Description of a Newly Isolated Blautia faecis Strain and Its Benefit in Mouse Models of Post-Influenza Secondary Enteric and Pulmonary Infections
Abstract
:1. Introduction
2. Material and Methods
2.1. Isolation of Extremely Oxygen-Sensitive Strains
2.2. Blautia Strains Used
2.3. Culture Conditions for Blautia hansenii DSM20583 and Blautia faecis DSM33383
2.4. Supernatant Collection from Bacterial Cultures
2.5. 16S rDNA Identification of EOS Strains by PCR from Pure Colony
2.6. Genomic DNA Extraction of Blautia faecis DSM33383 16S rDNA for Sequencing and Phylogenetic Analysis
2.7. Measurement of SCFA Concentrations in Bacterial Supernatant
2.8. Epithelial Cell Culture and In Vitro Anti-Inflammatory Assays
2.9. IL-8 Cytokine Quantification
2.10. Mice and Ethics Statement
2.11. Infections Gastric Administration and Assessment of Bacterial Loads in Mice
2.12. Measurement of SCFA Concentrations in Cecal Content of Mice
2.13. RNA Manipulation Procedures and Real-Time Quantitative RT-PCR
2.14. Statistical Analysis
3. Results
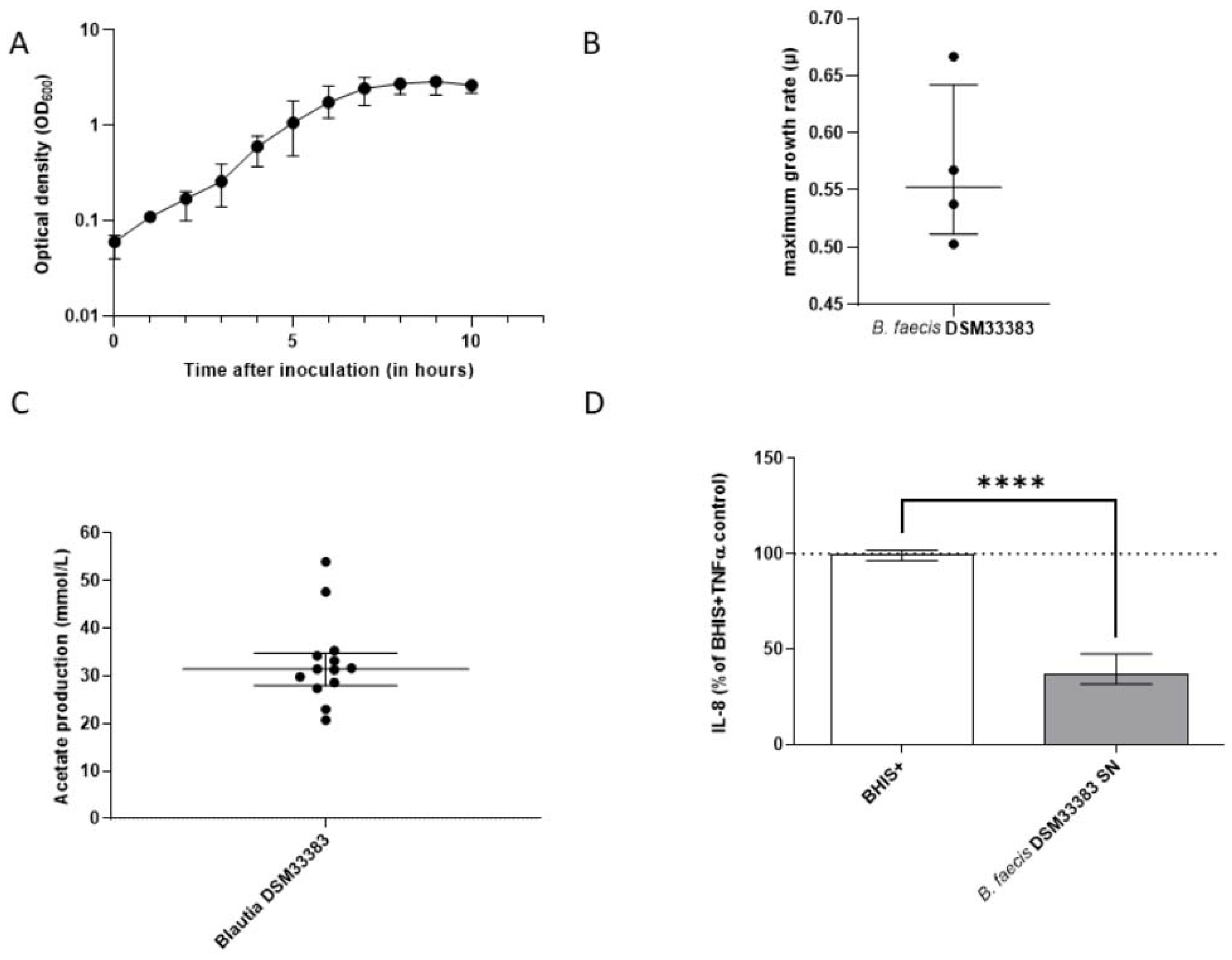
3.1. The Isolation of Extremely Oxygen-Sensitive Strains and Identification of a High-Acetate-Producing EOS Strain-Blautia faecis DSM33383
3.2. The Characteristics of Blautia faecis DSM33383
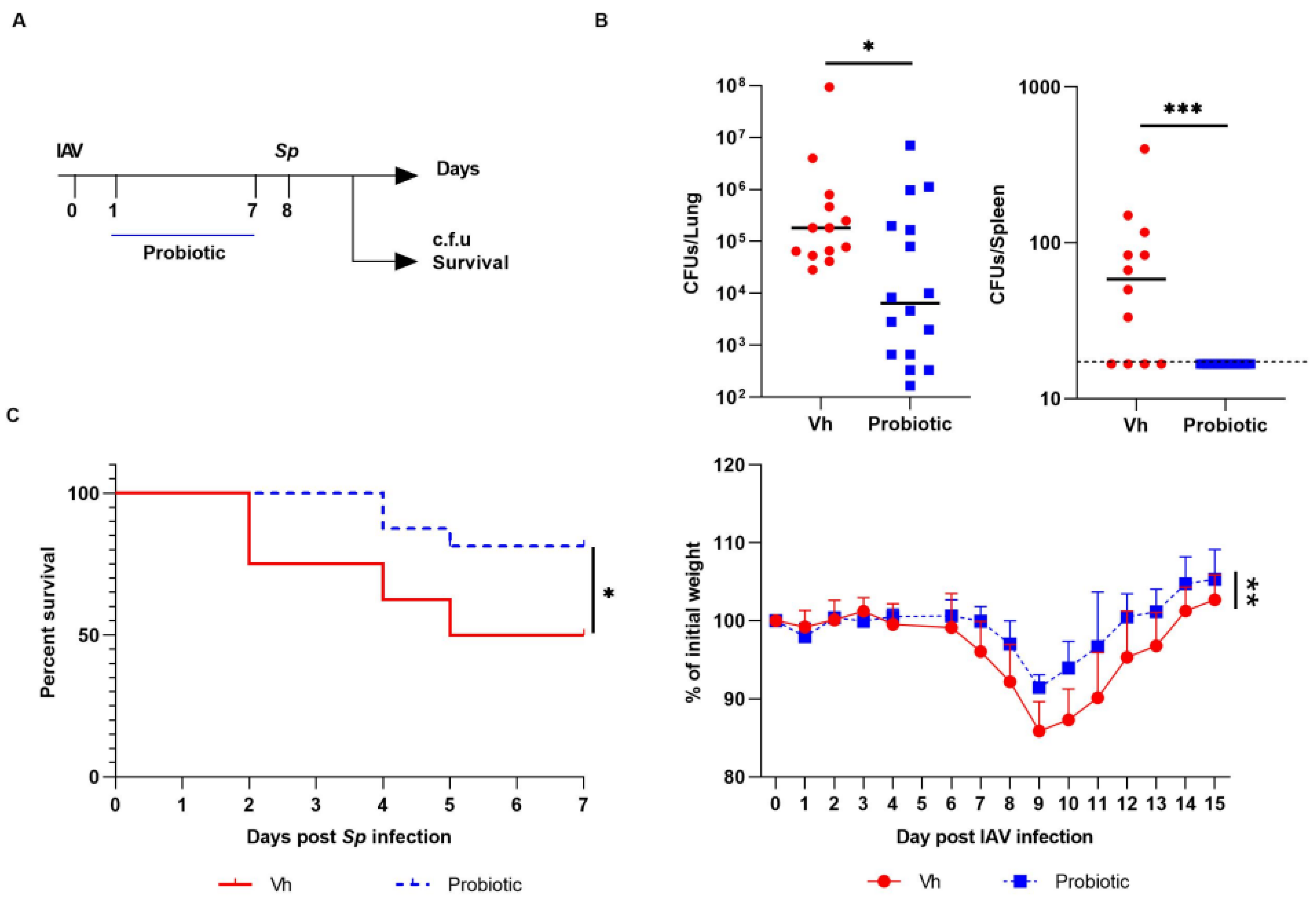
3.3. Blautia faecis DSM33383 Administration during Influenza Protects against Lung Bacterial Superinfection with Streptococcus pneumoniae
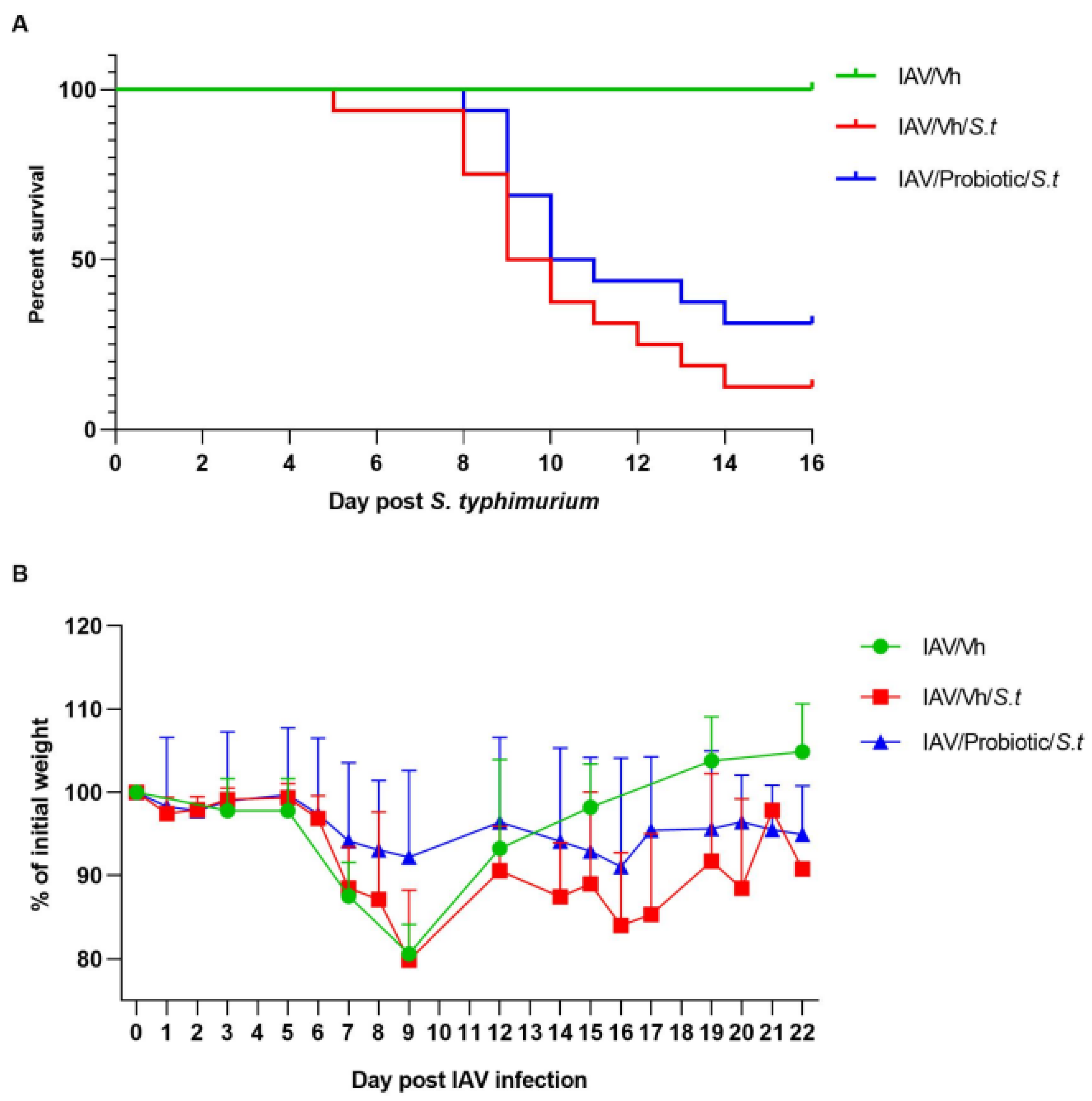
3.4. Blautia faecis DSM33383 Administration Tends to Reduce Secondary Salmonella enterica serovar Typhimurium Infection Post-Influenza
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lagkouvardos, I.; Overmann, J.; Clavel, T. Cultured microbes represent a substantial fraction of the human and mouse gut microbiota. Gut Microbes 2017, 8, 493–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef] [PubMed]
- Le Roy, T.; Moens de Hase, E.; Van Hul, M.; Paquot, A.; Pelicaen, R.; Regnier, M.; Depommier, C.; Druart, C.; Everard, A.; Maiter, D.; et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 2022, 71, 534–543. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.; Miquel, S.; Chain, F.; Natividad, J.M.; Jury, J.; Lu, J.; Sokol, H.; Theodorou, V.; Bercik, P.; Verdu, E.F.; et al. Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 2015, 15, 67. [Google Scholar] [CrossRef] [Green Version]
