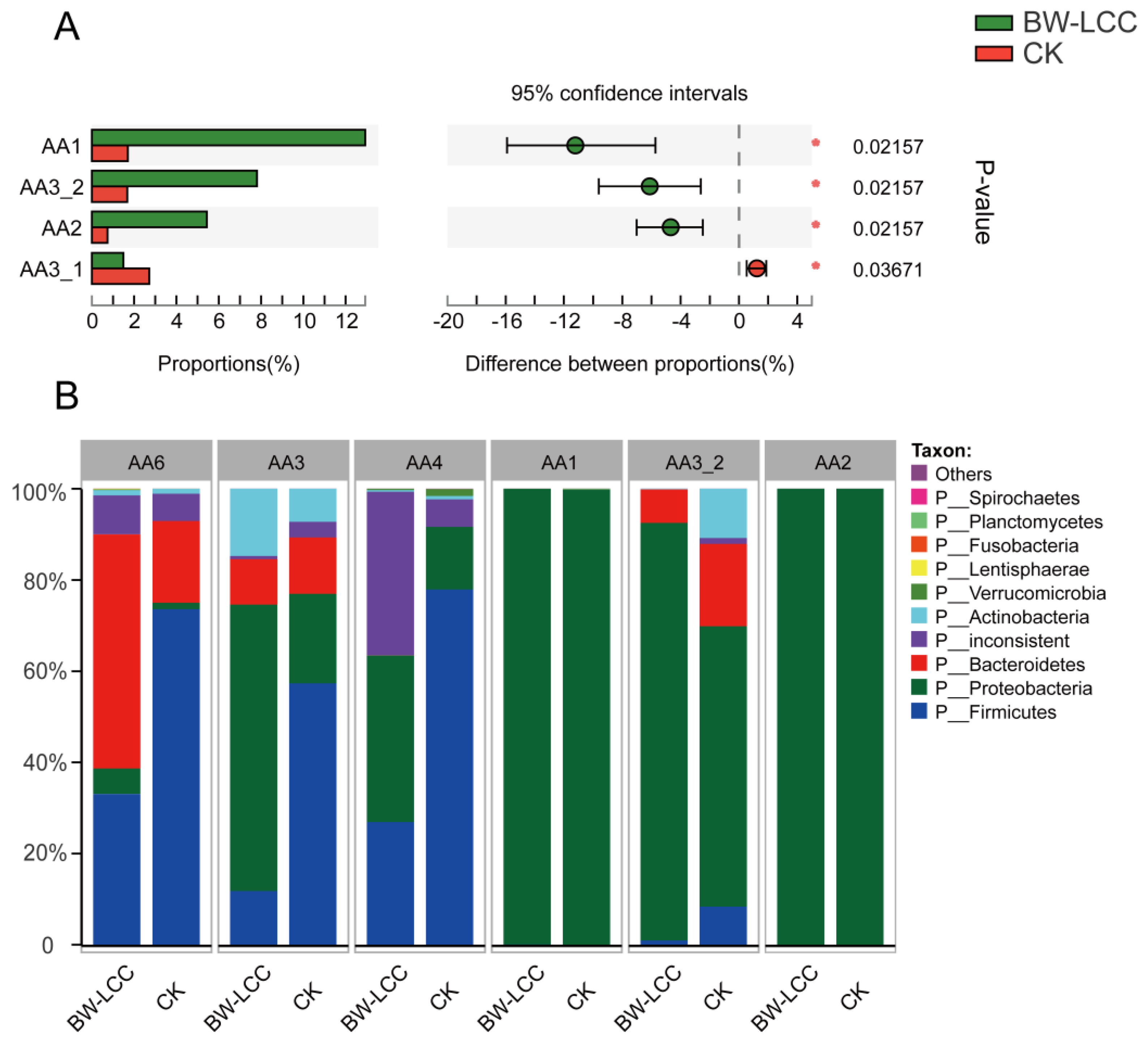
3.3.2. Effect of BW-LCC on Gut Microbiota Composition
3.4. Effect of BW-LCC on CAZymes
3.4.1. BW-LCC Alters the Relative Abundance of CAZymes
3.4.2. The Role of BW-LCC in Causing the Enrichment of Lignin-Degrading Enzymes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, J.L.; Bäckhed, F. Diet–Microbiota Interactions as Moderators of Human Metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Liu, Z.; Zhu, C.; Mou, H.; Kong, Q. Nondigestible Carbohydrates, Butyrate, and Butyrate-Producing Bacteria. Crit. Rev. Food Sci. Nutr. 2019, 59, S130–S152. [Google Scholar] [CrossRef]
- Jawhara, M.; Sørensen, S.B.; Heitmann, B.L.; Andersen, V. Biomarkers of Whole-Grain and Cereal-Fiber Intake in Human Studies: A Systematic Review of the Available Evidence and Perspectives. Nutrients 2019, 11, E2994. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Tang, Q.; Fu, C.; Regenstein, J.; Huang, J.; Wang, L. Effects of Different Particle-Sized Insoluble Dietary Fibre from Citrus Peel on Adsorption and Activity Inhibition of Pancreatic Lipase. Food Chem. 2023, 398, 133834. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Tan, J.K.; Macia, L.; Mackay, C.R. Dietary Fiber and SCFAs in the Regulation of Mucosal Immunity. J. Allergy Clin. Immunol. 2022, S0091-6749(22)01565-2. [Google Scholar] [CrossRef]
- Appunni, S.; Rubens, M.; Ramamoorthy, V.; Tonse, R.; Saxena, A.; McGranaghan, P.; Kaiser, A.; Kotecha, R. Emerging Evidence on the Effects of Dietary Factors on the Gut Microbiome in Colorectal Cancer. Front. Nutr. 2021, 8, 718389. [Google Scholar] [CrossRef]
- Chen, M.; Fan, B.; Liu, S.; Imam, K.M.S.U.; Xie, Y.; Wen, B.; Xin, F. The in vitro Effect of Fibers with Different Degrees of Polymerization on Human Gut Bacteria. Front. Microbiol. 2020, 11, 819. [Google Scholar] [CrossRef]
- Chen, M.; Liu, S.; Imam, K.M.S.U.; Sun, L.; Wang, Y.; Gu, T.; Wen, B.; Xin, F. The Effect of Xylooligosaccharide, Xylan, and Whole Wheat Bran on the Human Gut Bacteria. Front. Microbiol. 2020, 11, 568457. [Google Scholar] [CrossRef]
- La Rosa, S.L.; Kachrimanidou, V.; Buffetto, F.; Pope, P.B.; Pudlo, N.A.; Martens, E.C.; Rastall, R.A.; Gibson, G.R.; Westereng, B. Wood-Derived Dietary Fibers Promote Beneficial Human Gut Microbiota. mSphere 2019, 4, e00554-18. [Google Scholar] [CrossRef]
- Russell, W.R.; Scobbie, L.; Chesson, A.; Richardson, A.J.; Stewart, C.S.; Duncan, S.H.; Drew, J.E.; Duthie, G.G. Anti-Inflammatory Implications of the Microbial Transformation of Dietary Phenolic Compounds. Nutr. Cancer 2008, 60, 636–642. [Google Scholar] [CrossRef]
- Sakagami, H.; Kawano, M.; Thet, M.M.; Hashimoto, K.; Satoh, K.; Kanamoto, T.; Terakubo, S.; Nakashima, H.; Haishima, Y.; Maeda, Y.; et al. Anti-HIV and Immunomodulation Activities of Cacao Mass Lignin-Carbohydrate Complex. In Vivo 2011, 25, 229–236. [Google Scholar]
