High-Fat Diet Augments the Effect of Alcohol on Skeletal Muscle Mitochondrial Dysfunction in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Alcohol and Dietary Treatments
2.3. Tissue Collection
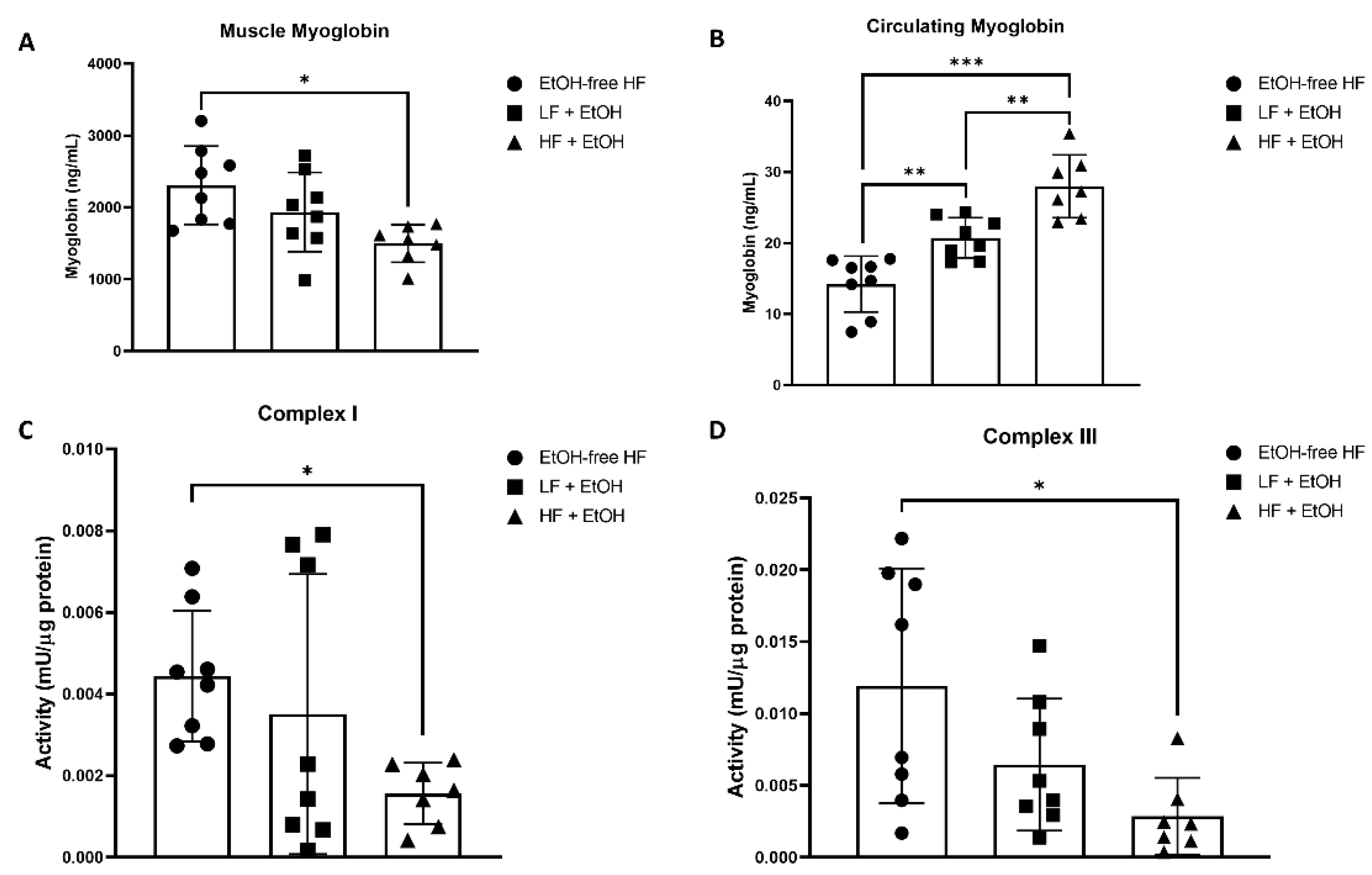
2.4. Myoglobin Concentration
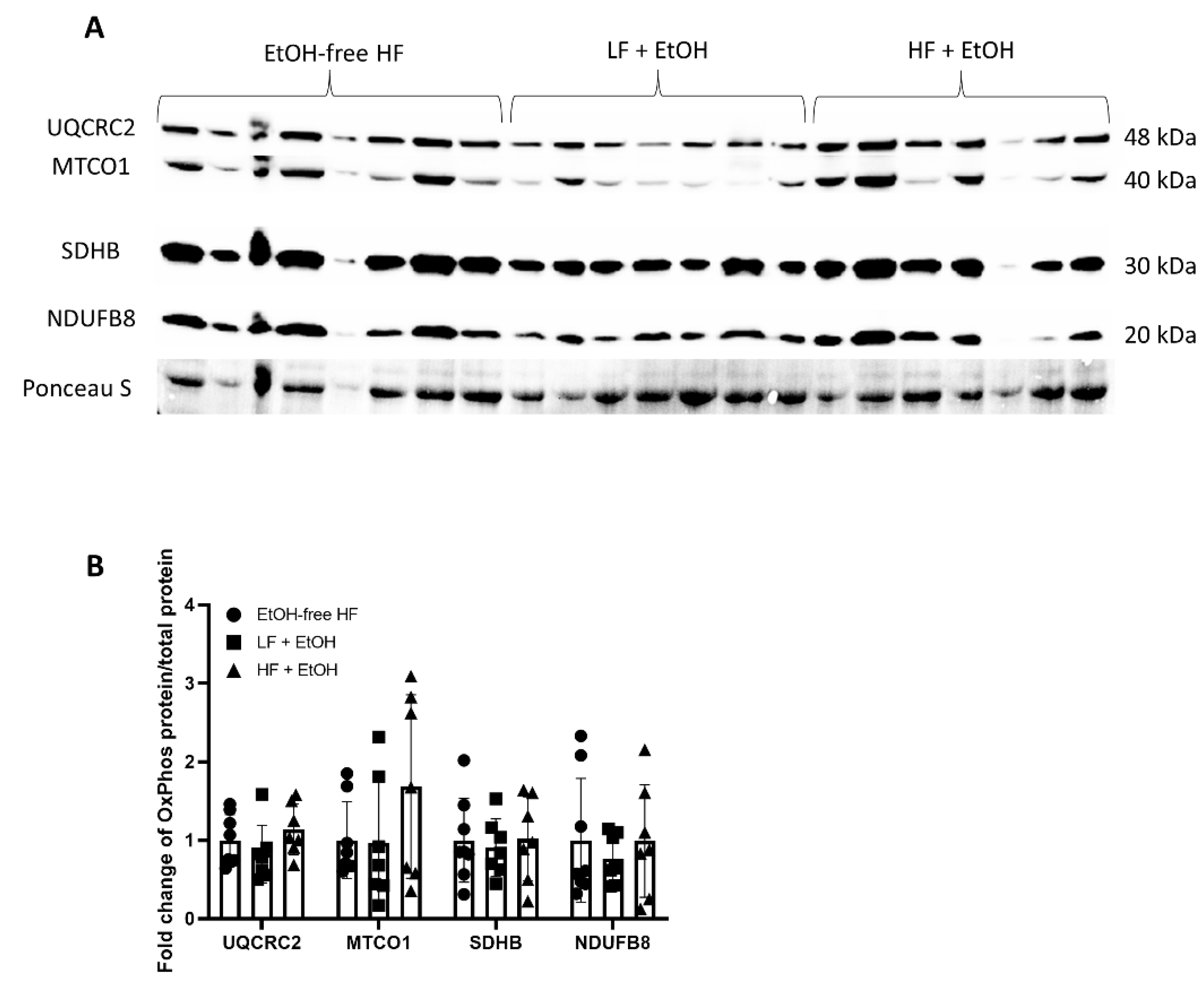
2.5. Electron Transport Chain Complex Activity
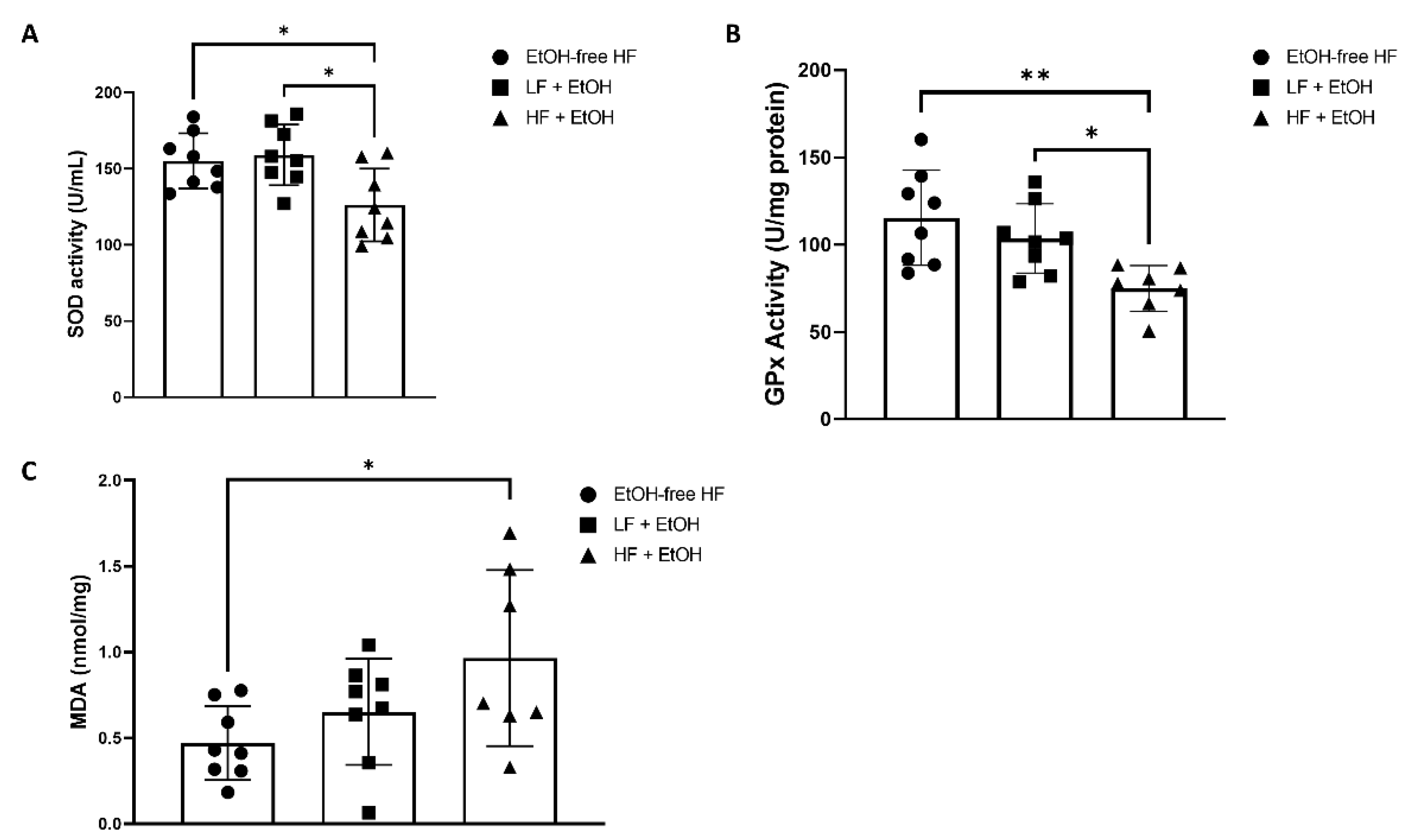
2.6. Oxidative Stress and Antioxidant Markers