- Miquel, S.; Leclerc, M.; Martin, R.; Chain, F.; Lenoir, M.; Raguideau, S.; Hudault, S.; Bridonneau, C.; Northen, T.; Bowen, B.; et al. Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio 2015, 6, e00300-15. [Google Scholar] [CrossRef] [Green Version]
- Quevrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermudez-Humaran, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [Green Version]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut Bifidobacteria Populations in Human Health and Aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [Green Version]
- Ohira, H.; Tsutsui, W.; Fujioka, Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J. Atheroscler. Thromb. 2017, 24, 660–672. [Google Scholar] [CrossRef] [Green Version]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [Green Version]
- Bloemen, J.G.; Venema, K.; van de Poll, M.C.; Olde Damink, S.W.; Buurman, W.A.; Dejong, C.H. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin. Nutr. 2009, 28, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Pomare, E.W.; Branch, W.J.; Cummings, J.H. Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J. Clin. Investig. 1985, 75, 1448–1454. [Google Scholar] [CrossRef] [Green Version]
- Bach Knudsen, K.E.; Laerke, H.N.; Hedemann, M.S.; Nielsen, T.S.; Ingerslev, A.K.; Gundelund Nielsen, D.S.; Theil, P.K.; Purup, S.; Hald, S.; Schioldan, A.G.; et al. Impact of Diet-Modulated Butyrate Production on Intestinal Barrier Function and Inflammation. Nutrients 2018, 10, 1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrzosek, L.; Miquel, S.; Noordine, M.L.; Bouet, S.; Joncquel Chevalier-Curt, M.; Robert, V.; Philippe, C.; Bridonneau, C.; Cherbuy, C.; Robbe-Masselot, C.; et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013, 11, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weitkunat, K.; Stuhlmann, C.; Postel, A.; Rumberger, S.; Fankhanel, M.; Woting, A.; Petzke, K.J.; Gohlke, S.; Schulz, T.J.; Blaut, M.; et al. Short-chain fatty acids and inulin, but not guar gum, prevent diet-induced obesity and insulin resistance through differential mechanisms in mice. Sci. Rep. 2017, 7, 6109. [Google Scholar] [CrossRef]
