Phenolic Extract from Extra Virgin Olive Oil Induces Different Anti-Proliferative Pathways in Human Bladder Cancer Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Phenolic Compound Extraction from Extra Virgin Olive Oil
2.3. HPLC-UV-DAD Analysis
2.4. Cell Culture and Viability Assay
2.5. Autophagy Detection
2.5.1. Measurement of Autophagic Vacuoles
2.5.2. Immunoblottings
2.6. Apoptotic Assays
2.6.1. Hoechst Staining
2.6.2. Annexin V Detection
2.6.3. Caspases Assay
2.7. Intracellular ROS Measurement
2.8. Glutathione Determination
2.9. Statistical Analysis
3. Results
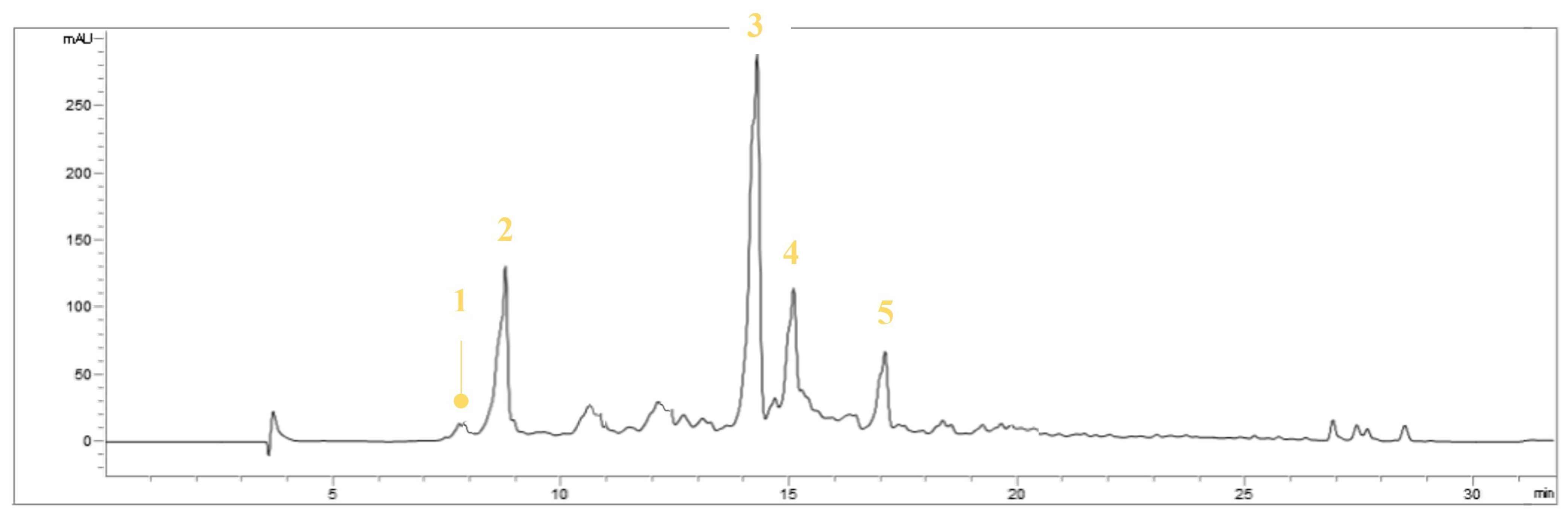
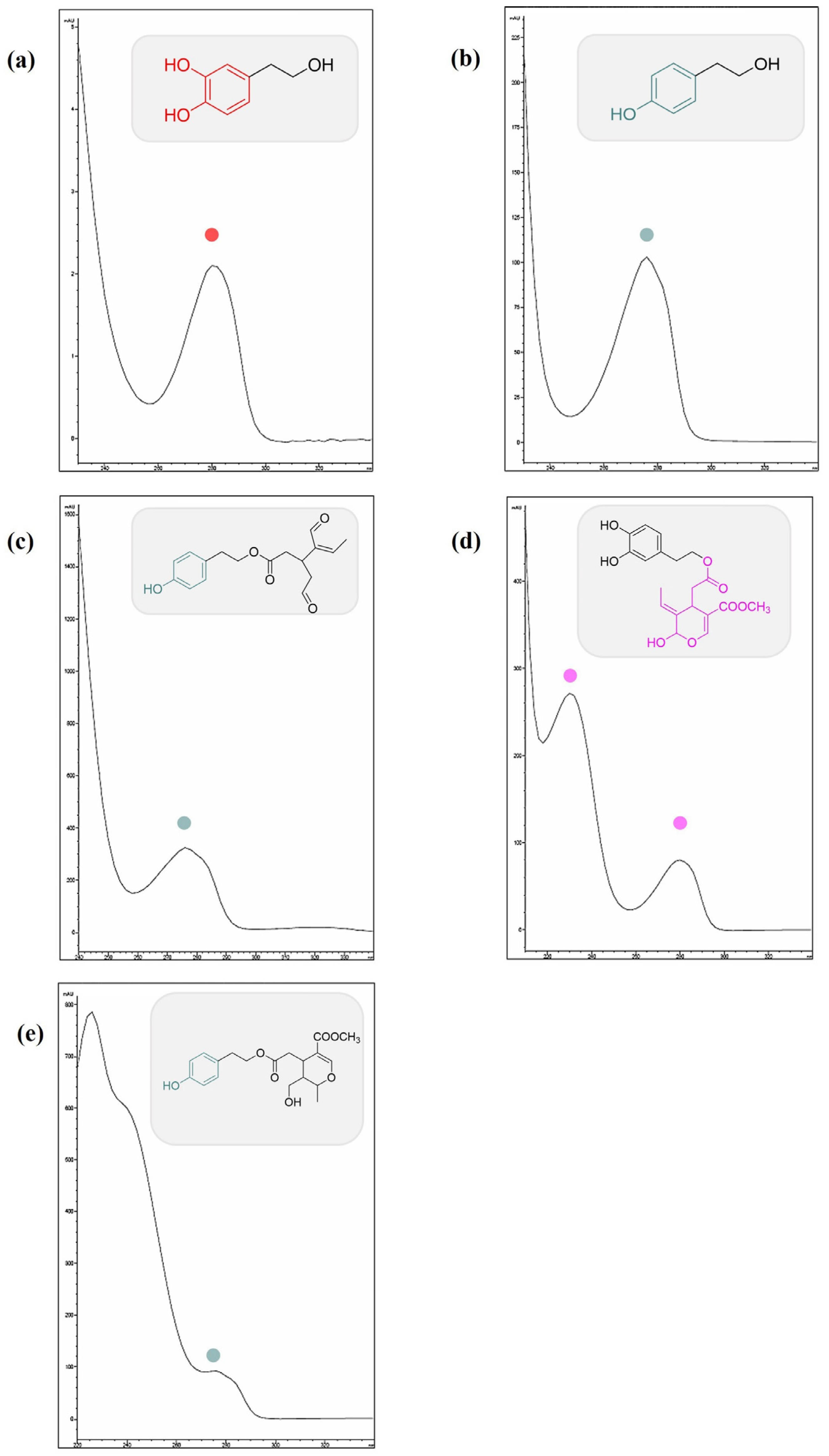
3.1. Identification of the Compounds in EVOOE by HPLC-UV-DAD
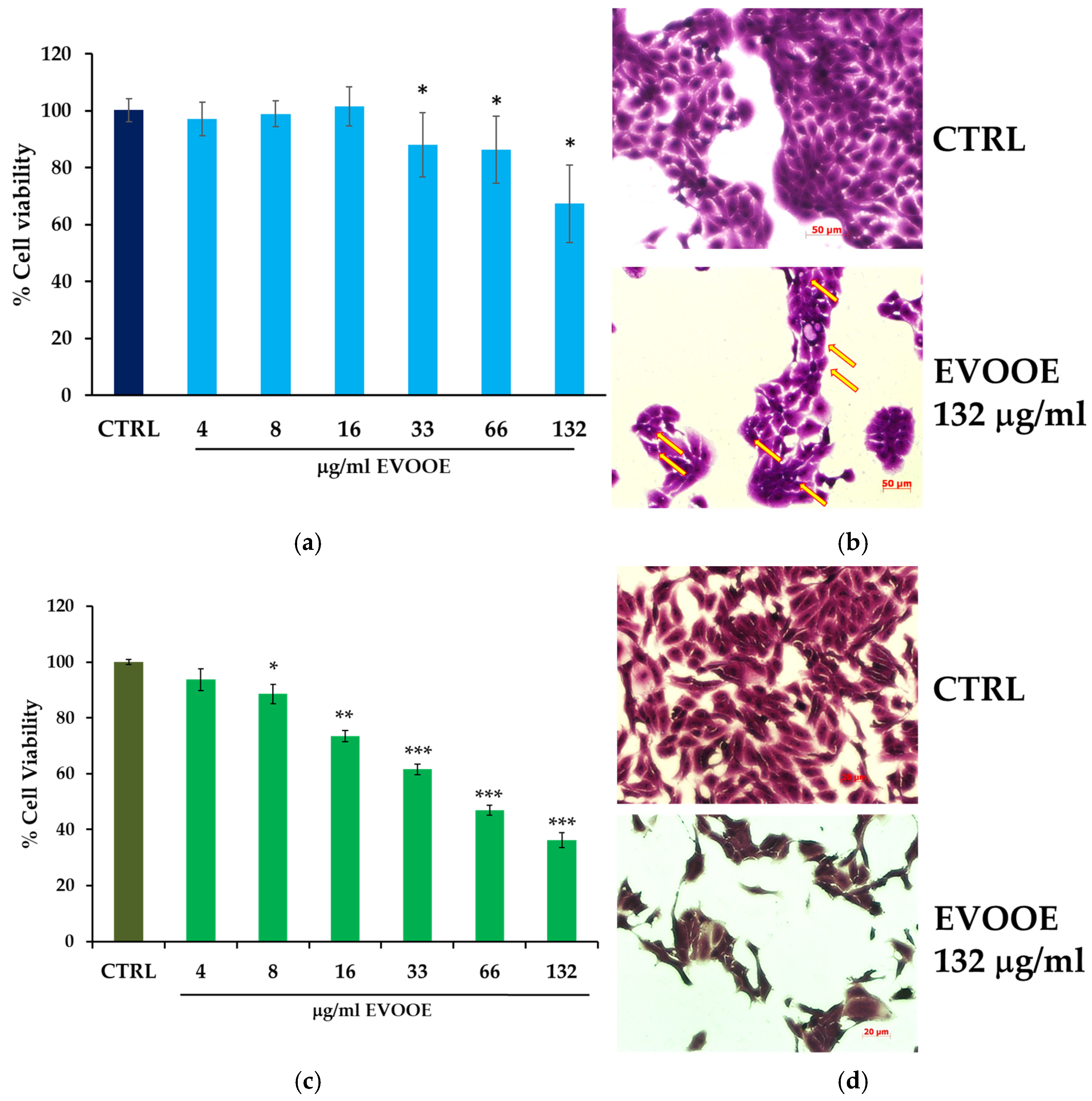
3.2. Extra Virgin Olive Oil Phenolic Extract Stimulation Reduces Cell Viability in RT112 and J82 Bladder Cell Lines
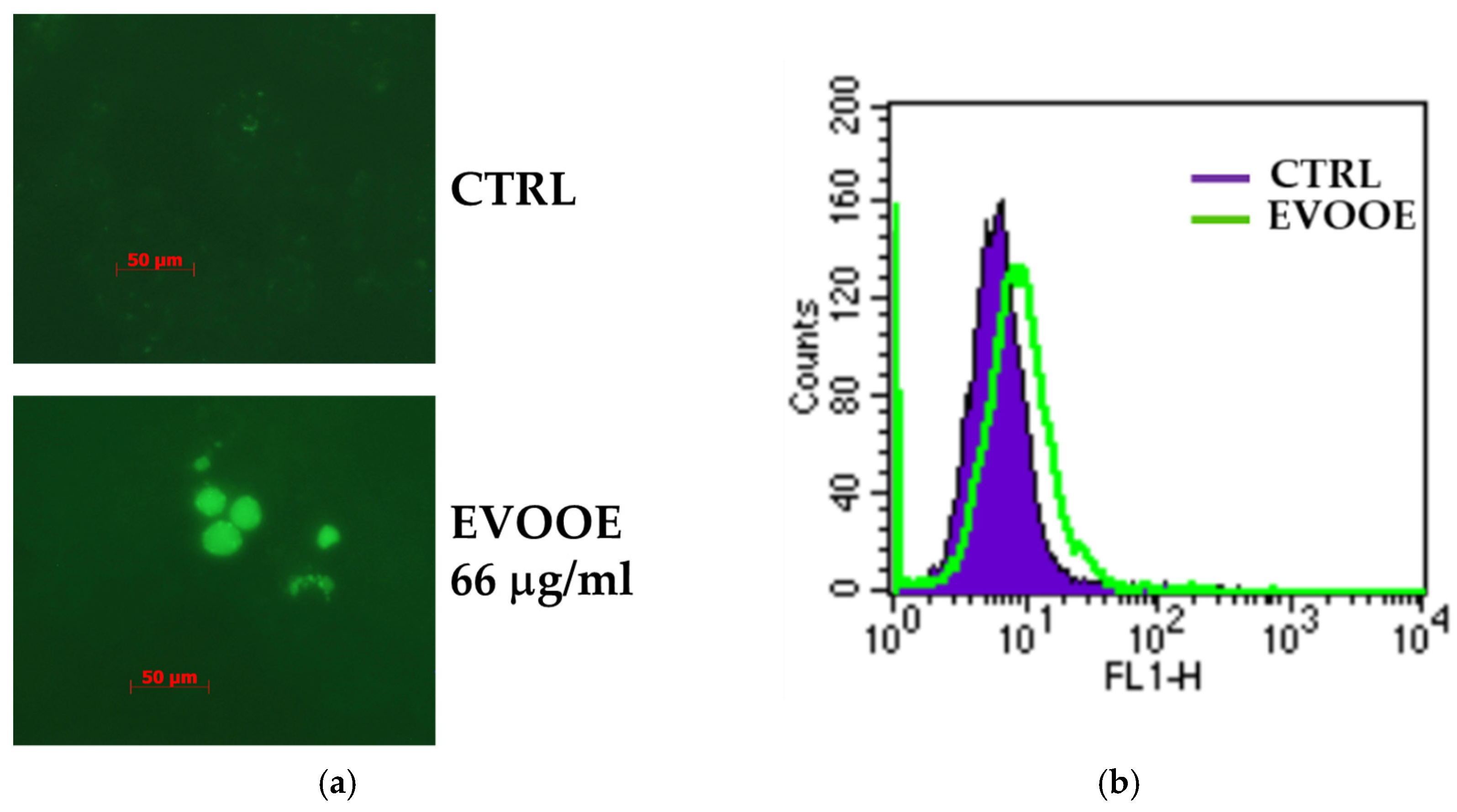
3.3. Extra Virgin Olive Oil Phenolic Extract Induces Autophagy in RT112 Cell Line
3.4. Pro-Apoptotic Effects of Extra Virgin Olive Oil Phenolic Extract in J82 Cell Line
3.5. Antioxidant Activity of EVOOE in Bladder Cancer Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lenis, A.T.; Lec, P.M.; Chamie, K.; Mshs, M.D. Bladder Cancer: A Review. JAMA 2020, 324, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Research Fund; American Institute for Cancer Research. Continuous Update Project Report: Diet, Nutrition, Physical Activity and Bladder Cancer. Available online: https://www.wcrf.org/bladder-cancer-2015 (accessed on 1 September 2022).
- Hong, M.; He, G.; Goh, S.; Low, A.W.X.; Tay, K.J.; Lim, T.K.H.; Yeong, J.; Khor, L.Y.; Lim, T.S. Biomarkers for Precision Urothelial Carcinoma Diagnosis: Current Approaches and the Application of Single-Cell Technologies. Cancers 2021, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- EAU Pocket Guidelines; EAU: Arnhem, The Netherlands, 2022.
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Dugue, P.A.; Hodge, A.M.; Brinkman, M.T.; Bassett, J.K.; Shivappa, N.; Hebert, J.R.; Hopper, J.L.; English, D.R.; Milne, R.L.; Giles, G.G. Association between selected dietary scores and the risk of urothelial cell carcinoma: A prospective cohort study. Int. J. Cancer 2016, 139, 1251–1260. [Google Scholar] [CrossRef]
- Buckland, G.; Ros, M.M.; Roswall, N.; Bueno-de-Mesquita, H.B.; Travier, N.; Tjonneland, A.; Kiemeney, L.A.; Sacerdote, C.; Tumino, R.; Ljungberg, B.; et al. Adherence to the Mediterranean diet and risk of bladder cancer in the EPIC cohort study. Int. J. Cancer 2014, 134, 2504–2511. [Google Scholar] [CrossRef]
