Body Mass Index and the Risk of Atrial Fibrillation: A Mendelian Randomization Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Instrumental Variable Selection
2.3. Statistical Analysis
2.3.1. Mendelian Randomization Analyses
2.3.2. Sensitivity Analyses
3. Results
3.1. Validity of Instrumental Variables
3.2. Mendelian Randomization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef] [PubMed]
- Hindricks, G.; Potpara, T.; Dagres, N.; Arbelo, E.; Bax, J.J.; Blomstrom-Lundqvist, C.; Boriani, G.; Castella, M.; Dan, G.A.; Dilaveris, P.E.; et al. 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): The Task Force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC) Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur. Heart J. 2021, 42, 373–498. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, R.B.; Yin, X.; Gona, P.; Larson, M.G.; Beiser, A.S.; McManus, D.D.; Newton-Cheh, C.; Lubitz, S.A.; Magnani, J.W.; Ellinor, P.T.; et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: A cohort study. Lancet 2015, 386, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Krijthe, B.P.; Kunst, A.; Benjamin, E.J.; Lip, G.Y.; Franco, O.H.; Hofman, A.; Witteman, J.C.; Stricker, B.H.; Heeringa, J. Projections on the number of individuals with atrial fibrillation in the European Union, from 2000 to 2060. Eur. Heart J. 2013, 34, 2746–2751. [Google Scholar] [CrossRef] [Green Version]
- Ball, J.; Carrington, M.J.; McMurray, J.J.; Stewart, S. Atrial fibrillation: Profile and burden of an evolving epidemic in the 21st century. Int. J. Cardiol. 2013, 167, 1807–1824. [Google Scholar] [CrossRef]
- Kim, Y.G.; Han, K.D.; Choi, J.I.; Boo, K.Y.; Kim, D.Y.; Oh, S.K.; Lee, K.N.; Shim, J.; Kim, J.S.; Kim, Y.H. The impact of body weight and diabetes on new-onset atrial fibrillation: A nationwide population based study. Cardiovasc. Diabetol. 2019, 18, 128. [Google Scholar] [CrossRef] [Green Version]
- Asad, Z.; Abbas, M.; Javed, I.; Korantzopoulos, P.; Stavrakis, S. Obesity is associated with incident atrial fibrillation independent of gender: A meta-analysis. J. Cardiovasc. Electrophysiol. 2018, 29, 725–732. [Google Scholar] [CrossRef]
