Astaxanthin Attenuates the Changes in the Expression of MicroRNAs Involved in the Activation of Hepatic Stellate Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Primary Mouse HSC Isolation and Culture
2.2. ASTX Treatment
2.3. miRNA Array and Quantitative Real-Time PCR (qRT-PCR)
2.4. RNA Sequencing and Identification of Target Genes
2.5. Target Gene Analysis by Reverse Transcription and qRT-PCR
2.6. Statistical Analysis
3. Results
3.1. The Expression of miRNAs Involved in Fibrosis Was Measured in qHSC, aHSC, and aHSC Treated with ASTX
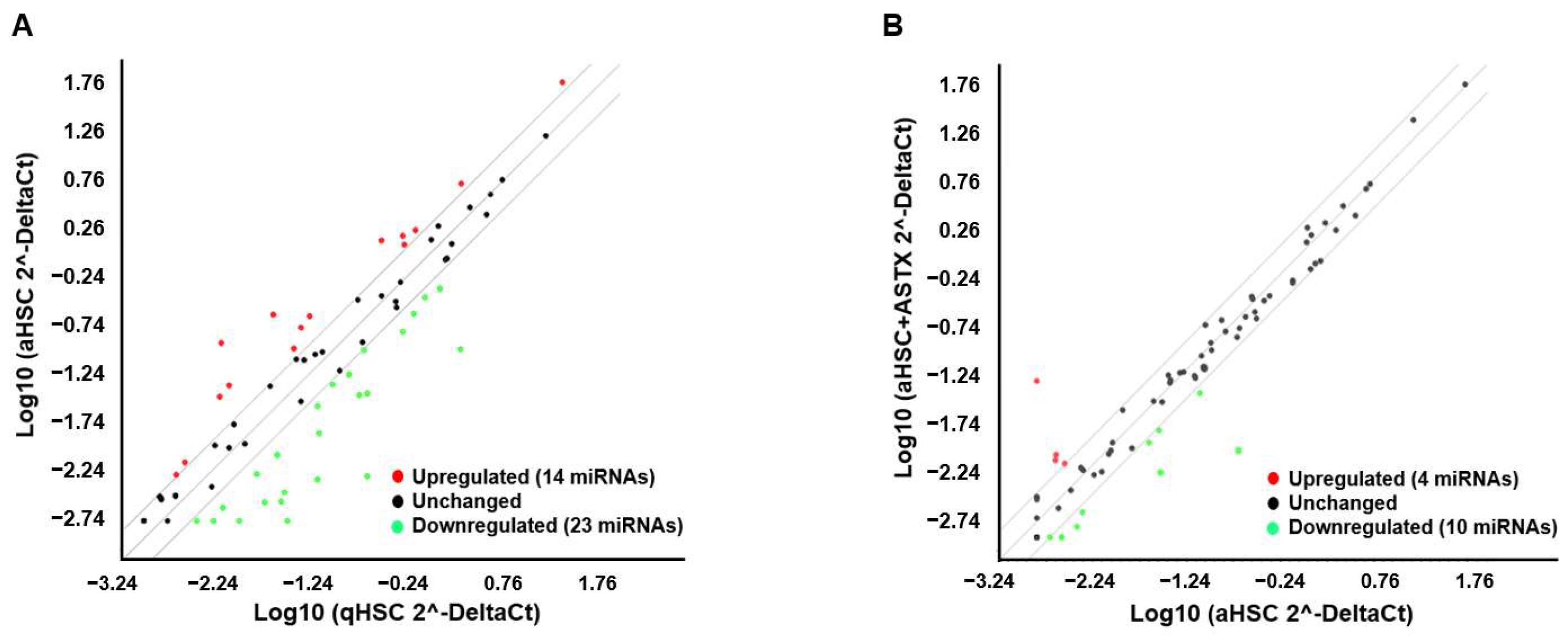
3.2. miRNAs Were Identified Whose Expression Was Altered in aHSC Compared to qHSC, Which Was Attenuated by ASTX
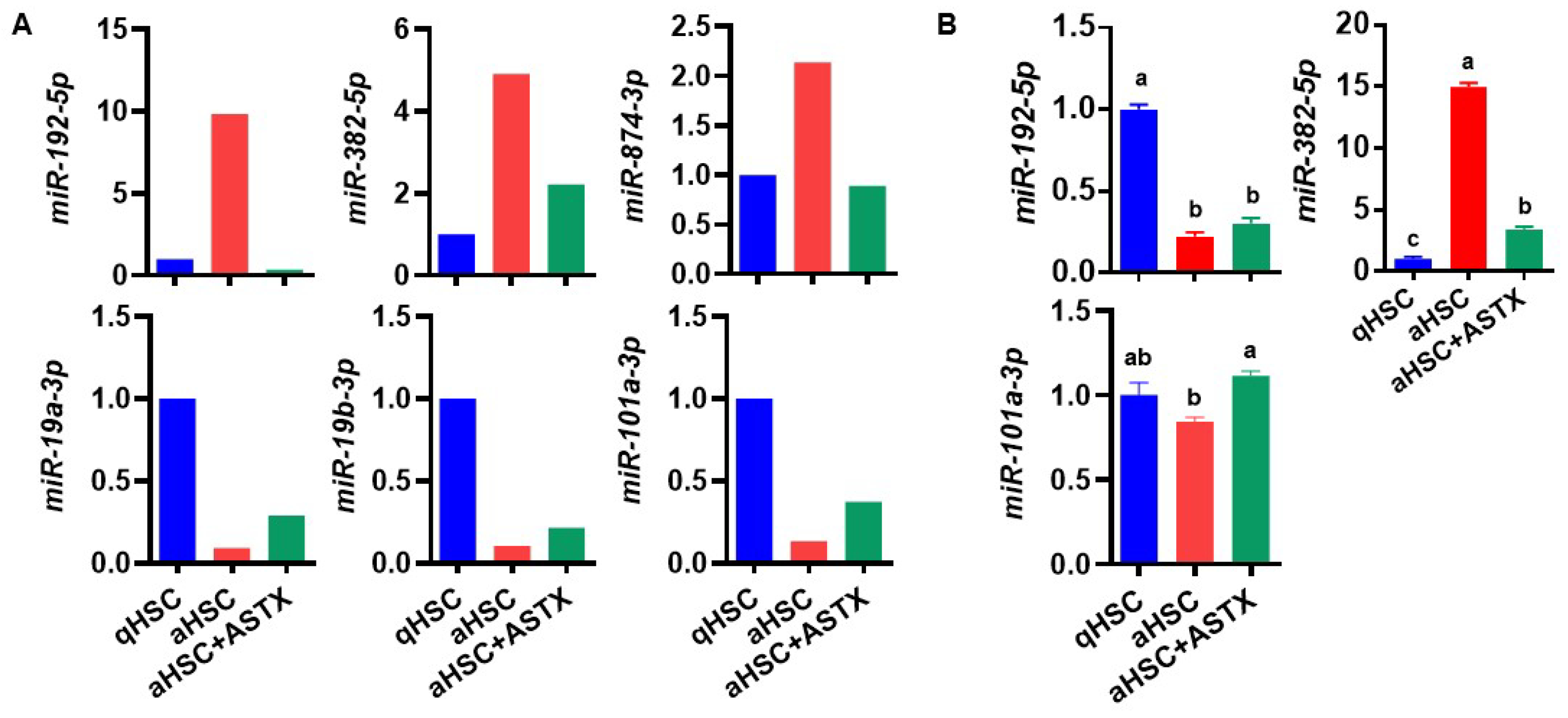
3.3. ASTX Attenuated the Changes in the Expression of miRNAs during HSC Activation
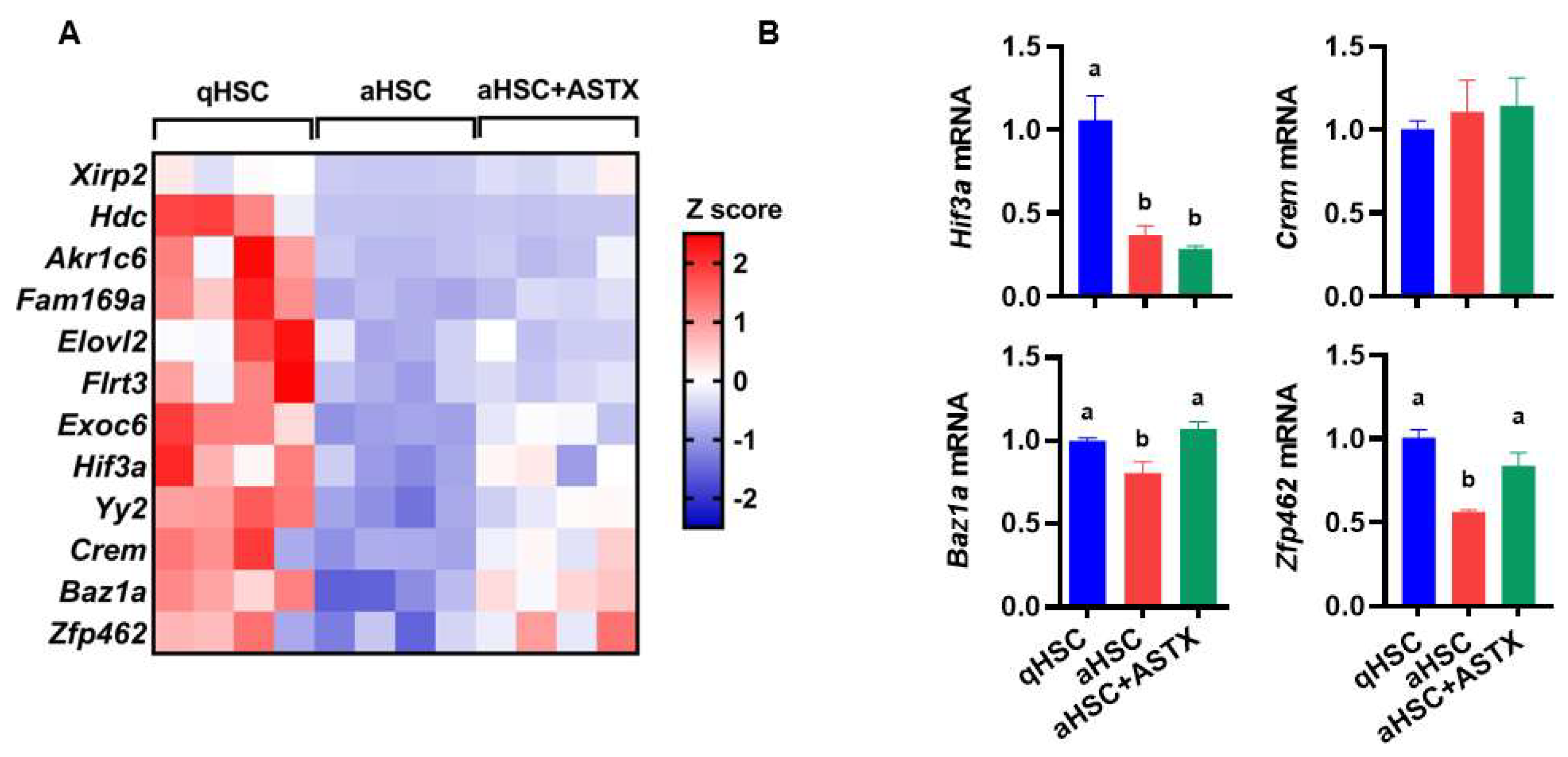
3.4. The Expression of Potential Target Genes of miR-382-5p Showed Drastic Differences between qHSC and aHSC, Which Were Attenuated by ASTX
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Schuppan, D.; Ruehl, M.; Somasundaram, R.; Hahn, E.G. Matrix as a modulator of hepatic fibrogenesis. In Seminars in Liver Disease; Thieme Medical Publishers, Inc.: New York, NY, USA, 2001; pp. 351–372. [Google Scholar]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Hepatic stellate cells: Protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar] [CrossRef] [PubMed]
- Wake, K. “Sternzellen” in the liver: Perisinusoidal cells with special reference to storage of vitamin A. Am. J. Anat. 1971, 132, 429–462. [Google Scholar] [CrossRef] [PubMed]
- Puche, J.E.; Saiman, Y.; Friedman, S.L. Hepatic stellate cells and liver fibrosis. Compr. Physiol. 2013, 3, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 631. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Isakova, A.; Fehlmann, T.; Keller, A.; Quake, S.R. A mouse tissue atlas of small noncoding RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 25634–25645. [Google Scholar] [CrossRef]