- Bravi, F.; Spei, M.E.; Polesel, J.; Di Maso, M.; Montella, M.; Ferraroni, M.; Serraino, D.; Libra, M.; Negri, E.; La Vecchia, C.; et al. Mediterranean Diet and Bladder Cancer Risk in Italy. Nutrients 2018, 10, 1061. [Google Scholar] [CrossRef]
- Witlox, W.J.A.; van Osch, F.H.M.; Brinkman, M.; Jochems, S.; Goossens, M.E.; Weiderpass, E.; White, E.; van den Brandt, P.A.; Giles, G.G.; Milne, R.L.; et al. An inverse association between the Mediterranean diet and bladder cancer risk: A pooled analysis of 13 cohort studies. Eur. J. Nutr. 2020, 59, 287–296. [Google Scholar] [CrossRef]
- Aveta, A.; Cacciapuoti, C.; Barone, B.; Di Zazzo, E.; Del Giudice, F.; Maggi, M.; Ferro, M.; Terracciano, D.; Busetto, G.M.; Lucarelli, G.; et al. The Impact of Meat Intake on Bladder Cancer Incidence: Is It Really a Relevant Risk? Cancers 2022, 14, 4775. [Google Scholar] [CrossRef]
- Grosso, G.; La Vignera, S.; Condorelli, R.A.; Godos, J.; Marventano, S.; Tieri, M.; Ghelfi, F.; Titta, L.; Lafranconi, A.; Gambera, A.; et al. Total, red and processed meat consumption and human health: An umbrella review of observational studies. Int. J. Food Sci. Nutr. 2022, 73, 726–737. [Google Scholar] [CrossRef]
- Brinkman, M.T.; Buntinx, F.; Kellen, E.; Van Dongen, M.C.; Dagnelie, P.C.; Muls, E.; Zeegers, M.P. Consumption of animal products, olive oil and dietary fat and results from the Belgian case-control study on bladder cancer risk. Eur. J. Cancer 2011, 47, 436–442. [Google Scholar] [CrossRef]
- Serreli, G.; Deiana, M. Extra Virgin Olive Oil Polyphenols: Modulation of Cellular Pathways Related to Oxidant Species and Inflammation in Aging. Cells 2020, 9, 478. [Google Scholar] [CrossRef]
- Serreli, G.; Deiana, M. Biological Relevance of Extra Virgin Olive Oil Polyphenols Metabolites. Antioxidants 2018, 7, 170. [Google Scholar] [CrossRef]
- Russo, G.L.; Siani, A.; Fogliano, V.; Geleijnse, J.M.; Giacco, R.; Giampaoli, S.; Iacoviello, L.; Kromhout, D.; Lionetti, L.; Naska, A.; et al. The Mediterranean diet from past to future: Key concepts from the second “Ancel Keys” International Seminar. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 717–732. [Google Scholar] [CrossRef]
- Dais, P.; Hatzakis, E. Quality assessment and authentication of virgin olive oil by NMR spectroscopy: A critical review. Anal. Chim. Acta 2013, 765, 1–27. [Google Scholar] [CrossRef]
- Cicerale, S.; Conlan, X.A.; Sinclair, A.J.; Keast, R.S. Chemistry and health of olive oil phenolics. Crit. Rev. Food Sci. Nutr. 2009, 49, 218–236. [Google Scholar] [CrossRef]
- Garcia-Oliveira, P.; Jimenez-Lopez, C.; Lourenco-Lopes, C.; Chamorro, F.; Pereira, A.G.; Carrera-Casais, A.; Fraga-Corral, M.; Carpena, M.; Simal-Gandara, J.; Prieto, M.A. Evolution of Flavors in Extra Virgin Olive Oil Shelf-Life. Antioxidants 2021, 10, 368. [Google Scholar] [CrossRef]