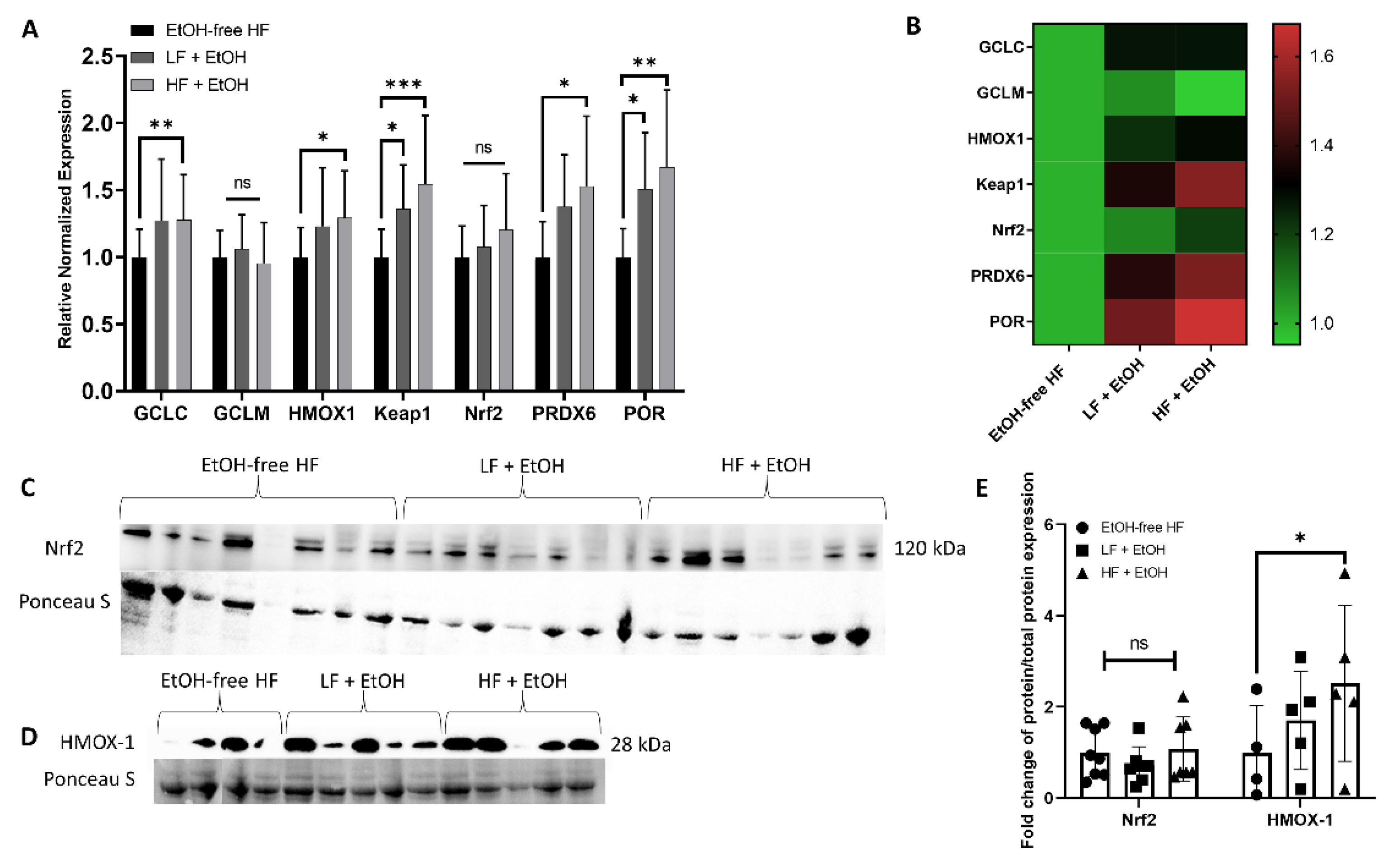
2.7. Gene Expression
2.8. Protein Expression
2.9. Statistical Analysis
3. Results
3.1. Alcohol Consumption, Body Weights, and Muscle Weights
3.2. Myoglobin Concentrations