- Yekta, S.; Shih, I.-h.; Bartel, D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004, 304, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.H. Extracellular/circulating microRNAs: Release mechanisms, functions and challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Li, Y.; Kowdley, K.V. MicroRNAs in common human diseases. Genom. Proteom. Bioinform. 2012, 10, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Li, B.; Xin, X.; Xu, M.; Ji, G.; Yu, H. MicroRNA-34a Promotes Hepatic Stellate Cell Activation via Targeting ACSL1. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2015, 21, 3008–3015. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, C.; Xu, Z.; Xia, P.; Dong, P.; Chen, B.; Yu, F. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol. Cell. Biochem. 2015, 398, 1–9. [Google Scholar] [CrossRef]
- Wei, J.; Feng, L.; Li, Z.; Xu, G.; Fan, X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed. Pharmacother. = Biomed. Pharmacother. 2013, 67, 387–392. [Google Scholar] [CrossRef]
- Sun, X.; He, Y.; Ma, T.T.; Huang, C.; Zhang, L.; Li, J. Participation of miR-200a in TGF-beta1-mediated hepatic stellate cell activation. Mol. Cell. Biochem. 2014, 388, 11–23. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Huang, C.; Sun, X.; Long, X.R.; Lv, X.W.; Li, J. MicroRNA-146a modulates TGF-beta1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell. Signal. 2012, 24, 1923–1930. [Google Scholar] [CrossRef]
- Sekiya, Y.; Ogawa, T.; Yoshizato, K.; Ikeda, K.; Kawada, N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem. Biophys. Res. Commun. 2011, 412, 74–79. [Google Scholar] [CrossRef]
- Wang, J.; Chu, E.S.; Chen, H.Y.; Man, K.; Go, M.Y.; Huang, X.R.; Lan, H.Y.; Sung, J.J.; Yu, J. microRNA-29b prevents liver fibrosis by attenuating hepatic stellate cell activation and inducing apoptosis through targeting PI3K/AKT pathway. Oncotarget 2015, 6, 7325–7338. [Google Scholar] [CrossRef]