- Cicerale, S.; Lucas, L.J.; Keast, R.S. Antimicrobial, antioxidant and anti-inflammatory phenolic activities in extra virgin olive oil. Curr. Opin. Biotechnol. 2012, 23, 129–135. [Google Scholar] [CrossRef]
- Pourkerman, M.; Rashidkhani, B.; Moslehi, N. Correlating Dietary Pattern and Bladder Cancer Risk Using Principal Component and Reduced Rank Regression Analyses. Nutr. Cancer 2022, 74, 2955–2963. [Google Scholar] [CrossRef]
- Casaburi, I.; Puoci, F.; Chimento, A.; Sirianni, R.; Ruggiero, C.; Avena, P.; Pezzi, V. Potential of olive oil phenols as chemopreventive and therapeutic agents against cancer: A review of in vitro studies. Mol. Nutr. Food Res. 2013, 57, 71–83. [Google Scholar] [CrossRef]
- Hashim, Y.Z.; Eng, M.; Gill, C.I.; McGlynn, H.; Rowland, I.R. Components of olive oil and chemoprevention of colorectal cancer. Nutr. Rev. 2005, 63, 374–386. [Google Scholar] [CrossRef]
- Johnson, I.T. Phytochemicals and cancer. Proc. Nutr. Soc. 2007, 66, 207–215. [Google Scholar] [CrossRef]
- Izuegbuna, O.O. Polyphenols: Chemoprevention and therapeutic potentials in hematological malignancies. Front. Nutr. 2022, 9, 1008893. [Google Scholar] [CrossRef]
- Owen, R.W.; Giacosa, A.; Hull, W.E.; Haubner, R.; Wurtele, G.; Spiegelhalder, B.; Bartsch, H. Olive-oil consumption and health: The possible role of antioxidants. Lancet Oncol. 2000, 1, 107–112. [Google Scholar] [CrossRef]
- Centrone, M.; D’Agostino, M.; Difonzo, G.; De Bruno, A.; Di Mise, A.; Ranieri, M.; Montemurro, C.; Valenti, G.; Poiana, M.; Caponio, F.; et al. Antioxidant Efficacy of Olive By-Product Extracts in Human Colon HCT8 Cells. Foods 2020, 10, 11. [Google Scholar] [CrossRef]
- Emma, M.R.; Augello, G.; Di Stefano, V.; Azzolina, A.; Giannitrapani, L.; Montalto, G.; Cervello, M.; Cusimano, A. Potential Uses of Olive Oil Secoiridoids for the Prevention and Treatment of Cancer: A Narrative Review of Preclinical Studies. Int. J. Mol. Sci. 2021, 22, 1234. [Google Scholar] [CrossRef] [PubMed]
- Asryants, R.A.; Duszenkova, I.V.; Nagradova, N.K. Determination of Sepharose-bound protein with Coomassie brilliant blue G-250. Anal. Biochem. 1985, 151, 571–574. [Google Scholar] [CrossRef]
- Russo, G.L.; Spagnuolo, C.; Russo, M.; Tedesco, I.; Moccia, S.; Cervellera, C. Mechanisms of aging and potential role of selected polyphenols in extending healthspan. Biochem. Pharmacol. 2020, 173, 113719. [Google Scholar] [CrossRef]
- Zamora-Ros, R.; Sacerdote, C.; Ricceri, F.; Weiderpass, E.; Roswall, N.; Buckland, G.; St-Jules, D.E.; Overvad, K.; Kyro, C.; Fagherazzi, G.; et al. Flavonoid and lignan intake in relation to bladder cancer risk in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Br. J. Cancer 2014, 111, 1870–1880. [Google Scholar] [CrossRef]
