Mechanism of Astragalus membranaceus Alleviating Acquired Hyperlipidemia Induced by High-Fat Diet through Regulating Lipid Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Acquisition of Active Components and Targets of AM
2.2. Microarray Analysis (Acquisition of Acquired Hyperlipidemia/Obesity-Related Targets)
2.3. Acquisition of Common Drug and Disease Targets
2.4. Enrichment Analysis Using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
2.5. Visualization of Network Pharmacology and Construction of a Protein-Protein Interaction (PPI) Network
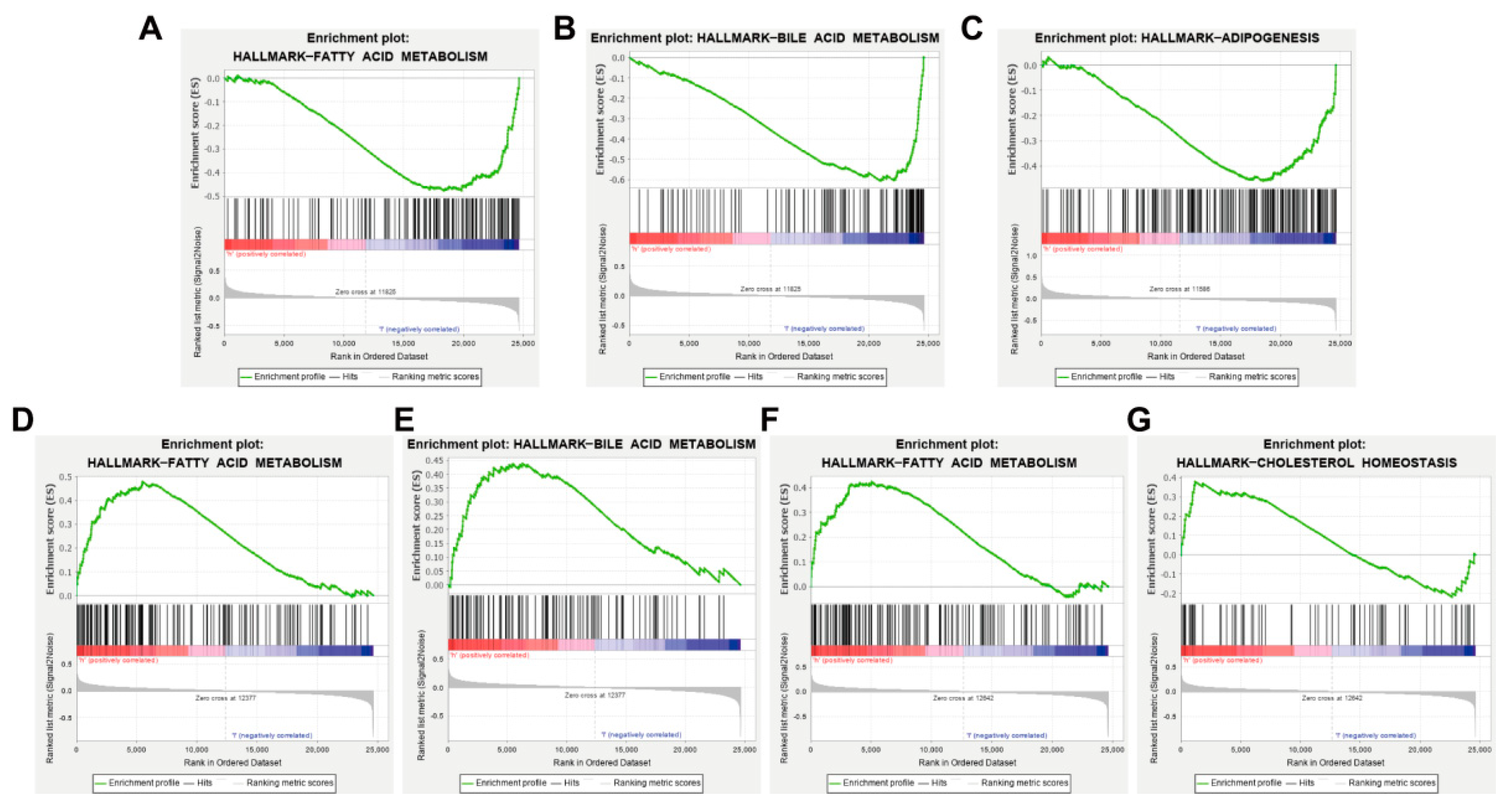
2.6. Gene Set Enrichment Analysis (GSEA) of Key Targets
2.7. Chemicals and Reagents
2.8. Animals and Diet
2.9. Collection of Serum and Tissue
2.10. Serum Biochemical Analyses
2.11. Liver Tissue and WAT Biochemical Analyses
2.12. Western Blot Analysis
2.13. Statistical Analysis
3. Results
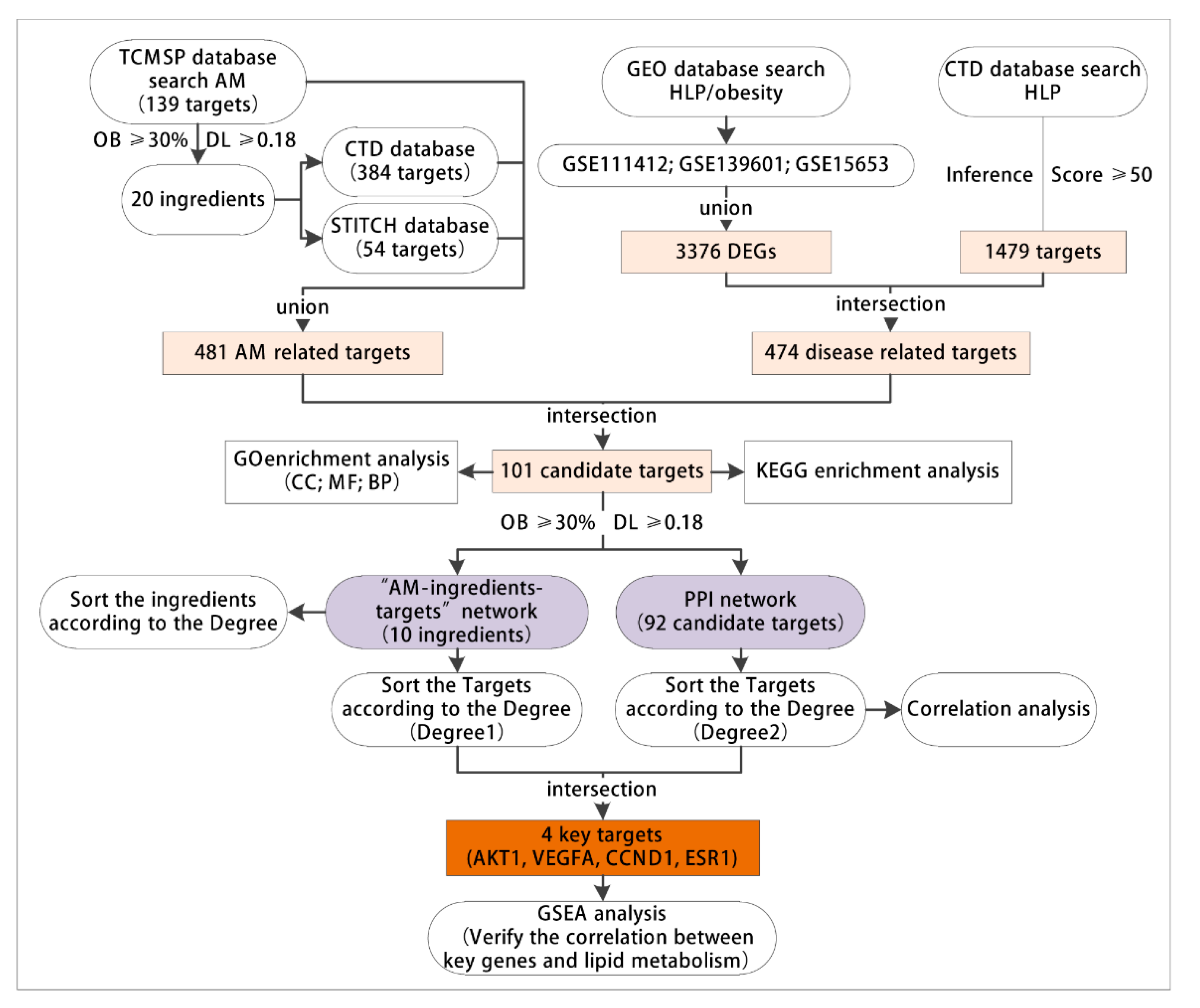
3.1. Network Pharmacology and Bioinformatics Analysis Process
3.2. AM Exerted Pharmaceutical Effects by Regulating 481 Related Targets through 20 Active Components
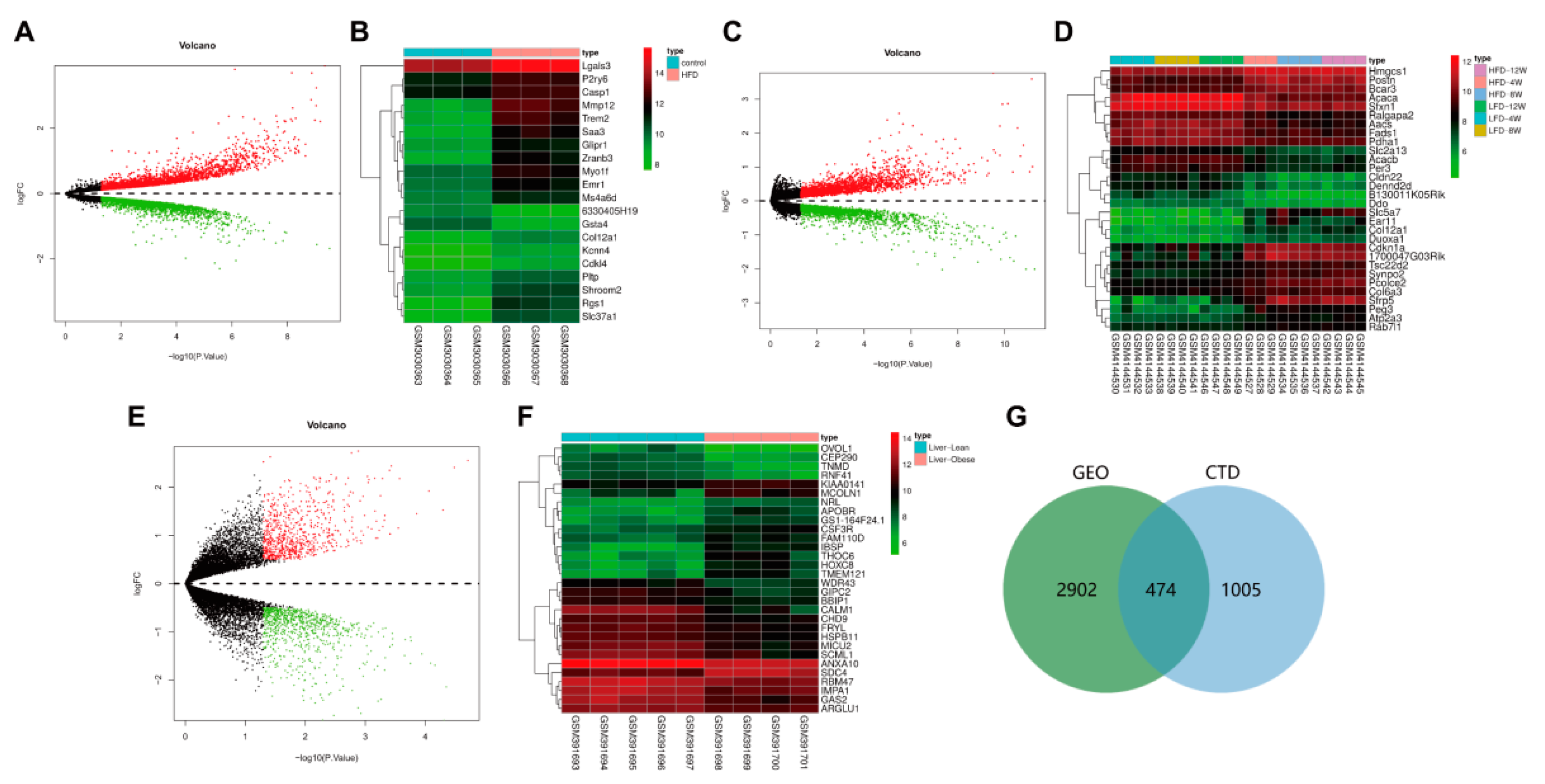
3.3. 474 Targets Were Closely Related to the Occurrence and Development of Hyperlipidemia Induced by HFD
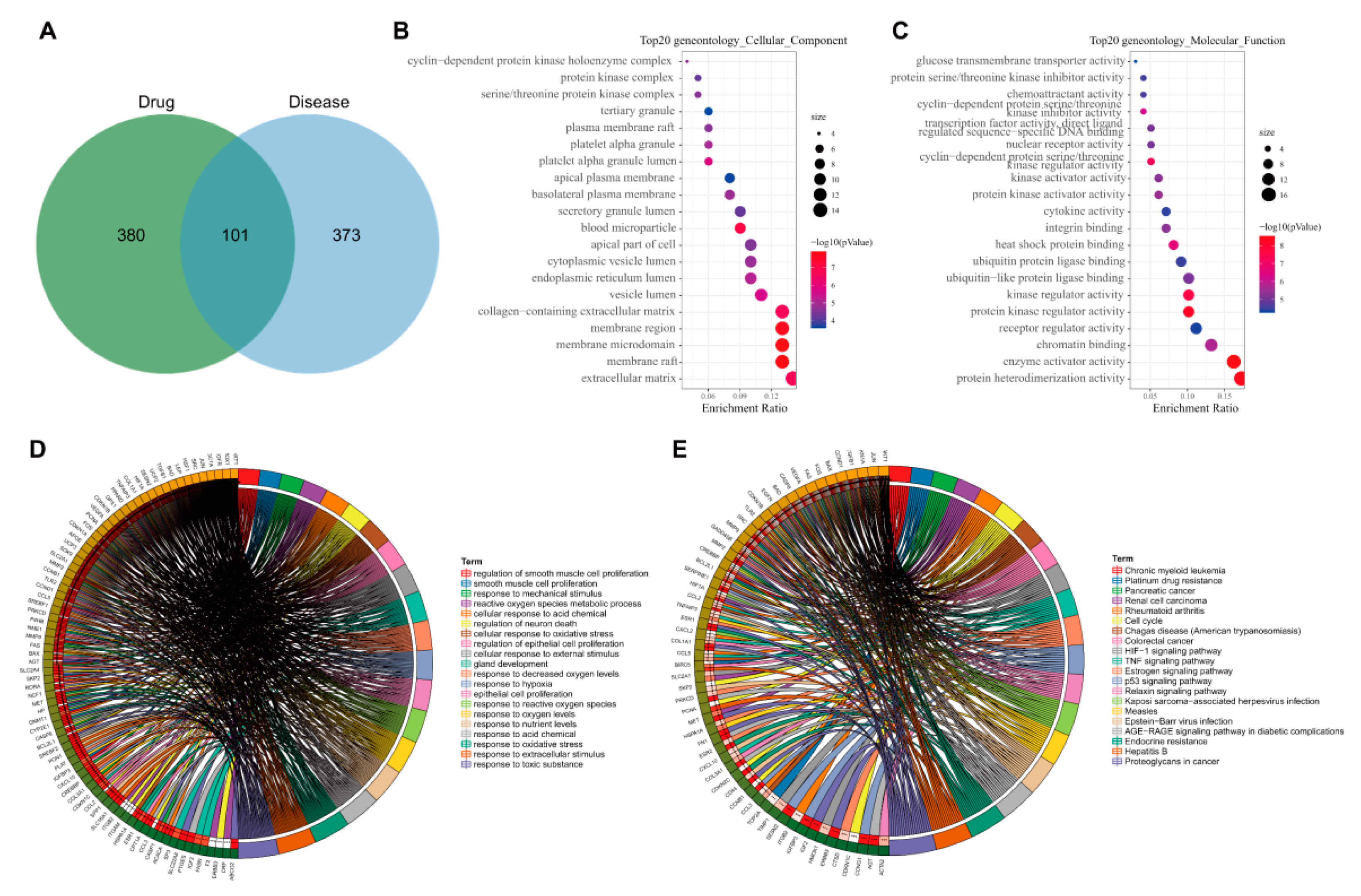
3.4. GO and KEGG Enrichment Analyses of 101 Common Targets of AM and Acquired Hyperlipidemia
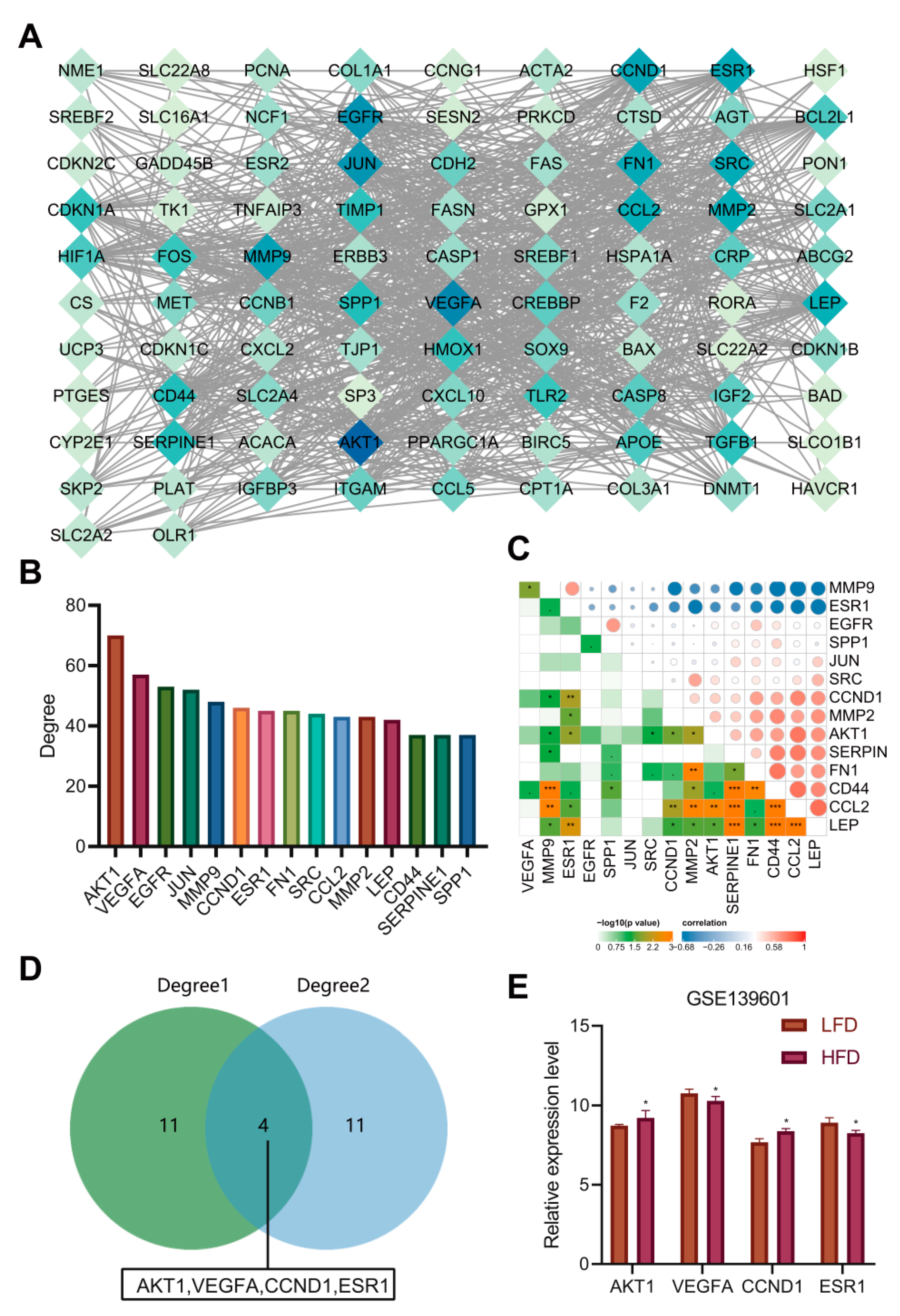
3.5. Construction of Network Relationship of “AM-Component-Target”
3.6. AKT1, VEGFA, CCND1 and ESR1 Were the Key Targets of AM in the Treatment of Acquired Hyperlipidemia
3.7. AKT1, VEGFA, CCND1 and ESR1 Are Closely Related to Abnormal Lipid Metabolism
3.8. AM Reduced the Weight Gain Caused by HFD and Alleviated Dyslipidemia
3.9. AM Inhibited Lipogenesis, Promoted Lipolysis and Lipid β-Oxidation in Liver Tissue and WAT of Mice Fed with HFD