- Lee, H.; Choi, E.K.; Lee, S.H.; Han, K.D.; Rhee, T.M.; Park, C.S.; Lee, S.R.; Choe, W.S.; Lim, W.H.; Kang, S.H.; et al. Atrial fibrillation risk in metabolically healthy obesity: A nationwide population-based study. Int. J. Cardiol. 2017, 240, 221–227. [Google Scholar] [CrossRef]
- Nystrom, P.K.; Carlsson, A.C.; Leander, K.; de Faire, U.; Hellenius, M.L.; Gigante, B. Obesity, metabolic syndrome and risk of atrial fibrillation: A Swedish, prospective cohort study. PLoS ONE 2015, 10, e0127111. [Google Scholar] [CrossRef] [Green Version]
- Carreras-Torres, R.; Johansson, M.; Haycock, P.C.; Relton, C.L.; Davey Smith, G.; Brennan, P.; Martin, R.M. Role of obesity in smoking behaviour: Mendelian randomisation study in UK Biobank. BMJ 2018, 361, k1767. [Google Scholar] [CrossRef] [Green Version]
- Katan, M.B. Commentary: Mendelian Randomization, 18 years on. Int. J. Epidemiol. 2004, 33, 10–11. [Google Scholar] [CrossRef]
- Smith, G.D.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Davey Smith, G.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef] [Green Version]
- Georgiopoulos, G.; Ntritsos, G.; Stamatelopoulos, K.; Tsioufis, C.; Aimo, A.; Masi, S.; Evangelou, E. The relationship between blood pressure and risk of atrial fibrillation: A Mendelian randomization study. Eur. J. Prev. Cardiol. 2021, 28, 1617. [Google Scholar] [CrossRef]
- Yuan, S.; Carter, P.; Bruzelius, M.; Vithayathil, M.; Kar, S.; Mason, A.M.; Lin, A.; Burgess, S.; Larsson, S.C. Effects of tumour necrosis factor on cardiovascular disease and cancer: A two-sample Mendelian randomization study. EBioMedicine 2020, 59, 102956. [Google Scholar] [CrossRef]
- Wang, T.; Xu, L. Circulating Vitamin E Levels and Risk of Coronary Artery Disease and Myocardial Infarction: A Mendelian Randomization Study. Nutrients 2019, 11, 2153. [Google Scholar] [CrossRef] [Green Version]
- Allara, E.; Morani, G.; Carter, P.; Gkatzionis, A.; Zuber, V.; Foley, C.N.; Rees, J.M.B.; Mason, A.M.; Bell, S.; Gill, D.; et al. Genetic Determinants of Lipids and Cardiovascular Disease Outcomes: A Wide-Angled Mendelian Randomization Investigation. Circ. Genom. Precis. Med. 2019, 12, e002711. [Google Scholar] [CrossRef]