- Higuera-Ciapara, I.; Felix-Valenzuela, L.; Goycoolea, F. Astaxanthin: A review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 2006, 46, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kim, B.; Park, Y.-K.; Koo, S.I.; Lee, J.-Y. Astaxanthin prevents TGFβ1-induced pro-fibrogenic gene expression by inhibiting Smad3 activation in hepatic stellate cells. Biochim. Et Biophys. Acta 2015, 1850, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bae, M.; Park, Y.-K.; Lee, Y.; Pham, T.X.; Rudraiah, S.; Manautou, J.; Koo, S.I.; Lee, J.-Y. Histone deacetylase 9 plays a role in the antifibrogenic effect of astaxanthin in hepatic stellate cells. J. Nutr. Biochem. 2017, 40, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bae, M.; Kim, B.; Park, Y.-K.; Koo, S.I.; Lee, J.-Y. Astaxanthin prevents and reverses the activation of mouse primary hepatic stellate cells. J. Nutr. Biochem. 2016, 29, 21–26. [Google Scholar] [CrossRef]
- Shen, M.; Chen, K.; Lu, J.; Cheng, P.; Xu, L.; Dai, W.; Wang, F.; He, L.; Zhang, Y.; Chengfen, W.; et al. Protective effect of astaxanthin on liver fibrosis through modulation of TGF-beta1 expression and autophagy. Mediat. Inflamm. 2014, 2014, 954502. [Google Scholar] [CrossRef]
- Kim, B.; Farruggia, C.; Ku, C.S.; Pham, T.X.; Yang, Y.; Bae, M.; Wegner, C.J.; Farrell, N.J.; Harness, E.; Park, Y.K.; et al. Astaxanthin inhibits inflammation and fibrosis in the liver and adipose tissue of mouse models of diet-induced obesity and nonalcoholic steatohepatitis. J. Nutr. Biochem. 2016, 43, 27–35. [Google Scholar] [CrossRef]
- Bae, M.; Lee, Y.; Park, Y.-K.; Shin, D.-G.; Joshi, P.; Hong, S.-H.; Alder, N.; Koo, S.I.; Lee, J.-Y. Astaxanthin attenuates the increase in mitochondrial respiration during the activation of hepatic stellate cells. J. Nutr. Biochem. 2019, 71, 82–89. [Google Scholar] [CrossRef]
- Bae, M.; Lee, Y.; Pham, T.X.; Hu, S.; Park, Y.-K.; Lee, J.-Y. Astaxanthin inhibits the reduction of glycolysis during the activation of hepatic stellate cells. Life Sci. 2020, 256, 117926. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Park, Y.-K.; Rasmussen, H.E.; Ehlers, S.J.; Blobaum, K.R.; Lu, F.; Schlegal, V.L.; Carr, T.P.; Lee, J.-Y. Repression of proinflammatory gene expression by lipid extract of Nostoc commune var sphaeroides Kützing, a blue-green alga, via inhibition of nuclear factor-κB in RAW 264.7 macrophages. Nutr. Res. 2008, 28, 83–91. [Google Scholar] [CrossRef]