3.3. Electron Transport Chain Activity and Expression
3.4. Oxidative Stress and Antioxidant Markers
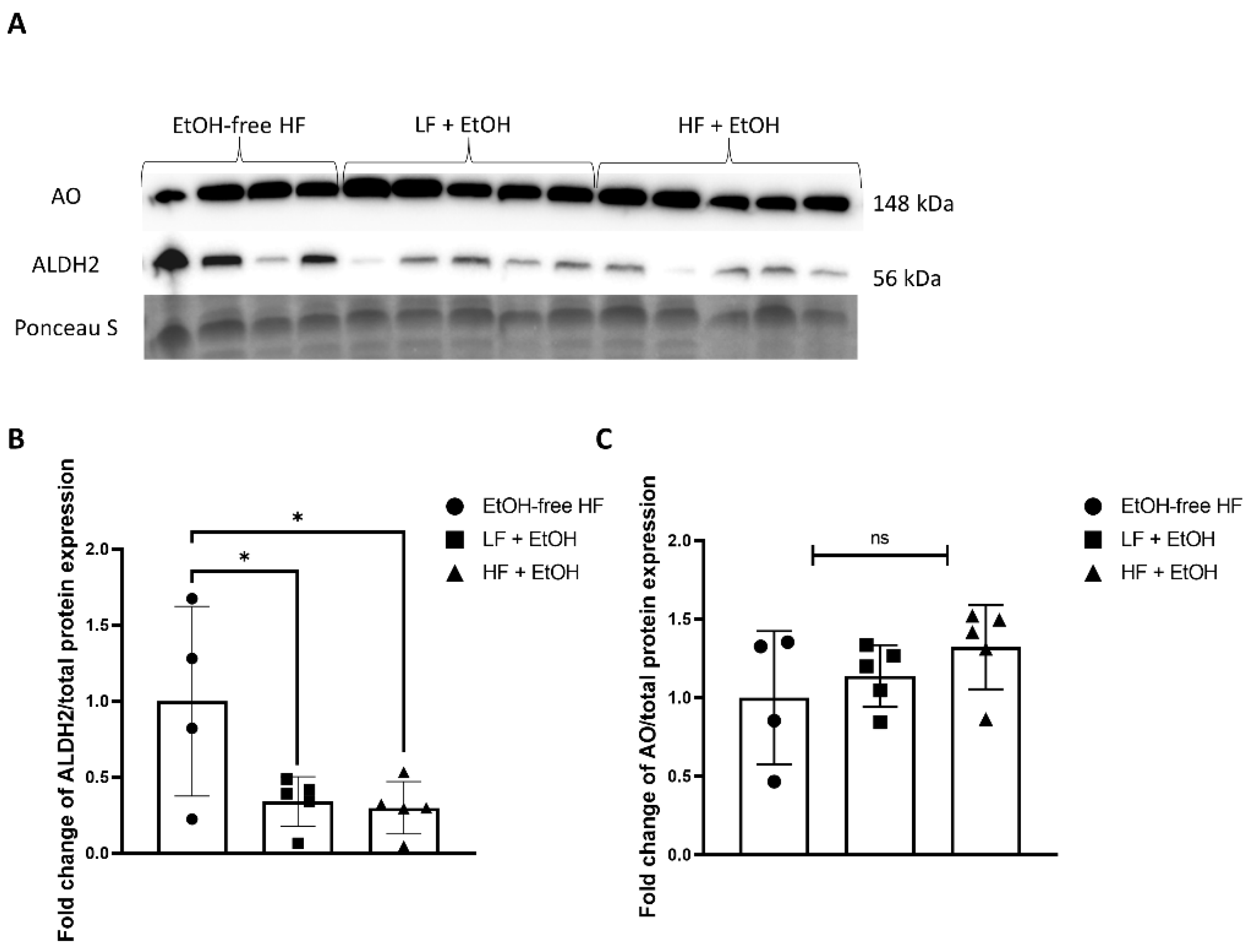
3.5. Alcohol-Associated Regulators of Mitochondrial Function
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boersma, P.; Villarroel, M.A.; Vahratian, A. Heavy Drinking among U.S. Adults, 2018; NCHS Data Brief; National Center for Health Statistics: Hyattsville, MD, USA, 2020; pp. 1–8.