- Coccia, A.; Bastianelli, D.; Mosca, L.; Monticolo, R.; Panuccio, I.; Carbone, A.; Calogero, A.; Lendaro, E. Extra virgin olive oil phenols suppress migration and invasion of T24 human bladder cancer cells through modulation of matrix metalloproteinase-2. Nutr. Cancer 2014, 66, 946–954. [Google Scholar] [CrossRef]
- Coccia, A.; Mosca, L.; Puca, R.; Mangino, G.; Rossi, A.; Lendaro, E. Extra-virgin olive oil phenols block cell cycle progression and modulate chemotherapeutic toxicity in bladder cancer cells. Oncol. Rep. 2016, 36, 3095–3104. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.D.; Shiao, C.K.; Lee, Y.C.; Shih, Y.W. Apigenin, a dietary flavonoid, inhibits proliferation of human bladder cancer T-24 cells via blocking cell cycle progression and inducing apoptosis. Cancer Cell Int. 2015, 15, 33. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yuan, M.; Li, S.; Thuan, U.T.; Nguyen, T.T.; Kang, T.W.; Liao, W.; Lian, S.; Jung, Y.D. Apigenin Suppresses the IL-1beta-Induced Expression of the Urokinase-Type Plasminogen Activator Receptor by Inhibiting MAPK-Mediated AP-1 and NF-kappaB Signaling in Human Bladder Cancer T24 Cells. J. Agric. Food Chem. 2018, 66, 7663–7673. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Naiki, T.; Naiki-Ito, A.; Suzuki, S.; Kato, H.; Nozaki, S.; Nagai, T.; Etani, T.; Nagayasu, Y.; Ando, R.; et al. Luteolin suppresses bladder cancer growth via regulation of mechanistic target of rapamycin pathway. Cancer Sci. 2020, 111, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Vasyutin, I.; Zerihun, L.; Ivan, C.; Atala, A. Bladder Organoids and Spheroids: Potential Tools for Normal and Diseased Tissue Modelling. Anticancer Res. 2019, 39, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Rieger, K.M.; Little, A.F.; Swart, J.M.; Kastrinakis, W.V.; Fitzgerald, J.M.; Hess, D.T.; Libertino, J.A.; Summerhayes, I.C. Human bladder carcinoma cell lines as indicators of oncogenic change relevant to urothelial neoplastic progression. Br. J. Cancer 1995, 72, 683–690. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu Reagent. Methods Enzymol. 1999, 229, 152–178. [Google Scholar]
- Marshall, C.J.; Franks, L.M.; Carbonell, A.W. Markers of neoplastic transformation in epithelial cell lines derived from human carcinomas. J. Natl. Cancer Inst. 1977, 58, 1743–1751. [Google Scholar] [CrossRef]
- O’Toole, C.; Price, Z.H.; Ohnuki, Y.; Unsgaard, B. Ultrastructure, karyology and immunology of a cell line originated from a human transitional-cell carcinoma. Br. J. Cancer 1978, 38, 64–76. [Google Scholar] [CrossRef]