- Rasmussen, H.E.; Blobaum, K.R.; Park, Y.-K.; Ehlers, S.J.; Lu, F.; Lee, J.-Y. Lipid extract of Nostoc commune var. sphaeroides Kützing, a blue-green alga, inhibits the activation of sterol regulatory element binding proteins in HepG2 cells. J. Nutr. 2008, 138, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, A.J.; Fang, Y.; Liu, Y.; Tian, Z.; Mladinov, D.; Matus, I.R.; Ding, X.; Greene, A.S.; Liang, M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: A novel role of miR-382. Nucleic Acids Res. 2010, 38, 8338–8347. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L.; Roll, F.J.; Boyles, J.; Bissell, D.M. Hepatic lipocytes: The principal collagen-producing cells of normal rat liver. Proc. Natl. Acad. Sci. USA 1985, 82, 8681–8685. [Google Scholar] [CrossRef] [PubMed]
- Racki, L.R.; Yang, J.G.; Naber, N.; Partensky, P.D.; Acevedo, A.; Purcell, T.J.; Cooke, R.; Cheng, Y.; Narlikar, G.J. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature 2009, 462, 1016–1021. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, D.; Yao, J.; Zhou, B.; Shen, T.; Qi, Y.; Ni, T.; Wei, G. Chromatin remodeling factor BAZ1A regulates cellular senescence in both cancer and normal cells. Life Sci. 2019, 229, 225–232. [Google Scholar] [CrossRef]
- Nakao, A.; Imamura, T.; Souchelnytskyi, S.; Kawabata, M.; Ishisaki, A.; Oeda, E.; Tamaki, K.; Hanai, J.i.; Heldin, C.H.; Miyazono, K. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997, 16, 5353–5362. [Google Scholar] [CrossRef] [PubMed]
- Weiss, K.; Wigby, K.; Fannemel, M.; Henderson, L.B.; Beck, N.; Ghali, N.; Study, D.; Anderlid, B.-M.; Lundin, J.; Hamosh, A. Haploinsufficiency of ZNF462 is associated with craniofacial anomalies, corpus callosum dysgenesis, ptosis, and developmental delay. Eur. J. Hum. Genet. 2017, 25, 946–951. [Google Scholar] [CrossRef]