3.10. AM Regulated the Expression of AKT1, VEGFA, CCND1 and ESR1 in the Liver and WAT of Mice Fed with HFD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- El-Tantawy, W.H.; Temraz, A. Natural products for controlling hyperlipidemia: Review. Arch. Physiol. Biochem. 2019, 125, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Schatz, U.; Julius, U. Current standards in diagnosis and therapy of hyperlipoproteinemia. Atheroscler. Suppl. 2013, 14, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.; Ma, J.; Su, X.; Zhong, Y. Emerging insights into the relationship between hyperlipidemia and the risk of diabetic retinopathy. Lipids Heal. Dis. 2020, 19, 241. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Xu, J.; Lian, F.; Yu, X.; Zhao, Y.; Xu, L.; Zhang, M.; Zhao, X.; Shen, J.; Wu, S.; et al. Structural Alteration of Gut Microbiota during the Amelioration of Human Type 2 Diabetes with Hyperlipidemia by Metformin and a Traditional Chinese Herbal Formula: A Multicenter, Randomized, Open Label Clinical Trial. mBio 2018, 9, e02392-17. [Google Scholar] [CrossRef] [Green Version]
- Guo, J. Research progress on prevention and treatment of glucolipid metabolic disease with integrated traditional Chinese and Western medicine. Chin. J. Integr. Med. 2017, 23, 403–409. [Google Scholar] [CrossRef]
- Wang, B.; Lin, L.; Ni, Q.; Su, C.-L. Chinese Medicine for Treating Diabetic Nephropathy. Chin. J. Integr. Med. 2011, 17, 794–800. [Google Scholar] [CrossRef]
- Zhou, P.; Xie, W.; He, S.; Sun, Y.; Meng, X.; Sun, G.; Sun, X. Ginsenoside Rb1 as an Anti-Diabetic Agent and Its Underlying Mechanism Analysis. Cells 2019, 8, 204. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Zhao, Y.; Du, L. Emerging approaches of traditional Chinese medicine formulas for the treatment of hyperlipidemia. J. Ethnopharmacol. 2012, 140, 345–367. [Google Scholar] [CrossRef]
- Tao, W.; Xu, X.; Wang, X.; Li, B.; Wang, Y.; Li, Y.; Yang, L. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J. Ethnopharmacol. 2013, 145, 1–10. [Google Scholar] [CrossRef]
- Zhang, H.W.; Lin, Z.X.; Xu, C.; Leung, C.; Chan, L.S. Astragalus (a traditional Chinese medicine) for treating chronic kidney disease. Cochrane Database Syst. Rev. 2014, 10, CD008369. [Google Scholar] [CrossRef]