- Russo, M.; Spagnuolo, C.; Volpe, S.; Tedesco, I.; Bilotto, S.; Russo, G.L. ABT-737 resistance in B-cells isolated from chronic lymphocytic leukemia patients and leukemia cell lines is overcome by the pleiotropic kinase inhibitor quercetin through Mcl-1 down-regulation. Biochem. Pharmacol. 2013, 85, 927–936. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Francioso, A.; Federico, R.; Maggiore, A.; Fontana, M.; Boffi, A.; D’Erme, M.; Mosca, L. Green Route for the Isolation and Purification of Hyrdoxytyrosol, Tyrosol, Oleacein and Oleocanthal from Extra Virgin Olive Oil. Molecules 2020, 25, 3654. [Google Scholar] [CrossRef] [PubMed]
- Peyrot des Gachons, C.; Beauchamp, G.K.; Stern, R.M.; Koch, K.L.; Breslin, P.A. Bitter taste induces nausea. Curr. Biol. CB 2011, 21, R247–R248. [Google Scholar] [CrossRef] [PubMed]
- Zun-Qiu, W.; Gui-Zhou, Y.; Qing-Ping, Z.; You-Jun, J.; Kai-Yu, T.; Hua-Ping, C.; Ze-Shen, Y.; Qian-Ming, H. Purification, dynamic changes and antioxidant activities of oleuropein in olive (Olea europaea L.). J. Food Biochem. 2015, 39, 566–574. [Google Scholar] [CrossRef]
- Briante, R.; La Cara, F.; Febbraio, F.; Patumi, M.; Nucci, R. Bioactive derivatives from oleuropein by a biotransformation on Olea europaea leaf extracts. J. Biotechnol. 2002, 93, 109–119. [Google Scholar] [CrossRef]
- Papanikolaou, C.; Melliou, E.; Magiatis, P. Olive Oil Phenols. In Functional Foods; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Rios, J.J.; Gil, M.J.; Gutierrez-Rosales, F. Solid-phase extraction gas chromatography-ion trap-mass spectrometry qualitative method for evaluation of phenolic compounds in virgin olive oil and structural confirmation of oleuropein and ligstroside aglycons and their oxidation products. J. Chromatogr. A 2005, 1093, 167–176. [Google Scholar] [CrossRef]
- Negro, C.; Aprile, A.; Luvisi, A.; Nicoli, F.; Nutricati, E.; Vergine, M.; Miceli, A.; Blando, F.; Sabella, E.; De Bellis, L. Phenolic Profile and Antioxidant Activity of Italian Monovarietal Extra Virgin Olive Oils. Antioxidants 2019, 8, 161. [Google Scholar] [CrossRef]
- Pedan, V.; Popp, M.; Rohn, S.; Nyfeler, M.; Bongartz, A. Characterization of Phenolic Compounds and Their Contribution to Sensory Properties of Olive Oil. Molecules 2019, 24, 2041. [Google Scholar] [CrossRef]
- Baiano, A.; Gambacorta, G.; Terracone, C.; Previtali, M.A.; Lamacchia, C.; La Notte, E. Changes in phenolic content and antioxidant activity of Italian extra-virgin olive oils during storage. J. Food Sci. 2009, 74, C177–C183. [Google Scholar] [CrossRef]
- Phenol-Explorer. Available online: http://phenol-explorer.eu/contents/food/822 (accessed on 23 September 2022).