- Laurent, A.; Massé, J.; Omilli, F.; Deschamps, S.; Richard-Parpaillon, L.; Chartrain, I.; Pellerin, I. ZFPIP/Zfp462 is maternally required for proper early Xenopus laevis development. Dev. Biol. 2009, 327, 169–176. [Google Scholar] [CrossRef]
- Massé, J.; Piquet-Pellorce, C.; Viet, J.; Guerrier, D.; Pellerin, I.; Deschamps, S. ZFPIP/Zfp462 is involved in P19 cell pluripotency and in their neuronal fate. Exp. Cell Res. 2011, 317, 1922–1934. [Google Scholar] [CrossRef]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Gan, J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE 2014, 9, e105192. [Google Scholar] [CrossRef]
- Pirola, C.J.; Fernandez Gianotti, T.; Castano, G.O.; Mallardi, P.; San Martino, J.; Mora Gonzalez Lopez Ledesma, M.; Flichman, D.; Mirshahi, F.; Sanyal, A.J.; Sookoian, S. Circulating microRNA signature in non-alcoholic fatty liver disease: From serum non-coding RNAs to liver histology and disease pathogenesis. Gut 2015, 64, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Saha, B.; Kodys, K.; Catalano, D.; Satishchandran, A.; Szabo, G. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J. Transl. Med. 2015, 13, 261. [Google Scholar] [CrossRef] [PubMed]
- Starkey Lewis, P.J.; Dear, J.; Platt, V.; Simpson, K.J.; Craig, D.G.; Antoine, D.J.; French, N.S.; Dhaun, N.; Webb, D.J.; Costello, E.M.; et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011, 54, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, C.H.; Lee, S.-W. Exosomal transmission of microRNA from HCV replicating cells stimulates transdifferentiation in hepatic stellate cells. Mol. Ther. -Nucleic Acids 2019, 14, 483–497. [Google Scholar] [CrossRef]
- Chung, A.C.; Huang, X.R.; Meng, X.; Lan, H.Y. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J. Am. Soc. Nephrol. 2010, 21, 1317–1325. [Google Scholar] [CrossRef]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef]
- Coll, M.; El Taghdouini, A.; Perea, L.; Mannaerts, I.; Vila-Casadesus, M.; Blaya, D.; Rodrigo-Torres, D.; Affo, S.; Morales-Ibanez, O.; Graupera, I.; et al. Integrative miRNA and Gene Expression Profiling Analysis of Human Quiescent Hepatic Stellate Cells. Sci. Rep. 2015, 5, 11549. [Google Scholar] [CrossRef]