- Auyeung, K.K.; Han, Q.; Ko, J. Astragalus membranaceus: A Review of its Protection Against Inflammation and Gastrointestinal Cancers. Am. J. Chin. Med. 2016, 44, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.-Q.; Yu, F.; Wang, N.; Wu, Y.; Zou, F.; Wu, K.; Liu, M.; Ouyang, J.-P. Hypoglycemic effect of polysaccharide enriched extract of Astragalus membranaceus in diet induced insulin resistant C57BL/6J mice and its potential mechanism. Phytomedicine 2009, 16, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Su, Y.; Zhang, J.; Zhang, Y.; Li, Y.; Han, Y.; Dong, X.; Li, W.; Li, W. Astragaloside IV alleviates liver injury in type 2 diabetes due to promotion of AMPK/mTOR-mediated autophagy. Mol. Med. Rep. 2021, 23, 437. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Xia, R.; Yang, S.; Liu, L.; Zhang, J.; Feng, K.; Shang, Y.; Qu, J.; Li, L.; Chen, N.; et al. Formononetin attenuates atherosclerosis via regulating interaction between KLF4 and SRA in apoE(-/-) mice. Theranostics 2020, 10, 1090–1106. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Tang, K.; Wu, S.; Liu, L.-J.; Qiang, C.; Lin, X.; Liu, B. Astragalus Polysaccharides Lowers Plasma Cholesterol through Mechanisms Distinct from Statins. PLoS ONE 2011, 6, e27437. [Google Scholar] [CrossRef] [Green Version]
- Syed, A.A.; Reza, M.I.; Shafiq, M.; Kumariya, S.; Singh, P.; Husain, A.; Hanif, K.; Gayen, J.R. Naringin ameliorates type 2 diabetes mellitus-induced steatohepatitis by inhibiting RAGE/NF-kappaB mediated mitochondrial apoptosis. Life Sci. 2020, 257, 118118. [Google Scholar] [CrossRef] [PubMed]
- Barile, G.R.; Pachydaki, S.I.; Tari, S.R.; Lee, S.E.; Donmoyer, C.M.; Ma, W.; Rong, L.L.; Buciarelli, L.G.; Wendt, T.; Horig, H.; et al. The RAGE axis in early diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2916–2924. [Google Scholar] [CrossRef] [Green Version]