- Pierce, B.L.; Burgess, S. Efficient design for Mendelian randomization studies: Subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 2013, 178, 1177–1184. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [Green Version]
- Yengo, L.; Sidorenko, J.; Kemper, K.E.; Zheng, Z.; Wood, A.R.; Weedon, M.N.; Frayling, T.M.; Hirschhorn, J.; Yang, J.; Visscher, P.M.; et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Nielsen, J.B.; Thorolfsdottir, R.B.; Fritsche, L.G.; Zhou, W.; Skov, M.W.; Graham, S.E.; Herron, T.J.; McCarthy, S.; Schmidt, E.M.; Sveinbjornsson, G.; et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 2018, 50, 1234–1239. [Google Scholar] [CrossRef]
- Larsson, S.C. Mendelian randomization as a tool for causal inference in human nutrition and metabolism. Curr. Opin. Lipidol. 2021, 32, 1–8. [Google Scholar] [CrossRef]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, R.M. Influence of Inflammation and Atherosclerosis in Atrial Fibrillation. Curr. Atheroscler. Rep. 2017, 19, 2. [Google Scholar] [CrossRef]
- Kallistratos, M.S.; Poulimenos, L.E.; Manolis, A.J. Atrial fibrillation and arterial hypertension. Pharmacol. Res. 2018, 128, 322–326. [Google Scholar] [CrossRef]
- Chung, M.K.; Eckhardt, L.L.; Chen, L.Y.; Ahmed, H.M.; Gopinathannair, R.; Joglar, J.A.; Noseworthy, P.A.; Pack, Q.R.; Sanders, P.; Trulock, K.M.; et al. Lifestyle and Risk Factor Modification for Reduction of Atrial Fibrillation: A Scientific Statement from the American Heart Association. Circulation 2020, 141, e750–e772. [Google Scholar] [CrossRef]
- Yao, Y.S.; Liu, F.; Wang, Y.Y.; Liu, Z.Z. Lipid levels and risk of new-onset atrial fibrillation: A systematic review and dose-response meta-analysis. Clin. Cardiol. 2020, 43, 935–943. [Google Scholar] [CrossRef]