- Long, L.H.; Clement, M.V.; Halliwell, B. Artifacts in cell culture: Rapid generation of hydrogen peroxide on addition of (-)-epigallocatechin, (−)-epigallocatechin gallate, (+)-catechin, and quercetin to commonly used cell culture media. Biochem. Biophys. Res. Commun. 2000, 273, 50–53. [Google Scholar] [CrossRef]
- Jiang, Z.Y.; Hunt, J.V.; Wolff, S.P. Ferrous ion oxidation in the presence of xylenol orange for detection of lipid hydroperoxide in low density lipoprotein. Anal. Biochem. 1992, 202, 384–389. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Gewirtz, D.A. The four faces of autophagy: Implications for cancer therapy. Cancer Res. 2014, 74, 647–651. [Google Scholar] [CrossRef]
- Gewirtz, D.A. Cytoprotective and nonprotective autophagy in cancer therapy. Autophagy 2013, 9, 1263–1265. [Google Scholar] [CrossRef]
- Patel, N.H.; Sohal, S.S.; Manjili, M.H.; Harrell, J.C.; Gewirtz, D.A. The Roles of Autophagy and Senescence in the Tumor Cell Response to Radiation. Radiat. Res. 2020, 194, 103–115. [Google Scholar] [CrossRef]
- Julien, O.; Wells, J.A. Caspases and their substrates. Cell Death Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef]
- Benvenuto, M.; Albonici, L.; Focaccetti, C.; Ciuffa, S.; Fazi, S.; Cifaldi, L.; Miele, M.T.; De Maio, F.; Tresoldi, I.; Manzari, V.; et al. Polyphenol-Mediated Autophagy in Cancer: Evidence of In Vitro and In Vivo Studies. Int. J. Mol. Sci. 2020, 21, 6635. [Google Scholar] [CrossRef]
- Sharma, A.; Kaur, M.; Katnoria, J.K.; Nagpal, A.K. Polyphenols in Food: Cancer Prevention and Apoptosis Induction. Curr. Med. Chem. 2018, 25, 4740–4757. [Google Scholar] [CrossRef]
- Tedesco, I.; Russo, M.; Bilotto, S.; Spagnuolo, C.; Scognamiglio, A.; Palumbo, R.; Nappo, A.; Iacomino, G.; Moio, L.; Russo, G.L. Dealcoholated red wine induces autophagic and apoptotic cell death in an osteosarcoma cell line. Food Chem. Toxicol. 2013, 60, 377–384. [Google Scholar] [CrossRef]
- Li, F.; Guo, H.; Yang, Y.; Feng, M.; Liu, B.; Ren, X.; Zhou, H. Autophagy modulation in bladder cancer development and treatment (Review). Oncol. Rep. 2019, 42, 1647–1655. [Google Scholar] [CrossRef]
- Hoyer-Hansen, M.; Jaattela, M. Autophagy: An emerging target for cancer therapy. Autophagy 2008, 4, 574–580. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, E.; Bai, L.; Li, Y. Autophagy in Cancer Immunotherapy. Cells 2022, 11, 2996. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Hohn, A.; Kruger, K.; Skowron, M.A.; Bormann, S.; Schumacher, L.; Schulz, W.A.; Hoffmann, M.J.; Niegisch, G.; Fritz, G. Distinct mechanisms contribute to acquired cisplatin resistance of urothelial carcinoma cells. Oncotarget 2016, 7, 41320–41335. [Google Scholar] [CrossRef]
- Russo, G.L.; Tedesco, I.; Spagnuolo, C.; Russo, M. Antioxidant polyphenols in cancer treatment: Friend, foe or foil? Semin. Cancer Biol. 2017, 46, 1–13. [Google Scholar] [CrossRef]
- Linder, B.; Kogel, D. Autophagy in Cancer Cell Death. Biology 2019, 8, 82. [Google Scholar] [CrossRef]