- Al-Ani, B.; AlZamil, N.M.; Hewett, P.W.; Al-Hashem, F.; Bin-Jaliah, I.; Shatoor, A.S.; Kamar, S.S.; Latif, N.S.A.; Haidara, M.A.; Dawood, A.F. Metformin ameliorates ROS-p53-collagen axis of fibrosis and dyslipidemia in type 2 diabetes mellitus-induced left ventricular injury. Arch. Physiol. Biochem. 2021, 2021, 1–7. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Xue, B.-H.; Wang, X.-D.; Yu, W.-L. Negative Regulation of SIRT1 by IRF9 Involved in Hyperlipidemia Acute Pancreatitis Associated with Kidney Injury. Am. J. Dig. Dis. 2021, 66, 1063–1071. [Google Scholar] [CrossRef]
- Wan, M.; Easton, R.M.; Gleason, C.E.; Monks, B.R.; Ueki, K.; Kahn, C.R.; Birnbaum, M.J. Loss of Akt1 in Mice Increases Energy Expenditure and Protects against Diet-Induced Obesity. Mol. Cell. Biol. 2012, 32, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Bujaldon, E.; Cornide-Petronio, M.E.; Gulfo, J.; Rotondo, F.; Avalos de Leon, C.; Negrete-Sanchez, E.; Gracia-Sancho, J.; Novials, A.; Jimenez-Castro, M.B.; Peralta Uroz, C. Relevance of VEGFA in rat livers subjected to partial hepatectomy under ischemia-reperfusion. J. Mol. Med. 2019, 97, 1299–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FatimaL, A.; Campello, R.S.; Santos, R.D.S.; Freitas, H.S.; Frank, A.P.; Machado, U.F.; Clegg, D.J. Estrogen receptor 1 (ESR1) regulates VEGFA in adipose tissue. Sci. Rep. 2017, 7, 16716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Moore, T.M.; Drew, B.G.; Ribas, V.; Wanagat, J.; Civelek, M.; Segawa, M.; Wolf, D.M.; Norheim, F.; Seldin, M.M.; et al. Estrogen receptor α controls metabolism in white and brown adipocytes by regulating Polg1 and mitochondrial remodeling. Sci. Transl. Med. 2020, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Liang, J.; Sharabi, K.; Hatting, M.; Perry, E.A.; Tavares, C.D.; Goyal, L.; Srivastava, A.; Bilodeau, M.; Zhu, A.X.; et al. Obesity/Type 2 Diabetes-Associated Liver Tumors Are Sensitive to Cyclin D1 Deficiency. Cancer Res. 2020, 80, 3215–3221. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, Z.; Huang, L.; Zheng, S.; Wang, D.; Chen, S.; Zhang, H.; Yang, S. Review of the Botanical Characteristics, Phytochemistry, and Pharmacology of Astragalus membranaceus (Huangqi). Phytother. Res. 2014, 28, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Bułdak, Ł.; Marek, B.; Kajdaniuk, D.; Urbanek, A.; Janyga, S.; Bołdys, A.; Basiak, M.; Maligłówka, M.; Okopień, B. Endocrine diseases as causes of secondary hyperlipidemia. Endokrynol. Polska 2019, 70, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, D.H.; Choi, S.; Lee, J.S.; Jang, D.S.; Kang, K.S. Identification and Isolation of Active Compounds from Astragalus membranaceus that Improve Insulin Secretion by Regulating Pancreatic beta-Cell Metabolism. Biomolecules 2019, 9, 618. [Google Scholar] [CrossRef] [Green Version]
- Jin, Q.; Hao, X.-F.; Xie, L.-K.; Xu, J.; Sun, M.; Yuan, H.; Wang, S.-H.; Wu, G.-P.; Miao, M.-L. A Network Pharmacology to Explore the Mechanism of Astragalus Membranaceus in the Treatment of Diabetic Retinopathy. Evid.-Based Complement. Altern. Med. 2020, 2020, 8878569. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, W.; Wei, Q.; Chen, P.; Li, Y. Cytoprotective effects of the medicinal herb Astragalus membranaceus on lipopolysaccharide-exposed cells. Mol. Med. Rep. 2018, 18, 4321–4327. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Yao, L.; Liu, Y.; Pan, Z.; Peng, S.; Wan, G.; Cheng, J.; Wang, J.; Cao, W. Astragaloside IV regulates NF-kappaB-mediated cellular senescence and apoptosis of hepatic stellate cells to suppress PDGF-BB-induced activation. Exp. Ther. Med. 2019, 18, 3741–3750. [Google Scholar]
- Jaishree, V.; Narsimha, S. Swertiamarin and quercetin combination ameliorates hyperglycemia, hyperlipidemia and oxidative stress in streptozotocin-induced type 2 diabetes mellitus in wistar rats. Biomed. Pharmacother. 2020, 130, 110561. [Google Scholar] [CrossRef] [PubMed]
- Eisvand, F.; Razavi, B.M.; Hosseinzadeh, H. The effects of Ginkgo biloba on metabolic syndrome: A review. Phytother. Res. 2020, 34, 1798–1811. [Google Scholar] [CrossRef] [PubMed]