- Guo, C.J.; Pan, Q.; Cheng, T.; Jiang, B.; Chen, G.Y.; Li, D.G. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009, 276, 5163–5176. [Google Scholar] [CrossRef]
- Leong, K.W.; Cheng, C.W.; Wong, C.M.; Ng, I.O.; Kwong, Y.L.; Tse, E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget 2017, 8, 11343–11355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Y.; Li, X.; Liang, X.; Wang, L.; Song, J.; Zhang, X.; Zhang, C.; Niu, J.; Zhang, P.; et al. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis. 2018, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Guan, L.Y.; Ye, Y.S.; Liu, H.Y.; Li, R. MiR-874 inhibits metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting SOX12. Am. J. Cancer Res. 2017, 7, 1310–1321. [Google Scholar] [PubMed]
- Tu, X.; Zhang, H.; Zhang, J.; Zhao, S.; Zheng, X.; Zhang, Z.; Zhu, J.; Chen, J.; Dong, L.; Zang, Y.; et al. MicroRNA-101 suppresses liver fibrosis by targeting the TGFbeta signalling pathway. J. Pathol. 2014, 234, 46–59. [Google Scholar] [CrossRef]
- Lakner, A.M.; Steuerwald, N.M.; Walling, T.L.; Ghosh, S.; Li, T.; McKillop, I.H.; Russo, M.W.; Bonkovsky, H.L.; Schrum, L.W. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology 2012, 56, 300–310. [Google Scholar] [CrossRef]
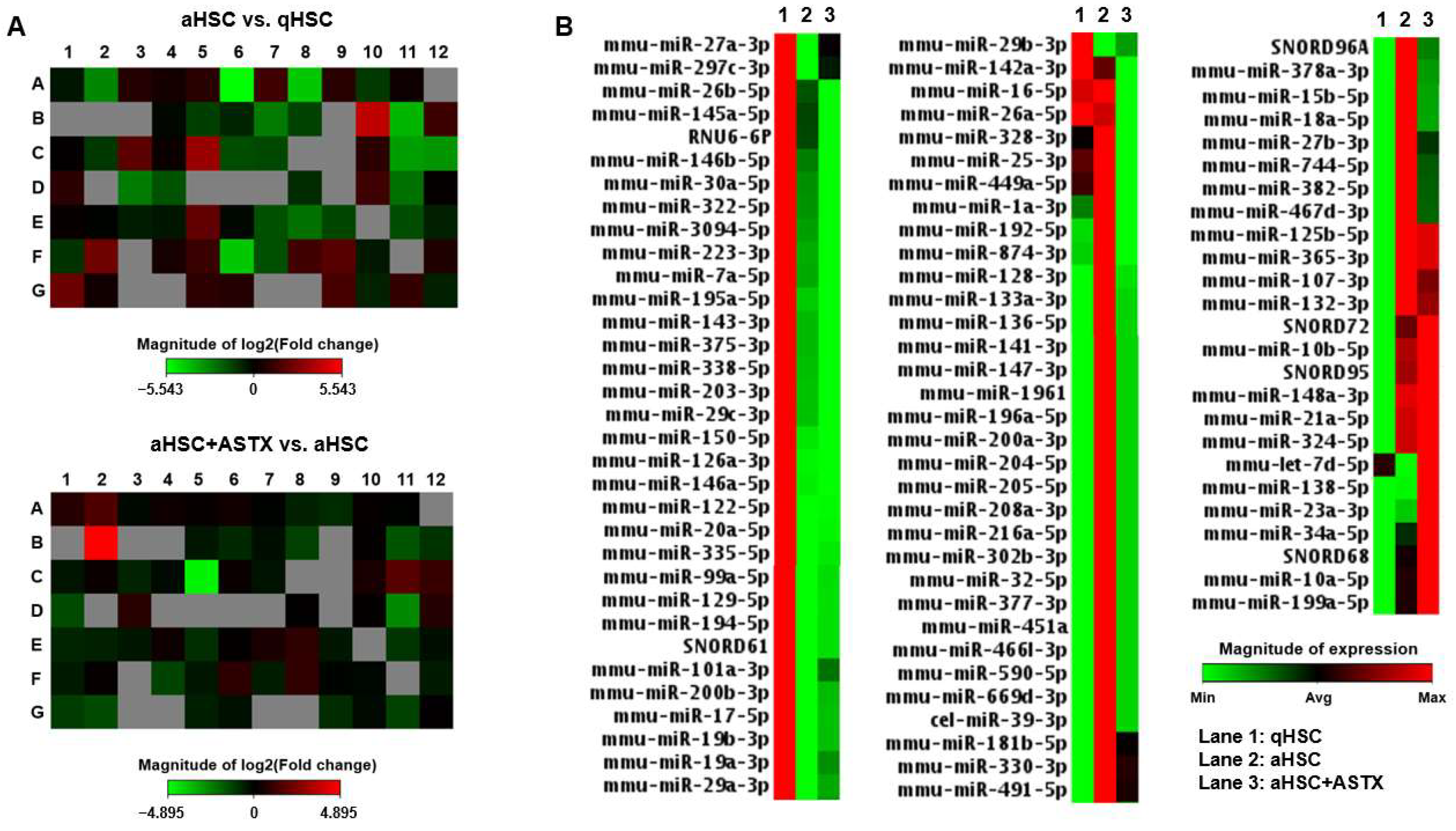
| Overexpressed | Fold Change | Underexpressed | Fold Change |
|---|---|---|---|
| miRNAs | (aHSC vs. qHSC) | miRNAs | (aHSC vs. qHSC) |
| mmu-miR-148a-3p | 17.35 | mmu-miR-122-5p | 0.02 |
| mmu-miR-192-5p | 9.83 | mmu-miR-126a-3p | 0.05 |
| mmu-miR-324-5p | 5.23 | mmu-miR-335-5p | 0.05 |
| mmu-miR-382-5p | 4.91 | mmu-miR-150-5p | 0.07 |
| mmu-miR-27b-3p | 4.31 | mmu-miR-19a-3p | 0.09 |
| mmu-miR-181b-5p | 3.91 | mmu-miR-19b-3p | 0.11 |
| mmu-miR-365-3p | 3.7 | mmu-miR-101a-3p | 0.13 |
| mmu-miR-744-5p | 2.9 | mmu-miR-200b-3p | 0.16 |
| mmu-miR-34a-5p | 2.63 | mmu-miR-146a-5p | 0.16 |
| mmu-miR-21a-5p | 2.58 | mmu-miR-223-3p | 0.18 |
| mmu-miR-125b-5p | 2.53 | mmu-miR-29b-3p | 0.19 |
| mmu-miR-15b-5p | 2.44 | mmu-miR-203-3p | 0.29 |
| mmu-miR-330-3p | 2.32 | mmu-miR-29a-3p | 0.3 |