- Sakagami, H.; Ikeda, M.; Unten, S.; Takeda, K.; Murayama, J.; Hamada, A.; Kimura, K.; Komatsu, N.; Konno, K. Antitumor Activity of Polysaccharide Fractions from Pine Cone Extract of Pinus Parviflora Sieb. et Zucc. Anticancer Res. 1987, 7, 1153–1159. [Google Scholar]
- Wang, H.; Huang, X.; Tan, H.; Chen, X.; Chen, C.; Nie, S. Interaction between Dietary Fiber and Bifidobacteria in Promoting Intestinal Health. Food Chem. 2022, 393, 133407. [Google Scholar] [CrossRef]
- Niemi, P.; Aura, A.-M.; Maukonen, J.; Smeds, A.I.; Mattila, I.; Niemelä, K.; Tamminen, T.; Faulds, C.B.; Buchert, J.; Poutanen, K. Interactions of a Lignin-Rich Fraction from Brewer’s Spent Grain with Gut Microbiota in Vitro. J. Agric. Food Chem. 2013, 61, 6754–6762. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, L.; Shi, C.; Shi, Q.; Ma, F.; Zhang, X.; Yu, W.; Yu, H. Structural Characterization of Lignin-Carbohydrate Complexes (LCCs) and Their Biotransformation by Intestinal Microbiota In Vitro. J. Agric. Food Chem. 2021, 69, 12880–12890. [Google Scholar] [CrossRef]
- Zhang, M.; Chekan, J.R.; Dodd, D.; Hong, P.-Y.; Radlinski, L.; Revindran, V.; Nair, S.K.; Mackie, R.I.; Cann, I. Xylan Utilization in Human Gut Commensal Bacteria Is Orchestrated by Unique Modular Organization of Polysaccharide-Degrading Enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, E3708–E3717. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The Abundance and Variety of Carbohydrate-Active Enzymes in the Human Gut Microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the Enzymatic Repertoire of the CAZy Database to Integrate Auxiliary Redox Enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Baky, M.H.; Salah, M.; Ezzelarab, N.; Shao, P.; Elshahed, M.S.; Farag, M.A. Insoluble Dietary Fibers: Structure, Metabolism, Interactions with Human Microbiome, and Role in Gut Homeostasis. Crit. Rev. Food Sci. Nutr. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-Active Enzymes (CAZymes) in the Gut Microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Zhu, W.; Chen, S.; Deng, T.; Ma, S.; Wang, H. A Novel Mechanocatalytical Reaction System Driven by Fluid Shear Force for the Mild and Rapid Pretreatment of Lignocellulosic Biomass. Waste Manag. 2022, 148, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Schwab, C.; Ruscheweyh, H.-J.; Bunesova, V.; Pham, V.T.; Beerenwinkel, N.; Lacroix, C. Trophic Interactions of Infant Bifidobacteria and Eubacterium Hallii during L-Fucose and Fucosyllactose Degradation. Front. Microbiol. 2017, 8, 95. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wu, J.; Zhang, H.; Perry, S.W.; Yin, B.; Tan, X.; Chai, T.; Liang, W.; Huang, Y.; Li, Y.; et al. The Gut Microbiome Modulates Gut–Brain Axis Glycerophospholipid Metabolism in a Region-Specific Manner in a Nonhuman Primate Model of Depression. Mol. Psychiatry 2021, 26, 2380–2392. [Google Scholar] [CrossRef]