- Hoo, R.L.; Wong, J.Y.; Qiao, C.; Xu, A.; Xu, H.; Lam, K.S. The effective fraction isolated from Radix Astragali alleviates glucose intolerance, insulin resistance and hypertriglyceridemia in db/db diabetic mice through its anti-inflammatory activity. Nutr. Metab. 2010, 7, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhao, H.; Li, X.; Wang, Q.; Yan, M.; Zhang, H.; Zhao, T.; Zhang, N.; Zhang, P.; Peng, L.; et al. Formononetin alleviates hepatic steatosis by facilitating TFEB-mediated lysosome biogenesis and lipophagy. J. Nutr. Biochem. 2019, 73, 108214. [Google Scholar] [CrossRef]
- Ressaissi, A.; Attia, N.; Fale, P.L.; Pacheco, R.; Victor, B.L.; Machuqueiro, M.; Serralheiro, M.L.M. Isorhamnetin derivatives and piscidic acid for hypercholesterolemia: Cholesterol permeability, HMG-CoA reductase inhibition, and docking studies. Arch. Pharm. Res. 2017, 40, 1278–1286. [Google Scholar] [CrossRef]
- Li, M.; Wang, W.; Xue, J.; Gu, Y.; Lin, S. Meta-analysis of the clinical value of Astragalus membranaceus in diabetic nephropathy. J. Ethnopharmacol. 2011, 133, 412–419. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, J.; Yang, S.; Hua, Y.; Su, J.; Shang, Y.; Wang, Z.; Feng, K.; Zhang, J.; Yang, X.; et al. Astragalus Flavone Ameliorates Atherosclerosis and Hepatic Steatosis Via Inhibiting Lipid-Disorder and Inflammation in apoE(-/-) Mice. Front. Pharmacol. 2020, 11, 610550. [Google Scholar] [CrossRef]
- Nie, T.; Zhao, S.; Mao, L.; Yang, Y.; Sun, W.; Lin, X.; Liu, S.; Li, K.; Sun, Y.; Li, P.; et al. The natural compound, formononetin, extracted from Astragalus membranaceus increases adipocyte thermogenesis by modulating PPARgamma activity. Br. J. Pharmacol. 2018, 175, 1439–1450. [Google Scholar] [CrossRef] [Green Version]
- Araki, K.; Miyoshi, Y. Mechanism of resistance to endocrine therapy in breast cancer: The important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer 2017, 25, 392–401. [Google Scholar] [CrossRef]
- Peng, J.; Li, Q.; Li, K.; Zhu, L.; Lin, X.; Lin, X.; Shen, Q.; Li, G.; Xie, X. Quercetin Improves Glucose and Lipid Metabolism of Diabetic Rats: Involvement of Akt Signaling and SIRT1. J. Diabetes Res. 2017, 2017, 3417306. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-Y.; Han, H.-F.; He, Z.-Y.; Xu, X.