- Ishizaka, S.; Kikuchi, E.; Higashino, T.; Kinoshita, K.; Tsujii, T. Effects of acetate on the immune system of mice. Int. J. Immunopharmacol. 1990, 12, 135–143. [Google Scholar] [CrossRef]
- Qiu, J.; Villa, M.; Sanin, D.E.; Buck, M.D.; O’Sullivan, D.; Ching, R.; Matsushita, M.; Grzes, K.M.; Winkler, F.; Chang, C.H.; et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep. 2019, 27, 2063–2074.e2065. [Google Scholar] [CrossRef] [Green Version]
- Balmer, M.L.; Ma, E.H.; Bantug, G.R.; Grahlert, J.; Pfister, S.; Glatter, T.; Jauch, A.; Dimeloe, S.; Slack, E.; Dehio, P.; et al. Memory CD8(+) T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity 2016, 44, 1312–1324. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal. Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef] [Green Version]
- Vieira, A.T.; Galvao, I.; Macia, L.M.; Sernaglia, E.M.; Vinolo, M.A.; Garcia, C.C.; Tavares, L.P.; Amaral, F.A.; Sousa, L.P.; Martins, F.S.; et al. Dietary fiber and the short-chain fatty acid acetate promote resolution of neutrophilic inflammation in a model of gout in mice. J. Leukoc. Biol. 2017, 101, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.K.; Gu, M.J.; Ko, K.H.; Bae, S.; Kim, G.; Jin, G.D.; Kim, E.B.; Kong, Y.Y.; Park, T.S.; Park, B.C.; et al. Regulation of CD4(+)CD8(-)CD25(+) and CD4(+)CD8(+)CD25(+) T cells by gut microbiota in chicken. Sci. Rep. 2018, 8, 8627. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.G.; Sencio, V.; Trottein, F. Short-Chain Fatty Acids as a Potential Treatment for Infections: A Closer Look at the Lungs. Infect. Immun. 2021, 89, e0018821. [Google Scholar] [CrossRef] [PubMed]
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut through Type I Interferons. PLoS Pathog. 2016, 12, e1005572. [Google Scholar] [CrossRef]
- Sencio, V.; Barthelemy, A.; Tavares, L.P.; Machado, M.G.; Soulard, D.; Cuinat, C.; Queiroz-Junior, C.M.; Noordine, M.L.; Salome-Desnoulez, S.; Deryuter, L.; et al. Gut Dysbiosis during Influenza Contributes to Pulmonary Pneumococcal Superinfection through Altered Short-Chain Fatty Acid Production. Cell Rep. 2020, 30, 2934–2947.e2936. [Google Scholar] [CrossRef] [Green Version]