- Griswold, M.G.; Fullman, N.; Hawley, C.; Arian, N.; Zimsen, S.R.M.; Tymeson, H.D.; Venkateswaran, V.; Tapp, A.D.; Forouzanfar, M.H.; Salama, J.S.; et al. Alcohol use and burden for 195 countries and territories, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2018, 392, 1015–1035. [Google Scholar] [CrossRef] [Green Version]
- Evangelou, E.; Suzuki, H.; Bai, W.J.; Pazoki, R.; Gao, H.; Matthews, P.M.; Elliott, P. Alcohol consumption in the general population is associated with structural changes in multiple organ systems. eLife 2021, 10, e65325. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.; Jolley, S.E.; Molina, P.E. Alcoholic Myopathy: Pathophysiologic Mechanisms and Clinical Implications. Alcohol Res. 2017, 38, 207–217. [Google Scholar] [PubMed]
- Steiner, J.L.; Lang, C.H. Dysregulation of skeletal muscle protein metabolism by alcohol. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E699–E712. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.S.; Kumar, A.; Welch, N.; Sekar, J.; Mishra, S.; Bellar, A.; Gangadhariah, M.; Attaway, A.; Al Khafaji, H.; Wu, X.; et al. Multiomics-Identified Intervention to Restore Ethanol-Induced Dysregulated Proteostasis and Secondary Sarcopenia in Alcoholic Liver Disease. Cell. Physiol. Biochem. 2021, 55, 91–116. [Google Scholar] [CrossRef]
- Kumar, A.; Davuluri, G.; Welch, N.; Kim, A.; Gangadhariah, M.; Allawy, A.; Priyadarshini, A.; McMullen, M.R.; Sandlers, Y.; Willard, B.; et al. Oxidative stress mediates Ethanol-Induced skeletal muscle mitochondrial dysfunction and dysregulated protein synthesis and autophagy. Free Radic. Biol. Med. 2019, 145, 284–299. [Google Scholar] [CrossRef]
- Dekeyser, G.J.; Clary, C.R.; Otis, J.S. Chronic alcohol ingestion delays skeletal muscle regeneration following injury. Regen. Med. Res. 2013, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Otis, J.S.; Brown, L.A.; Guidot, D.M. Oxidant-induced atrogin-1 and transforming growth Factor-Beta1 precede Alcohol-Related myopathy in rats. Muscle Nerve 2007, 36, 842–848. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Sola, J.; Preedy, V.R.; Lang, C.H.; Gonzalez-Reimers, E.; Arno, M.; Lin, J.C.; Wiseman, H.; Zhou, S.; Emery, P.W.; Nakahara, T.; et al. Molecular and cellular events in alcohol-induced muscle disease. Alcohol. Clin. Exp. Res. 2007, 31, 1953–1962. [Google Scholar] [CrossRef]
- Trounce, I.; Byrne, E.; Dennett, X. Biochemical and morphological studies of skeletal muscle in experimental chronic alcoholic myopathy. Acta Neurol. Scand. 1990, 82, 386–391. [Google Scholar] [CrossRef]
- Mokdad, A.H.; Marks, J.S.; Stroup, D.F.; Gerberding, J.L. Actual causes of death in the United States, 2000. JAMA 2004, 291, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.H.; Zeng, L.M.; Zheng, C.B.; Song, B.; Li, F.N.; Kong, X.F.; Xu, K. Inflammatory Links Between High Fat Diets and Diseases. Front. Immunol. 2018, 9, 2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miotto, P.M.; LeBlanc, P.J.; Holloway, G.P. High-Fat Diet Causes Mitochondrial Dysfunction as a Result of Impaired ADP Sensitivity. Diabetes 2018, 67, 2199–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putti, R.; Migliaccio, V.; Sica, R.; Lionetti, L. Skeletal Muscle Mitochondrial Bioenergetics and Morphology in High Fat Diet Induced Obesity and Insulin Resistance: Focus on Dietary Fat Source. Front. Physiol. 2015, 6, 426. [Google Scholar] [CrossRef] [Green Version]