| mmu-miR-874-3p | 2.14 | mmu-miR-338-5p | 0.31 |
| mmu-miR-3094-5p | 0.31 | ||
| mmu-miR-194-5p | 0.32 | ||
| mmu-miR-195a-5p | 0.34 | ||
| mmu-miR-29c-3p | 0.35 | ||
| mmu-miR-146b-5p | 0.37 | ||
| mmu-miR-143-3p | 0.39 | ||
| mmu-miR-129-5p | 0.43 | ||
| mmu-miR-17-5p | 0.45 | ||
| mmu-miR-322-5p | 0.47 |
| miRNAs | Fold Change (aHSC + ASTX vs. aHSC) | |
|---|---|---|
| Overexpressed | mmu-miR-138-5p | 29.75 |
| mmu-miR-19a-3p | 3.17 | |
| mmu-miR-101a-3p | 2.80 | |
| mmu-miR-19b-3p | 2.05 | |
| Underexpressed | mmu-miR-192-5p | 0.04 |
| mmu-miR-223-3p | 0.16 | |
| mmu-miR-150-5p | 0.33 | |
| mmu-miR-449a-5p | 0.37 | |
| mmu-miR-1a-3p | 0.37 | |
| mmu-miR-328-3p | 0.41 | |
| mmu-miR-874-3p | 0.42 | |
| mmu-miR-146b-5p | 0.43 | |
| mmu-miR-382-5p | 0.45 | |
| mmu-miR-3094-5p | 0.50 |
| Gene | Full Name | Function |
|---|---|---|
| Xirp2 | Xin actin-binding repeat containing 2 | Xirp2 belongs to muscle-specific, actin-binding Xin gene family. It is expressed in cardiac and skeletal muscle interacting with filamentous actin and α-actinin via the actin-binding motif, the Xin repeat. |
| Hdc | Histidine decarboxylase | HDC catalyzes the decarboxylation of histidine to form histamine. |
| Akr1c6 | Aldo-keto reductase family 1, member C6 | Akr1c6 encodes estradiol 17 β-dehydrogenase 5, which catalyzes the reduction of 4-androstenedione, 5-α-androstane-3,17-dione, androsterone and dehydroepiandrosterone to testosterone, dihydrotestosterone, 5-α-androstane-3-α,17-β-diol, and 5-androstene-3-β,17-β-diol, respectively. |
| Fam169a | Family with sequence similarity 169, member A | Soluble lamina-associated protein of 75 kD. |
| Elovl2 | Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 | ELOVL2 is a condensing enzyme catalyzing the elongation of long-chain polyunsaturated fatty acids. |
| Flrt3 | Fibronectin leucine rich transmembrane protein 3 | FLRT3 is involved in cell–cell adhesion, cell migration, and axon guidance. |
| Exoc6 | Exocyst complex component 6 | EXOC6 is a component of the exocyst complex involved in vesicle trafficking, specifically the tethering of secretory vesicles to the plasma membrane during exocytosis. |