- Sencio, V.; Gallerand, A.; Gomes Machado, M.; Deruyter, L.; Heumel, S.; Soulard, D.; Barthelemy, J.; Cuinat, C.; Vieira, A.T.; Barthelemy, A.; et al. Influenza Virus Infection Impairs the Gut’s Barrier Properties and Favors Secondary Enteric Bacterial Infection through Reduced Production of Short-Chain Fatty Acids. Infect. Immun. 2021, 89, e0073420. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [Green Version]
- Dupraz, L.; Magniez, A.; Rolhion, N.; Richard, M.L.; Da Costa, G.; Touch, S.; Mayeur, C.; Planchais, J.; Agus, A.; Danne, C.; et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal gammadelta T cells. Cell Rep. 2021, 36, 109332. [Google Scholar] [CrossRef]
- Kechaou, N.; Chain, F.; Gratadoux, J.J.; Blugeon, S.; Bertho, N.; Chevalier, C.; Le Goffic, R.; Courau, S.; Molimard, P.; Chatel, J.M.; et al. Identification of one novel candidate probiotic Lactobacillus plantarum strain active against influenza virus infection in mice by a large-scale screening. Appl. Environ. Microbiol. 2013, 79, 1491–1499. [Google Scholar] [CrossRef] [Green Version]
- Barthelemy, A.; Ivanov, S.; Fontaine, J.; Soulard, D.; Bouabe, H.; Paget, C.; Faveeuw, C.; Trottein, F. Influenza A virus-induced release of interleukin-10 inhibits the anti-microbial activities of invariant natural killer T cells during invasive pneumococcal superinfection. Mucosal. Immunol. 2017, 10, 460–469. [Google Scholar] [CrossRef] [Green Version]
- Barthelemy, A.; Sencio, V.; Soulard, D.; Deruyter, L.; Faveeuw, C.; Le Goffic, R.; Trottein, F. Interleukin-22 Immunotherapy during Severe Influenza Enhances Lung Tissue Integrity and Reduces Secondary Bacterial Systemic Invasion. Infect. Immun. 2018, 86, e00706-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullers, J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014, 12, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; de Sousa e Melo, F.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016, 65, 575–583. [Google Scholar] [CrossRef] [Green Version]
- Thevaranjan, N.; Puchta, A.; Schulz, C.; Naidoo, A.; Szamosi, J.C.; Verschoor, C.P.; Loukov, D.; Schenck, L.P.; Jury, J.; Foley, K.P.; et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe 2017, 21, 455–466.e454. [Google Scholar] [CrossRef] [Green Version]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Saint-Criq, V.; Lugo-Villarino, G.; Thomas, M. Dysbiosis, malnutrition and enhanced gut-lung axis contribute to age-related respiratory diseases. Ageing Res. Rev. 2021, 66, 101235. [Google Scholar] [CrossRef]