- Carter, H.N.; Chen, C.C.; Hood, D.A. Mitochondria, muscle health, and exercise with advancing age. Physiology 2015, 30, 208–223. [Google Scholar] [CrossRef] [Green Version]
- Peterson, C.M.; Johannsen, D.L.; Ravussin, E. Skeletal muscle mitochondria and aging: A review. J. Aging Res. 2012, 2012, 194821. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, S.; Takada, S.; Furihata, T.; Fukushima, A.; Yokota, T.; Kinugawa, S. Mitochondrial respiration of complex II is not lower than that of complex I in mouse skeletal muscle. Biochem. Biophys. Rep. 2020, 21, 100717. [Google Scholar] [CrossRef]
- Nakamura, S.; Takamura, T.; Matsuzawa-Nagata, N.; Takayama, H.; Misu, H.; Noda, H.; Nabemoto, S.; Kurita, S.; Ota, T.; Ando, H.; et al. Palmitate induces insulin resistance in H4IIEC3 hepatocytes through reactive oxygen species produced by mitochondria. J. Biol. Chem. 2009, 284, 14809–14818. [Google Scholar] [CrossRef] [Green Version]
- McLennan, H.R.; Degli Esposti, M. The contribution of mitochondrial respiratory complexes to the production of reactive oxygen species. J. Bioenergy Biomembr. 2000, 32, 153–162. [Google Scholar] [CrossRef]
- Navarro-Yepes, J.; Burns, M.; Anandhan, A.; Khalimonchuk, O.; del Razo, L.M.; Quintanilla-Vega, B.; Pappa, A.; Panayiotidis, M.I.; Franco, R. Oxidative stress, redox signaling, and autophagy: Cell death versus survival. Antioxid. Redox Signal. 2014, 21, 66–85. [Google Scholar] [CrossRef] [Green Version]
- Wilburn, D.; Ismaeel, A.; Machek, S.; Fletcher, E.; Koutakis, P. Shared and distinct mechanisms of skeletal muscle atrophy: A narrative review. Ageing Res. Rev. 2021, 71, 101463. [Google Scholar] [CrossRef] [PubMed]
- Dionisi, T.; Addolorato, G. Effect of Alcohol Combined With High Fat Diet: Two Wrongs Don’t Make a Right but They Could Make a Good Excuse Comment. Alcohol Alcohol. 2021, 56, 348–350. [Google Scholar] [CrossRef]
- Steiner, J.L.; Lang, C.H. Alcohol, Adipose Tissue and Lipid Dysregulation. Biomolecules 2017, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Osna, N.A.; Ganesan, M.; Seth, D.; Wyatt, T.A.; Kidambi, S.; Kharbanda, K.K. Second hits exacerbate Alcohol-Related organ damage: An update. Alcohol Alcohol. 2021, 56, 8–16. [Google Scholar] [CrossRef]
- Ren, J. Interaction between High-Fat diet and alcohol dehydrogenase on Ethanol-Elicited cardiac depression in murine myocytes. Obesity 2007, 15, 2932–2941. [Google Scholar] [CrossRef] [Green Version]
- Rosa, D.F.; Sarandy, M.M.; Novaes, R.D.; da Matta, S.L.P.; Goncalves, R.V. Effect of a High-Fat diet and alcohol on cutaneous repair: A systematic review of murine experimental models. PLoS ONE 2017, 12, e0176240. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Portilla, M.; Montagud-Romero, S.; Navarrete, F.; Gasparyan, A.; Manzanares, J.; Minarro, J.; Rodriguez-Arias, M. Pairing Binge Drinking and a High-Fat Diet in Adolescence Modulates the Inflammatory Effects of Subsequent Alcohol Consumption in Mice. Int. J. Mol. Sci. 2021, 22, 5279. [Google Scholar] [CrossRef]
- Dwivedi, D.K.; Kumar, D.; Kwatra, M.; Pandey, S.N.; Choubey, P.; Lahkar, M.; Jangra, A. Voluntary alcohol consumption exacerbated high fat Diet-Induced cognitive deficits by NF-kappa B-calpain dependent apoptotic cell death in rat hippocampus: Ameliorative effect of melatonin. Biomed. Pharmacother. 2018, 108, 1393–1403. [Google Scholar] [CrossRef]