-G. Immune Mechanism, Gene Module, and Molecular Subtype Identification of Astragalus Membranaceus in the Treatment of Dilated Cardiomyopathy: An Integrated Bioinformatics Study. Evid.-Based Complement. Altern. Med. 2021, 2021, 2252832. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Fang, X.; Tajima, A.; Geng, X.; Ranganathan, S.; Dong, H.H.; Trucco, M.; Sperling, M.A. Evolution of Hepatic Steatosis to Fibrosis and Adenoma Formation in Liver-Specific Growth Hormone Receptor Knockout Mice. Front. Endocrinol. 2014, 5, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, P.K.-K.; Chan, J.Y.-W.; Kwok, H.-F.; Cheng, L.; Yu, H.; Lau, C.-P.; Leung, P.-C.; Fung, K.-P.; Lau, C.B.-S. Induction of Angiogenesis in Zebrafish Embryos and Proliferation of Endothelial Cells by an Active Fraction Isolated from the Root of Astragalus membranaceus using Bioassay-guided Fractionation. J. Tradit. Complement. Med. 2014, 4, 239–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, N.; Paterson, A.; Roshandel, D.; Raza, A.; Ajmal, M.; Waheed, N.K.; Azam, M.; Qamar, R. Association of IGF1 and VEGFA polymorphisms with diabetic retinopathy in Pakistani population. Geol. Rundsch. 2019, 57, 237–245. [Google Scholar] [CrossRef]
- Yao, J.; Qin, Z. Counteract of bone marrow of blotchy mice against the increases of plasma copper levels induced by high-fat diets in LDLR-/- mice. J. Trace Elem. Med. Biol. 2015, 31, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Swetzig, W.M.; Wang, J.; Das, G.M. Estrogen receptor alpha (ERalpha/ESR1) mediates the p53-independent overexpression of MDM4/MDMX and MDM2 in human breast cancer. Oncotarget 2016, 7, 16049–16069. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Huang, Y.; Zhao, S.; Guo, Q.; Zhou, J.; Han, W.; Xu, Y. Based on network pharmacology to explore the molecular mechanisms of Astragalus membranaceus for treating T2 diabetes mellitus. Ann. Transl. Med. 2019, 7, 633. [Google Scholar] [CrossRef]




| Mol ID | Molecule Name | OB (%) | DL |
|---|---|---|---|
| MOL000398 | Isoflavanone | 109.99 | 0.30 |
| MOL000378 | 7-O-methylisomucronulatol | 74.69 | 0.30 |
| MOL000392 | Formononetin | 69.67 | 0.21 |