- Ji, J.J.; Sun, Q.M.; Nie, D.Y.; Wang, Q.; Zhang, H.; Qin, F.F.; Wang, Q.S.; Lu, S.F.; Pang, G.M.; Lu, Z.G. Probiotics protect against RSV infection by modulating the microbiota-alveolar-macrophage axis. Acta Pharmacol. Sin. 2021, 42, 1630–1641. [Google Scholar] [CrossRef]
- Burapan, S.; Kim, M.; Han, J. Demethylation of Polymethoxyflavones by Human Gut Bacterium, Blautia sp. MRG-PMF1. J. Agric. Food Chem. 2017, 65, 1620–1629. [Google Scholar] [CrossRef]
- Burapan, S.; Kim, M.; Han, J. Curcuminoid Demethylation as an Alternative Metabolism by Human Intestinal Microbiota. J. Agric. Food Chem. 2017, 65, 3305–3310. [Google Scholar] [CrossRef]
- Kim, M.; Kim, N.; Han, J. Metabolism of Kaempferia parviflora polymethoxyflavones by human intestinal bacterium Bautia sp. MRG-PMF1. J. Agric. Food Chem. 2014, 62, 12377–12383. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.; Blanc, F.; Cherbuy, C.; Egidy, G.; Giuffra, E.; Lacroix-Lamande, S.; Wiedemann, A. Intestinal organoids in farm animals. Vet. Res. 2021, 52, 33. [Google Scholar] [CrossRef] [PubMed]
- Lukovac, S.; Belzer, C.; Pellis, L.; Keijser, B.J.; de Vos, W.M.; Montijn, R.C.; Roeselers, G. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio 2014, 5, e01438-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, I.A.; Arnold, J.W.; Samsa, L.A.; Gaynor, L.; DiSalvo, M.; Cocchiaro, J.L.; Carroll, I.; Azcarate-Peril, M.A.; Rawls, J.F.; Allbritton, N.L.; et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 301–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Bhatia, R.; Khare, P.; Sharma, S.; Rajarammohan, S.; Bishnoi, M.; Bhadada, S.K.; Sharma, S.S.; Kaur, J.; Kondepudi, K.K. Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate induced colitis and associated gut microbial dysbiosis in mice. Sci. Rep. 2020, 10, 18597. [Google Scholar] [CrossRef]