- Rencoret, J.; Marques, G.; Gutiérrez, A.; Nieto, L.; Jiménez-Barbero, J.; Martínez, Á.T.; del Río, J.C. Isolation and Structural Characterization of the Milled-Wood Lignin from Paulownia Fortunei Wood. Ind. Crops Prod. 2009, 30, 137–143. [Google Scholar] [CrossRef]
- Ralph, J.; Lapierre, C.; Boerjan, W. Lignin Structure and Its Engineering. Curr. Opin. Biotechnol. 2019, 56, 240–249. [Google Scholar] [CrossRef]
- Sette, M.; Wechselberger, R.; Crestini, C. Elucidation of Lignin Structure by Quantitative 2D NMR. Chemistry 2011, 17, 9529–9535. [Google Scholar] [CrossRef]
- Lee, H.-S.; Park, J.; Yeon, Y.J. Biocatalytic Valorization of Lignin Subunit: Screening a Carboxylic Acid Reductase with High Substrate Preference to Syringyl Functional Group. Enzym. Microb. Technol. 2022, 161, 110099. [Google Scholar] [CrossRef]
- Xu, K.; Cui, X.; Ren, X.; Meng, J.; Fu, X.; Xia, Q. Discovery of Natural Polyphenols from the Wild Vegetable Suaeda salsa L. with Potential Cardioprotective Functions. Food Chem. 2023, 405, 134968. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Al-Lahham, S.H.; Peppelenbosch, M.P.; Roelofsen, H.; Vonk, R.J.; Venema, K. Biological Effects of Propionic Acid in Humans; Metabolism, Potential Applications and Underlying Mechanisms. Biochim. Biophys. Acta 2010, 1801, 1175–1183. [Google Scholar] [CrossRef]
- Dou, Z.; Chen, C.; Fu, X. Bioaccessibility, Antioxidant Activity and Modulation Effect on Gut Microbiota of Bioactive Compounds from Moringa Oleifera Lam. Leaves during Digestion and Fermentation in vitro. Food Funct. 2019, 10, 5070–5079. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Dong, R.; Liu, S.; Xie, J.; Chen, Y.; Zheng, Y.; Zhang, X.; Zhao, E.; Wang, Z.; Xu, H.; Yu, Q. The Recovery, Catabolism and Potential Bioactivity of Polyphenols from Carrot Subjected to in vitro Simulated Digestion and Colonic Fermentation. Food Res. Int. 2021, 143, 110263. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Cameron, E.A.; Martens, E.C. How Glycan Metabolism Shapes the Human Gut Microbiota. Nat. Rev. Microbiol. 2012, 10, 323–335. [Google Scholar] [CrossRef]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; Qin, H. Probiotics Improve Gut Microbiota Dysbiosis in Obese Mice Fed a High-Fat or High-Sucrose Diet. Nutrition 2019, 60, 175–184. [Google Scholar] [CrossRef]