| MOL000433 | Folsaeure (FA) | 68.96 | 0.71 |
| MOL000438 | (3R)-3-(2-hydroxy-3,4-dimethoxyphenyl) chroman-7-ol | 67.67 | 0.26 |
| MOL000380 | (6aR,11aR)-9,10-dimethoxy-6a,11a-dihydro-6H-benzofurano[3,2-c] chromen-3-ol | 64.26 | 0.42 |
| MOL000211 | Mairin | 55.38 | 0.78 |
| MOL000371 | 3,9-di-O-methylnissolin | 53.74 | 0.48 |
| MOL000239 | Jaranol | 50.83 | 0.29 |
| MOL000354 | Isorhamnetin | 49.60 | 0.31 |
| MOL000439 | Isomucronulatol-7,2′-di-O-glucosiole | 49.28 | 0.62 |
| MOL000417 | Calycosin | 47.75 | 0.24 |
| MOL000098 | Quercetin | 46.43 | 0.28 |
| MOL000422 | Kaempferol | 41.88 | 0.24 |
| MOL000374 | 5′-hydroxyiso-muronulatol-2′,5′-di-O-glucoside | 41.72 | 0.69 |
| MOL000442 | 1,7-Dihydroxy-3,9-dimethoxy pterocarpene | 39.05 | 0.48 |
| MOL000296 | Hederagenin | 36.91 | 0.75 |
| MOL000379 | 9,10-dimethoxypterocarpan-3-O-β-D-glucoside | 36.74 | 0.92 |
| MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | 36.23 | 0.78 |
| MOL000387 | Bifendate | 31.10 | 0.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Zheng, W.; Yang, J.; Ali, A.; Qin, H. Mechanism of Astragalus membranaceus Alleviating Acquired Hyperlipidemia Induced by High-Fat Diet through Regulating Lipid Metabolism. Nutrients 2022, 14, 954. https://doi.org/10.3390/nu14050954
Wang L, Zheng W, Yang J, Ali A, Qin H. Mechanism of Astragalus membranaceus Alleviating Acquired Hyperlipidemia Induced by High-Fat Diet through Regulating Lipid Metabolism. Nutrients. 2022; 14(5):954. https://doi.org/10.3390/nu14050954
Chicago/Turabian StyleWang, Ling, Wenya Zheng, Jinxin Yang, Anwar Ali, and Hong Qin. 2022. "Mechanism of Astragalus membranaceus Alleviating Acquired Hyperlipidemia Induced by High-Fat Diet through Regulating Lipid Metabolism" Nutrients 14, no. 5: 954. https://doi.org/10.3390/nu14050954
APA StyleWang, L., Zheng, W., Yang, J., Ali, A., & Qin, H. (2022). Mechanism of Astragalus membranaceus Alleviating Acquired Hyperlipidemia Induced by High-Fat Diet through Regulating Lipid Metabolism. Nutrients, 14(5), 954. https://doi.org/10.3390/nu14050954