- Fujiwara, S.; Seto, Y.; Kimura, A.; Hashiba, H. Intestinal transit of an orally administered streptomycin-rifampicin-resistant variant of Bifidobacterium longum SBT2928: Its long-term survival and effect on the intestinal microflora and metabolism. J. Appl. Microbiol. 2001, 90, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Maldonado-Gomez, M.X.; Martinez, I.; Bottacini, F.; O’Callaghan, A.; Ventura, M.; van Sinderen, D.; Hillmann, B.; Vangay, P.; Knights, D.; Hutkins, R.W.; et al. Stable Engraftment of Bifidobacterium longum AH1206 in the Human Gut Depends on Individualized Features of the Resident Microbiome. Cell Host Microbe 2016, 20, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Mining Lactobacillus and Bifidobacterium for organisms with long-term gut colonization potential. Clin. Nutr. 2020, 39, 1315–1323. [Google Scholar] [CrossRef]
- Wang, J.J.; Wang, J.; Pang, X.Y.; Zhao, L.P.; Tian, L.; Wang, X.P. Sex differences in colonization of gut microbiota from a man with short-term vegetarian and inulin-supplemented diet in germ-free mice. Sci. Rep. 2016, 6, 36137. [Google Scholar] [CrossRef]
- George, F.; Daniel, C.; Thomas, M.; Singer, E.; Guilbaud, A.; Tessier, F.J.; Revol-Junelles, A.M.; Borges, F.; Foligne, B. Occurrence and Dynamism of Lactic Acid Bacteria in Distinct Ecological Niches: A Multifaceted Functional Health Perspective. Front. Microbiol. 2018, 9, 2899. [Google Scholar] [CrossRef] [Green Version]
- Miquel, S.; Beaumont, M.; Martin, R.; Langella, P.; Braesco, V.; Thomas, M. A proposed framework for an appropriate evaluation scheme for microorganisms as novel foods with a health claim in Europe. Microb. Cell Fact. 2015, 14, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forssten, S.D.; Laitila, A.; Maukonen, J.; Ouwehand, A.C. Probiotic triangle of success; strain production, clinical studies and product development. FEMS Microbiol. Lett. 2020, 367, fnaa167. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquet, J.C.; Claus, S.P.; Cordaillat-Simmons, M.; Mazier, W.; Rawadi, G.; Rinaldi, L.; Elustondo, F.; Rouanet, A. Entering First-in-Human Clinical Study With a Single-Strain Live Biotherapeutic Product: Input and Feedback Gained From the EMA and the FDA. Front. Med. 2021, 8, 716266. [Google Scholar] [CrossRef]


| Strain | 16S rDNA NCBI Blast | Acetate Production (mM) | Butyrate Production (mM) | Propionate Production (mM) |
|---|---|---|---|---|
| C7 | Ruminococcus sp. (99%) | 4.9 ± 2.9 | nd | nd |
| D4 | Coprococcus eutactus (99%) | 3.4 ± 1.5 | 6.6 ± 3.3 | nd |