- Bowden, J.; Spiller, W.; Del Greco, M.F.; Sheehan, N.; Thompson, J.; Minelli, C.; Davey Smith, G. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int. J. Epidemiol. 2018, 47, 2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zheng, Z.; Zhang, F.; Wu, Y.; Trzaskowski, M.; Maier, R.; Robinson, M.R.; McGrath, J.J.; Visscher, P.M.; Wray, N.R.; et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018, 9, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.Y.; Wang, J.S.; Hemani, G.; Bowden, J.; Small, D.S. Statistical Inference in Two-Sample Summary-Data Mendelian Randomization Using Robust Adjusted Profile Score. Ann. Stat. 2020, 48, 1742–1769. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.A.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef] [Green Version]
- Kuchenhoff, H.; Mwalili, S.M.; Lesaffre, E. A general method for dealing with misclassification in regression: The misclassification SIMEX. Biometrics 2006, 62, 85–96. [Google Scholar] [CrossRef]
- Hatala, R.; Keitz, S.; Wyer, P.; Guyatt, G.; Evidence-Based Medicine Teaching Tips Working, G. Tips for learners of evidence-based medicine: 4. Assessing heterogeneity of primary studies in systematic reviews and whether to combine their results. CMAJ 2005, 172, 661–665. [Google Scholar] [CrossRef] [Green Version]
- Stock, J.H.; Wright, J.H.; Yogo, M. A survey of weak instruments and weak identification in generalized method of moments. J. Bus. Econ. Stat. 2002, 20, 518–529. [Google Scholar] [CrossRef]
- Andersen, K.; Rasmussen, F.; Neovius, M.; Tynelius, P.; Sundstrom, J. Body size and risk of atrial fibrillation: A cohort study of 1.1 million young men. J. Intern. Med. 2018, 283, 346–355. [Google Scholar] [CrossRef]
- Feng, T.; Vegard, M.; Strand, L.B.; Laugsand, L.E.; Morkedal, B.; Aune, D.; Vatten, L.; Ellekjaer, H.; Loennechen, J.P.; Mukamal, K.; et al. Weight and weight change and risk of atrial fibrillation: The HUNT study. Eur. Heart J. 2019, 40, 2859–2866. [Google Scholar] [CrossRef]
- Javed, S.; Gupta, D.; Lip, G.Y.H. Obesity and atrial fibrillation: Making inroads through fat. Eur. Heart J. Cardiovasc. Pharm. 2021, 7, 59–67. [Google Scholar] [CrossRef]