- Ma, C.; Lo, P.K.; Xu, J.; Li, M.; Jiang, Z.; Li, G.; Zhu, Q.; Li, X.; Leong, S.Y.; Li, Q. Molecular Mechanisms Underlying Lignocellulose Degradation and Antibiotic Resistance Genes Removal Revealed via Metagenomics Analysis during Different Agricultural Wastes Composting. Bioresour. Technol. 2020, 314, 123731. [Google Scholar] [CrossRef]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Basle, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.; Labourel, A.; et al. Complex Pectin Metabolism by Gut Bacteria Reveals Novel Catalytic Functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Bode, C.E.; Brabetz, W.; Brade, H. Cloning and Characterization of 3-Deoxy-D-Manno-Oct-2-Ulosonic Acid (Kdo) Transferase Genes (KdtA) from Acinetobacter Baumannii and Acinetobacter Haemolyticus. Eur. J. Biochem. 1998, 254, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Gibson, R.P.; Lloyd, R.M.; Charnock, S.J.; Davies, G.J. Characterization of Escherichia Coli OtsA, a Trehalose-6-Phosphate Synthase from Glycosyltransferase Family 20. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Rusche, K.M.; Shin, H.; Fierke, C.A.; Christianson, D.W. Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 8146–8150. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, T.; Stepnov, A.A.; Hegnar, O.A.; Eijsink, V.G.H. Sugar Oxidoreductases and LPMOs—Two Sides of the Same Polysaccharide Degradation Story? Carbohydr. Res. 2021, 505, 108350. [Google Scholar] [CrossRef]
- Yang, Y.; Ghatge, S.; Hur, H.-G. Improvement of Thermoalkaliphilic Laccase (CtLac) by a Directed Evolution and Application to Lignin Degradation. Appl. Microbiol. Biotechnol. 2023, 107, 273–286. [Google Scholar] [CrossRef]
- Bayrak, S.; Öztürk, C.; Demir, Y.; Alım, Z.; Küfrevioglu, Ö.İ. Purification of Polyphenol Oxidase from Potato and Investigation of the Inhibitory Effects of Phenolic Acids on Enzyme Activity. PPL 2020, 27, 187–192. [Google Scholar] [CrossRef]
- Wong, D.W.S. Structure and Action Mechanism of Ligninolytic Enzymes. Appl. Biochem. Biotechnol. 2009, 157, 174–209. [Google Scholar] [CrossRef]
- Koua, D.; Cerutti, L.; Falquet, L.; Sigrist, C.J.A.; Theiler, G.; Hulo, N.; Dunand, C. PeroxiBase: A Database with New Tools for Peroxidase Family Classification. Nucleic Acids Res. 2009, 37, D261–D266. [Google Scholar] [CrossRef]
- Bissaro, B.; Várnai, A.; Røhr, Å.K.; Eijsink, V.G.H. Oxidoreductases and Reactive Oxygen Species in Conversion of Lignocellulosic Biomass. Microbiol. Mol. Biol. Rev. 2018, 82, e00029-18. [Google Scholar] [CrossRef]
- Daou, M.; Bisotto, A.; Haon, M.; Oliveira Correia, L.; Cottyn, B.; Drula, E.; Garajová, S.; Bertrand, E.; Record, E.; Navarro, D.; et al. A Putative Lignin Copper Oxidase from Trichoderma Reesei. J. Fungi 2021, 7, 643. [Google Scholar] [CrossRef]