| E5 | Ruminococcus albus (94%) | 4.7 | nd | nd |
| E9 | Fusicatenibacter saccharivorans (99%) | 8.2 ± 5.2 | nd | nd |
| E12 | Eubacterium ventriosum (99%) | 2.3 ± 1.5 | 7.5 ± 2.6 | nd |
| F2 | Coprococcus comes (99%) | 7.2 ± 4.9 | 6.5 ± 3.9 | nd |
| MA4 | Anaerostipes hadrus (99%) | nd | 16.7 ± 6.6 | nd |
| MA8 | Fusicatenibacter saccharivorans (99%) | 4.8 ± 8.3 | nd | nd |
| MA9 | Ruminococcus sp. (97%) | 6.4 | nd | nd |
| MA11 | Roseburia faecis (99%) | nd | 11.2 ± 4.4 | nd |
| MB6 | Eubacterium ventriosum (99%) | 1.8 ± 0.7 | 5.2 ± 2.4 | nd |
| DSM33383 | Blautia sp (96%) | 31.3 ± 6.2 | nd | nd |
| MC1 | Eubacterium ventriosum (98%) | 1.6 ± 2.0 | 6.5 ± 4.5 | nd |
| MC4 | Roseburia sp. (91%) | nd | 10.6 | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verstraeten, S.; Sencio, V.; Raise, A.; Huillet, E.; Layec, S.; Deruyter, L.; Heumel, S.; Auger, S.; Robert, V.; Langella, P.; et al. Description of a Newly Isolated Blautia faecis Strain and Its Benefit in Mouse Models of Post-Influenza Secondary Enteric and Pulmonary Infections. Nutrients 2022, 14, 1478. https://doi.org/10.3390/nu14071478
Verstraeten S, Sencio V, Raise A, Huillet E, Layec S, Deruyter L, Heumel S, Auger S, Robert V, Langella P, et al. Description of a Newly Isolated Blautia faecis Strain and Its Benefit in Mouse Models of Post-Influenza Secondary Enteric and Pulmonary Infections. Nutrients. 2022; 14(7):1478. https://doi.org/10.3390/nu14071478
Chicago/Turabian StyleVerstraeten, Sophie, Valentin Sencio, Audrey Raise, Eugénie Huillet, Séverine Layec, Lucie Deruyter, Séverine Heumel, Sandrine Auger, Véronique Robert, Philippe Langella, and et al. 2022. "Description of a Newly Isolated Blautia faecis Strain and Its Benefit in Mouse Models of Post-Influenza Secondary Enteric and Pulmonary Infections" Nutrients 14, no. 7: 1478. https://doi.org/10.3390/nu14071478
APA StyleVerstraeten, S., Sencio, V., Raise, A., Huillet, E., Layec, S., Deruyter, L., Heumel, S., Auger, S., Robert, V., Langella, P., Beney, L., Trottein, F., & Thomas, M. (2022). Description of a Newly Isolated Blautia faecis Strain and Its Benefit in Mouse Models of Post-Influenza Secondary Enteric and Pulmonary Infections. Nutrients, 14(7), 1478. https://doi.org/10.3390/nu14071478