- Hatem, E.; El Banna, N.; Huang, M.E. Multifaceted Roles of Glutathione and Glutathione-Based Systems in Carcinogenesis and Anticancer Drug Resistance. Antioxid. Redox Signal. 2017, 27, 1217–1234. [Google Scholar] [CrossRef]
- Oyenihi, A.B.; Smith, C. Are polyphenol antioxidants at the root of medicinal plant anti-cancer success? J. Ethnopharmacol. 2019, 229, 54–72. [Google Scholar] [CrossRef]
- Moskaug, J.O.; Carlsen, H.; Myhrstad, M.C.; Blomhoff, R. Polyphenols and glutathione synthesis regulation. Am. J. Clin. Nutr. 2005, 81, 277S–283S. [Google Scholar] [CrossRef]
- Carlsen, H.; Myhrstad, M.C.; Thoresen, M.; Moskaug, J.O.; Blomhoff, R. Berry intake increases the activity of the gamma-glutamylcysteine synthetase promoter in transgenic reporter mice. J. Nutr. 2003, 133, 2137–2140. [Google Scholar] [CrossRef]
- Myhrstad, M.C.; Carlsen, H.; Nordstrom, O.; Blomhoff, R.; Moskaug, J.O. Flavonoids increase the intracellular glutathione level by transactivation of the gamma-glutamylcysteine synthetase catalytical subunit promoter. Free Radic. Biol. Med. 2002, 32, 386–393. [Google Scholar] [CrossRef]
- Tomasetti, M.; Santarelli, L.; Alleva, R.; Dong, L.F.; Neuzil, J. Redox-active and redox-silent compounds: Synergistic therapeutics in cancer. Curr. Med. Chem. 2015, 22, 552–568. [Google Scholar] [CrossRef]
- Cruz-Lozano, M.; Gonzalez-Gonzalez, A.; Marchal, J.A.; Munoz-Muela, E.; Molina, M.P.; Cara, F.E.; Brown, A.M.; Garcia-Rivas, G.; Hernandez-Brenes, C.; Lorente, J.A.; et al. Hydroxytyrosol inhibits cancer stem cells and the metastatic capacity of triple-negative breast cancer cell lines by the simultaneous targeting of epithelial-to-mesenchymal transition, Wnt/beta-catenin and TGFbeta signaling pathways. Eur. J. Nutr. 2019, 58, 3207–3219. [Google Scholar] [CrossRef]
- Bertelli, M.; Kiani, A.K.; Paolacci, S.; Manara, E.; Kurti, D.; Dhuli, K.; Bushati, V.; Miertus, J.; Pangallo, D.; Baglivo, M.; et al. Hydroxytyrosol: A natural compound with promising pharmacological activities. J. Biotechnol. 2020, 309, 29–33. [Google Scholar] [CrossRef]
- Karkovic Markovic, A.; Toric, J.; Barbaric, M.; Jakobusic Brala, C. Hydroxytyrosol, Tyrosol and Derivatives and Their Potential Effects on Human Health. Molecules 2019, 24, 2001. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H. Targeting oxidative stress in disease: Promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021, 20, 689–709. [Google Scholar] [CrossRef]
- De Stefanis, D.; Scime, S.; Accomazzo, S.; Catti, A.; Occhipinti, A.; Bertea, C.M.; Costelli, P. Anti-Proliferative Effects of an Extra-Virgin Olive Oil Extract Enriched in Ligstroside Aglycone and Oleocanthal on Human Liver Cancer Cell Lines. Cancers 2019, 11, 1640. [Google Scholar] [CrossRef]
- Ahmad Farooqi, A.; Fayyaz, S.; Silva, A.S.; Sureda, A.; Nabavi, S.F.; Mocan, A.; Nabavi, S.M.; Bishayee, A. Oleuropein and Cancer Chemoprevention: The Link is Hot. Molecules 2017, 22, 705. [Google Scholar] [CrossRef]