- Lavie, C.J.; Pandey, A.; Lau, D.H.; Alpert, M.A.; Sanders, P. Obesity and Atrial Fibrillation Prevalence, Pathogenesis, and Prognosis: Effects of Weight Loss and Exercise. J. Am. Coll. Cardiol. 2017, 70, 2022–2035. [Google Scholar] [CrossRef]
- Stritzke, J.; Markus, M.R.; Duderstadt, S.; Lieb, W.; Luchner, A.; Doring, A.; Keil, U.; Hense, H.W.; Schunkert, H.; Investigators, M.K. The aging process of the heart: Obesity is the main risk factor for left atrial enlargement during aging the MONICA/KORA (monitoring of trends and determinations in cardiovascular disease/cooperative research in the region of Augsburg) study. J. Am. Coll. Cardiol. 2009, 54, 1982–1989. [Google Scholar] [CrossRef]
- Munger, T.M.; Dong, Y.X.; Masaki, M.; Oh, J.K.; Mankad, S.V.; Borlaug, B.A.; Asirvatham, S.J.; Shen, W.K.; Lee, H.C.; Bielinski, S.J.; et al. Electrophysiological and hemodynamic characteristics associated with obesity in patients with atrial fibrillation. J. Am. Coll. Cardiol. 2012, 60, 851–860. [Google Scholar] [CrossRef] [Green Version]
- Alpert, M.A.; Omran, J.; Bostick, B.P. Effects of Obesity on Cardiovascular Hemodynamics, Cardiac Morphology, and Ventricular Function. Curr. Obes. Rep. 2016, 5, 424–434. [Google Scholar] [CrossRef]
- Wong, C.X.; Ganesan, A.N.; Selvanayagam, J.B. Epicardial fat and atrial fibrillation: Current evidence, potential mechanisms, clinical implications, and future directions. Eur. Heart J. 2017, 38, 1294–1302. [Google Scholar] [CrossRef] [Green Version]
- Hatem, S.N.; Sanders, P. Epicardial adipose tissue and atrial fibrillation. Cardiovasc. Res. 2014, 102, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Venteclef, N.; Guglielmi, V.; Balse, E.; Gaborit, B.; Cotillard, A.; Atassi, F.; Amour, J.; Leprince, P.; Dutour, A.; Clement, K.; et al. Human epicardial adipose tissue induces fibrosis of the atrial myocardium through the secretion of adipo-fibrokines. Eur. Heart J. 2015, 36, 795–805. [Google Scholar] [CrossRef] [Green Version]