| Hif3a | Hypoxia inducible factor 3, alpha subunit | HIF3A belongs to the transcription factor family of hypoxia-inducible factors, which regulate the cellular response to hypoxia. |
| Yy2 | Yy2 transcription factor | Yy2 acts as a multifunctional transcription factor regulating a large number of genes positively and negatively. It is involved in development and differentiation. |
| Crem | cAMP responsive element modulator | CREM is a component of cAMP-mediated signal transduction during various physiological processes, including spermatogenesis, cardiac function, and circadian rhythm. |
| Baz1a | Bromodomain adjacent to zinc finger domain 1A | BAZ1A is the accessory, noncatalytic subunit of the ATP-dependent chromatin assembly factor, which regulates spacing of nucleosomes using ATP to form evenly spaced nucleosomes along the chromatin. |
| Zfp462 | Zinc finger protein 462 | ZFP462 or ZNF462 belongs to C2H2-type zinc finger family of proteins. It is involved in transcription by regulating chromatin structure. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, M.; Kim, M.-B.; Lee, J.-Y. Astaxanthin Attenuates the Changes in the Expression of MicroRNAs Involved in the Activation of Hepatic Stellate Cells. Nutrients 2022, 14, 962. https://doi.org/10.3390/nu14050962
Bae M, Kim M-B, Lee J-Y. Astaxanthin Attenuates the Changes in the Expression of MicroRNAs Involved in the Activation of Hepatic Stellate Cells. Nutrients. 2022; 14(5):962. https://doi.org/10.3390/nu14050962
Chicago/Turabian StyleBae, Minkyung, Mi-Bo Kim, and Ji-Young Lee. 2022. "Astaxanthin Attenuates the Changes in the Expression of MicroRNAs Involved in the Activation of Hepatic Stellate Cells" Nutrients 14, no. 5: 962. https://doi.org/10.3390/nu14050962
APA StyleBae, M., Kim, M.-B., & Lee, J.-Y. (2022). Astaxanthin Attenuates the Changes in the Expression of MicroRNAs Involved in the Activation of Hepatic Stellate Cells. Nutrients, 14(5), 962. https://doi.org/10.3390/nu14050962