- Fayyaz, S.; Aydin, T.; Cakir, A.; Gasparri, M.L.; Panici, P.B.; Farooqi, A.A. Oleuropein Mediated Targeting of Signaling Network in Cancer. Curr. Top. Med. Chem. 2016, 16, 2477–2483. [Google Scholar] [CrossRef]
- Silva, S.; Garcia-Aloy, M.; Figueira, M.E.; Combet, E.; Mullen, W.; Bronze, M.R. High Resolution Mass Spectrometric Analysis of Secoiridoids and Metabolites as Biomarkers of Acute Olive Oil Intake-An Approach to Study Interindividual Variability in Humans. Mol. Nutr. Food Res. 2018, 62, 1700065. [Google Scholar] [CrossRef]
- John, B.A.; Said, N. Insights from animal models of bladder cancer: Recent advances, challenges, and opportunities. Oncotarget 2017, 8, 57766–57781. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, S.; Hirose, M.; Tsuda, H.; Shirai, T.; Hirao, K. Histological classification of urinary bladder cancers in rats induced by N-butyl-n-(4-hydroxybutyl)nitrosamine. Gan 1976, 67, 81–90. [Google Scholar] [PubMed]
- Lin, J.H.; Zhao, H.; Sun, T.T. A tissue-specific promoter that can drive a foreign gene to express in the suprabasal urothelial cells of transgenic mice. Proc. Natl. Acad. Sci. USA 1995, 92, 679–683. [Google Scholar] [CrossRef] [PubMed]


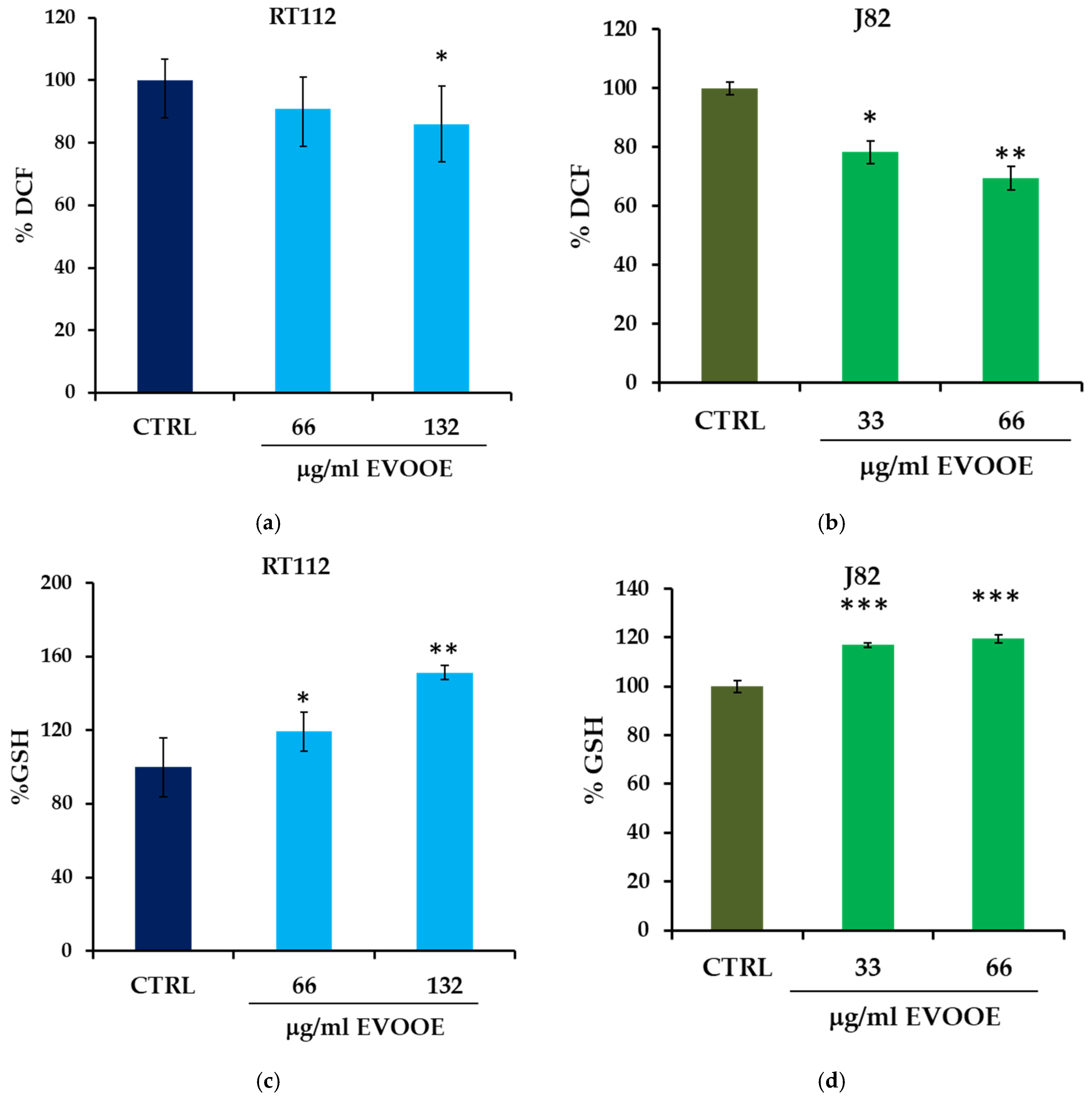
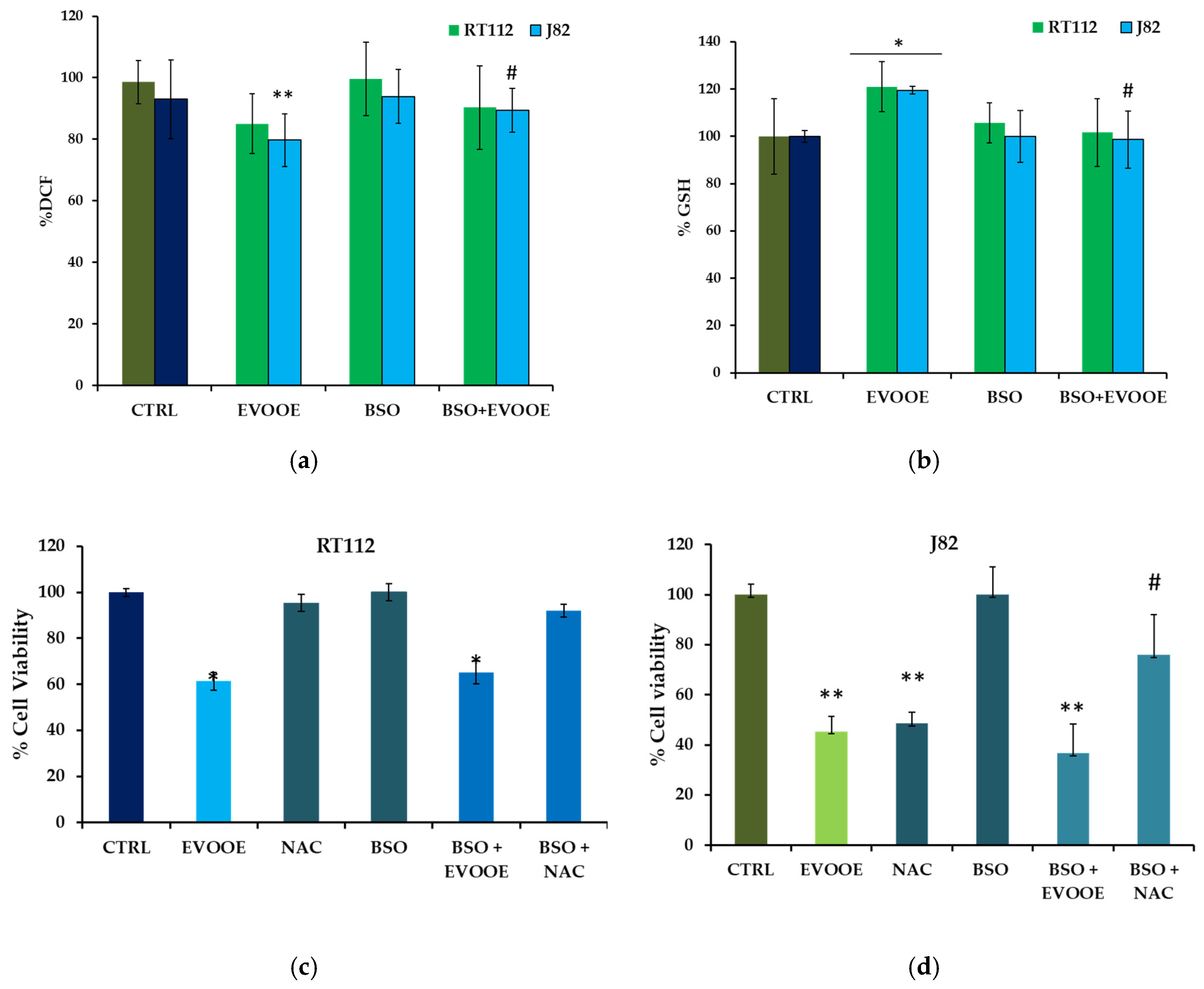
| Samples | % DCF Reduction (30 min) | % Cell Death (24 h) |
|---|---|---|
| Quercetin 30 μM | 34.3 ± 3.2 # | 31.1 ± 1.2 ** |
| Gallic Acid 30 μM | 49.9 ± 4.0 # | 1.8 ± 2.0 |
| Myricetin 30 μM | 46.0 ± 9.6 # | 1.7 ± 5.7 |
| Kaempferol 30 μM | 40.6 ± 8.1 # | 12.9 ± 4.5 * |
| Rutin 30 μM | 48.9 ± 5.3 # | 15.1 ± 4.5 * |
| Green Tea 5 μg/mL | 51.0 ± 6.2 # | 0.1 ± 2.4 |
| EVOOE 33 μg/mL | 57.9 ± 4.0 # | 43.7 ± 2.8 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spagnuolo, C.; Moccia, S.; Tedesco, I.; Crescente, G.; Volpe, M.G.; Russo, M.; Russo, G.L. Phenolic Extract from Extra Virgin Olive Oil Induces Different Anti-Proliferative Pathways in Human Bladder Cancer Cell Lines. Nutrients 2023, 15, 182. https://doi.org/10.3390/nu15010182
Spagnuolo C, Moccia S, Tedesco I, Crescente G, Volpe MG, Russo M, Russo GL. Phenolic Extract from Extra Virgin Olive Oil Induces Different Anti-Proliferative Pathways in Human Bladder Cancer Cell Lines. Nutrients. 2023; 15(1):182. https://doi.org/10.3390/nu15010182
Chicago/Turabian StyleSpagnuolo, Carmela, Stefania Moccia, Idolo Tedesco, Giuseppina Crescente, Maria Grazia Volpe, Maria Russo, and Gian Luigi Russo. 2023. "Phenolic Extract from Extra Virgin Olive Oil Induces Different Anti-Proliferative Pathways in Human Bladder Cancer Cell Lines" Nutrients 15, no. 1: 182. https://doi.org/10.3390/nu15010182
APA StyleSpagnuolo, C., Moccia, S., Tedesco, I., Crescente, G., Volpe, M. G., Russo, M., & Russo, G. L. (2023). Phenolic Extract from Extra Virgin Olive Oil Induces Different Anti-Proliferative Pathways in Human Bladder Cancer Cell Lines. Nutrients, 15(1), 182. https://doi.org/10.3390/nu15010182