- Thanassoulis, G.; Massaro, J.M.; O’Donnell, C.J.; Hoffmann, U.; Levy, D.; Ellinor, P.T.; Wang, T.J.; Schnabel, R.B.; Vasan, R.S.; Fox, C.S.; et al. Pericardial fat is associated with prevalent atrial fibrillation: The Framingham Heart Study. Circ. Arrhythm. Electrophysiol. 2010, 3, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Harada, M.; Van Wagoner, D.R.; Nattel, S. Role of inflammation in atrial fibrillation pathophysiology and management. Circ. J. 2015, 79, 495–502. [Google Scholar] [CrossRef] [Green Version]
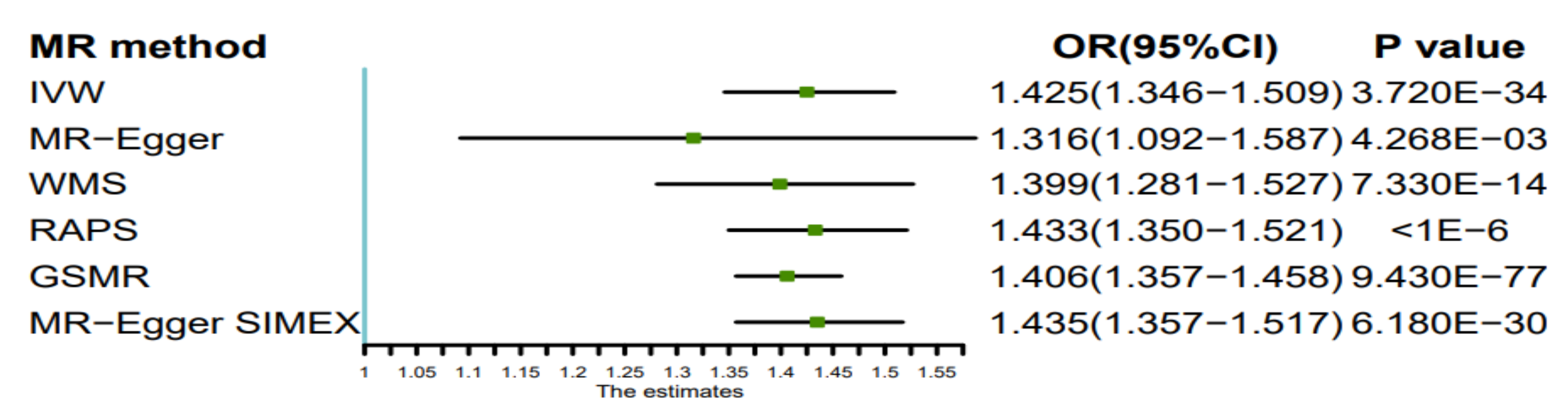
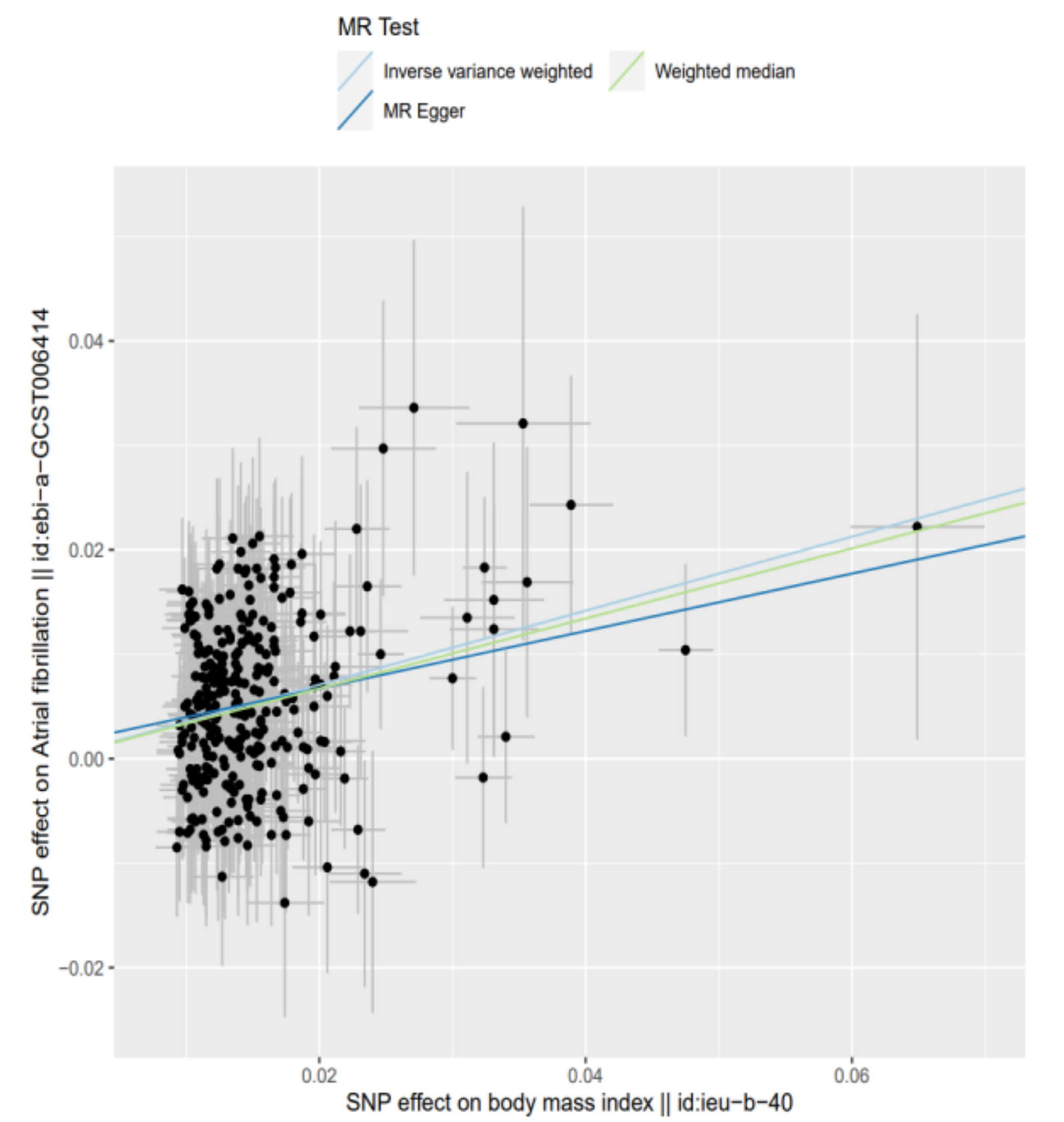
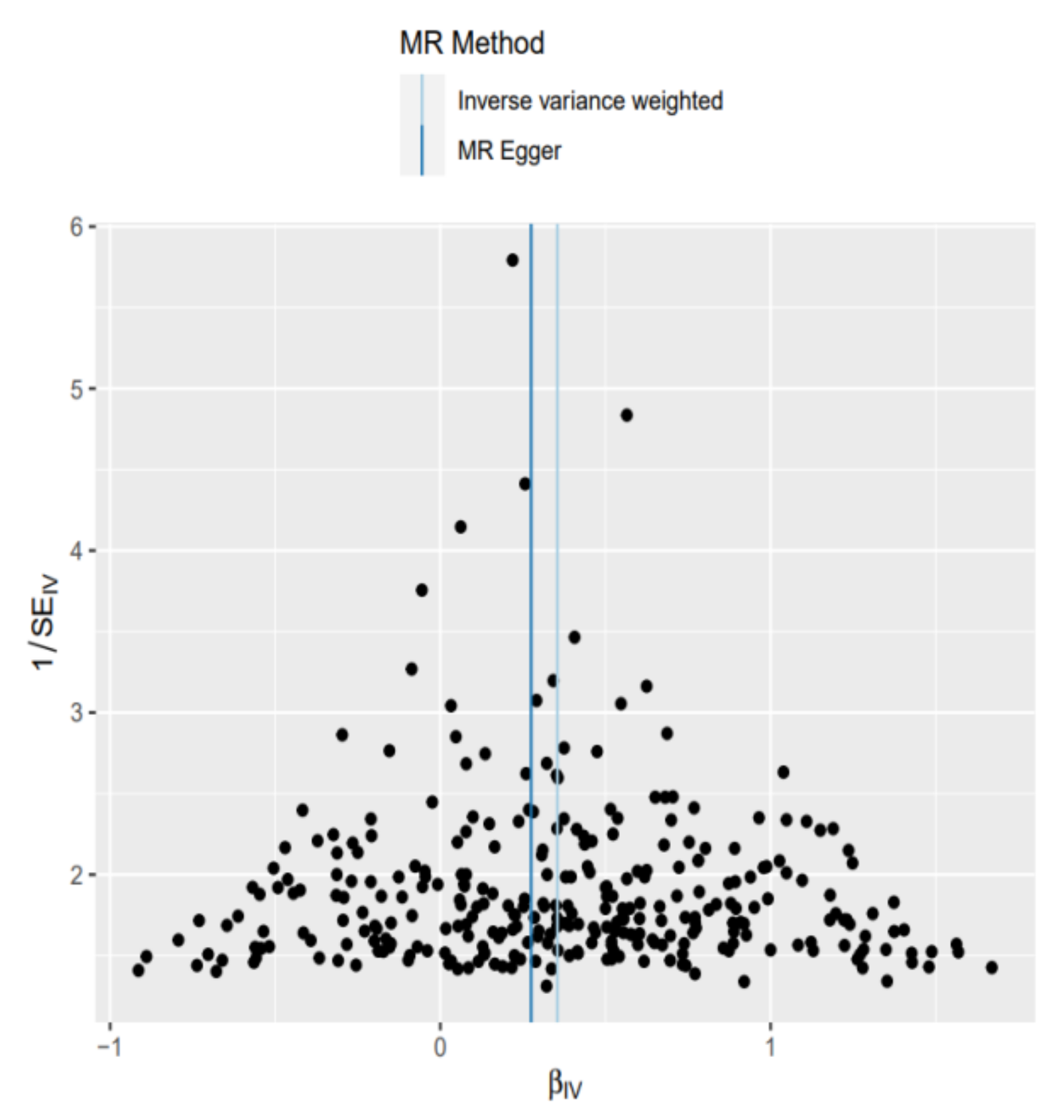
| MR Method | No. of SNPs | Beta | SE | P | OR (95%CI) |
|---|---|---|---|---|---|
| Inverse variance weighted | 303 | 0.354 | 0.029 | 3.720 × 10−34 | 1.425 (1.346–1.509) |
| MR-Egger | 303 | 0.275 | 0.095 | 4.268 × 10−3 | 1.316 (1.092–1.587) |
| Weighted median estimator | 303 | 0.336 | 0.045 | 7.330 × 10−14 | 1.399 (1.281–1.527) |
| RAPS | 303 | 0.360 | 0.030 | <1 × 10−6 | 1.433 (1.350–1.521) |
| GSMR | 303 | 0.341 | 0.018 | 9.430 × 10−77 | 1.406 (1.357–1.458) |
| MR-Egger SIMEX | 303 | 0.361 | 0.028 | 6.180 × 10−30 | 1.435 (1.357–1.517) |
| Cochran’s Q Test | I2 | MR-Egger | MR-Egger SIMEX | |||
|---|---|---|---|---|---|---|
| Q | p | Intercept (95%CI) | p | Intercept (95%CI) | p | |
| 277.28 | 0.84 | 8.91% | 0.0012(−0.0015–0.0040) | 0.38 | −0.0002 (−0.0010–0.0006) | 0.61 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, M.; Zhi, H.; Yang, S.; Yu, E.Y.-W.; Wang, L. Body Mass Index and the Risk of Atrial Fibrillation: A Mendelian Randomization Study. Nutrients 2022, 14, 1878. https://doi.org/10.3390/nu14091878
Ma M, Zhi H, Yang S, Yu EY-W, Wang L. Body Mass Index and the Risk of Atrial Fibrillation: A Mendelian Randomization Study. Nutrients. 2022; 14(9):1878. https://doi.org/10.3390/nu14091878
Chicago/Turabian StyleMa, Mi, Hong Zhi, Shengyi Yang, Evan Yi-Wen Yu, and Lina Wang. 2022. "Body Mass Index and the Risk of Atrial Fibrillation: A Mendelian Randomization Study" Nutrients 14, no. 9: 1878. https://doi.org/10.3390/nu14091878
APA StyleMa, M., Zhi, H., Yang, S., Yu, E. Y.-W., & Wang, L. (2022). Body Mass Index and the Risk of Atrial Fibrillation: A Mendelian Randomization Study. Nutrients, 14(9), 1878. https://doi.org/10.3390/nu14091878