- Mathieu, Y.; Piumi, F.; Valli, R.; Aramburu, J.C.; Ferreira, P.; Faulds, C.B.; Record, E. Activities of Secreted Aryl Alcohol Quinone Oxidoreductases from Pycnoporus Cinnabarinus Provide Insights into Fungal Degradation of Plant Biomass. Appl. Environ. Microbiol. 2016, 82, 2411–2423. [Google Scholar] [CrossRef]
- Vélez-Mercado, M.I.; Talavera-Caro, A.G.; Escobedo-Uribe, K.M.; Sánchez-Muñoz, S.; Luévanos-Escareño, M.P.; Hernández-Terán, F.; Alvarado, A.; Balagurusamy, N. Bioconversion of Lignocellulosic Biomass into Value Added Products under Anaerobic Conditions: Insight into Proteomic Studies. Int. J. Mol. Sci. 2021, 22, 12249. [Google Scholar] [CrossRef]
- Bissaro, B.; Røhr, Å.K.; Müller, G.; Chylenski, P.; Skaugen, M.; Forsberg, Z.; Horn, S.J.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Oxidative Cleavage of Polysaccharides by Monocopper Enzymes Depends on H2O2. Nat. Chem. Biol. 2017, 13, 1123–1128. [Google Scholar] [CrossRef]
- Jensen, K.A.; Ryan, Z.C.; Vanden Wymelenberg, A.; Cullen, D.; Hammel, K.E. An NADH:Quinone Oxidoreductase Active during Biodegradation by the Brown-Rot Basidiomycete Gloeophyllum Trabeum. Appl. Environ. Microbiol. 2002, 68, 2699–2703. [Google Scholar] [CrossRef]
- Demir, Y. Naphthoquinones, Benzoquinones, and Anthraquinones: Molecular Docking, ADME and Inhibition Studies on Human Serum Paraoxonase-1 Associated with Cardiovascular Diseases. Drug Dev. Res. 2020, 81, 628–636. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, Y.; Jiang, H.; Zang, H.; Wang, Y.; Sun, S.; Li, C. Efficient Vanillin Biosynthesis by Recombinant Lignin-Degrading Bacterium Arthrobacter sp. C2 and Its Environmental Profile via Life Cycle Assessment. Bioresour. Technol. 2022, 347, 126434. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Liu, S.; Wang, H.; Wang, Y.; Li, Z.; Gu, T.; Li, Y.; Xin, F.; Wen, B. In Vitro Fermentation of Beechwood Lignin–Carbohydrate Complexes Provides Evidence for Utilization by Gut Bacteria. Nutrients 2023, 15, 220. https://doi.org/10.3390/nu15010220
Ma X, Liu S, Wang H, Wang Y, Li Z, Gu T, Li Y, Xin F, Wen B. In Vitro Fermentation of Beechwood Lignin–Carbohydrate Complexes Provides Evidence for Utilization by Gut Bacteria. Nutrients. 2023; 15(1):220. https://doi.org/10.3390/nu15010220
Chicago/Turabian StyleMa, Xiaochen, Shujun Liu, Hongliang Wang, Yulu Wang, Zhen Li, Tianyi Gu, Yulong Li, Fengjiao Xin, and Boting Wen. 2023. "In Vitro Fermentation of Beechwood Lignin–Carbohydrate Complexes Provides Evidence for Utilization by Gut Bacteria" Nutrients 15, no. 1: 220. https://doi.org/10.3390/nu15010220
APA StyleMa, X., Liu, S., Wang, H., Wang, Y., Li, Z., Gu, T., Li, Y., Xin, F., & Wen, B. (2023). In Vitro Fermentation of Beechwood Lignin–Carbohydrate Complexes Provides Evidence for Utilization by Gut Bacteria. Nutrients, 15(1), 220. https://doi.org/10.3390/nu15010220