- Gabele, E.; Dostert, K.; Dorn, C.; Patsenker, E.; Stickel, F.; Hellerbrand, C. A new model of interactive effects of alcohol and High-Fat diet on hepatic fibrosis. Alcohol. Clin. Exp. Res. 2011, 35, 1361–1367. [Google Scholar] [CrossRef]
- Schonfeld, M.; O’Neil, M.; Villar, M.T.; Artigues, A.; Averilla, J.; Gunewardena, S.; Weinman, S.A.; Tikhanovich, I. A Western diet with alcohol in drinking water recapitulates features of alcohol-associated liver disease in mice. Alcohol. Clin. Exp. Res. 2021, 45, 1980–1993. [Google Scholar] [CrossRef]
- Guo, F.; Zheng, K.; Benede-Ubieto, R.; Cubero, F.J.; Nevzorova, Y.A. The Lieber-DeCarli Diet-A Flagship Model for Experimental Alcoholic Liver Disease. Alcohol. Clin. Exp. Res. 2018, 42, 1828–1840. [Google Scholar] [CrossRef]
- Lieber, C.S.; DeCarli, L.M. The feeding of alcohol in liquid diets: Two decades of applications and 1982 update. Alcohol. Clin. Exp. Res. 1982, 6, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Giampietro, O.; Clerico, A.; Buzzigoli, G.; Delchicca, M.G.; Boni, C.; Carpi, A. Detection of Hypothyroid Myopathy by Measurement of Various Serum Muscle Markers—Myoglobin, Creatine-Kinase, Lactate-Dehydrogenase and Their Isoenzymes—Correlations with Thyroid-Hormone Levels (Free and Total) and Clinical Usefulness. Horm. Res. 1984, 19, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Kagan, L.J. Myoglobinemia in inflammatory myopathies. JAMA 1977, 237, 1448–1452. [Google Scholar] [CrossRef]
- Melberg, A.; Holme, E.; Oldfors, A.; Lundberg, P.O. Rhabdomyolysis in autosomal dominant progressive external ophthalmoplegia. Neurology 1998, 50, 299–300. [Google Scholar] [CrossRef] [PubMed]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turk, B.R.; Theisen, B.E.; Nemeth, C.L.; Marx, J.S.; Shi, X.; Rosen, M.; Jones, R.O.; Moser, A.B.; Watkins, P.A.; Raymond, G.V.; et al. Antioxidant Capacity and Superoxide Dismutase Activity in Adrenoleukodystrophy. JAMA Neurol. 2017, 74, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Dworzanski, J.; Strycharz-Dudziak, M.; Kliszczewska, E.; Kielczykowska, M.; Dworzanska, A.; Drop, B.; Polz-Dacewicz, M. Glutathione peroxidase (GPx) and superoxide dismutase (SOD) activity in patients with diabetes mellitus type 2 infected with Epstein-Barr virus. PLoS ONE 2020, 15, e0230374. [Google Scholar] [CrossRef]
- Krejsa, C.M.; Franklin, C.C.; White, C.C.; Ledbetter, J.A.; Schieven, G.L.; Kavanagh, T.J. Rapid activation of glutamate cysteine ligase following oxidative stress. J. Biol. Chem. 2010, 285, 16116–16124. [Google Scholar] [CrossRef] [Green Version]
- Ham, M.; Kaunitz, J.D. Gastroduodenal mucosal defense. Curr. Opin. Gastroenterol. 2008, 24, 665–673. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Ji, P.; Zhang, L.; Bu, G.; Gu, H.; Wang, X.; Xiong, Y.; Zuo, B. The Expression of Porcine Prdx6 Gene Is Up-Regulated by C/EBPbeta and CREB. PLoS ONE 2015, 10, e0144851. [Google Scholar] [CrossRef] [Green Version]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, C.R.; Han, D.H.; Chen, M.; Terada, S.; Yasuda, T.; Wright, D.C.; Holloszy, J.O. High-Fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc. Natl. Acad. Sci. USA 2008, 105, 7815–7820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisner, V.; Lenaers, G.; Hajnoczky, G. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. J. Cell Biol. 2014, 205, 179–195. [Google Scholar] [CrossRef] [Green Version]
- Oh, K.H.; Sheoran, S.; Richmond, J.E.; Kim, H. Alcohol induces mitochondrial fragmentation and stress responses to maintain normal muscle function in Caenorhabditis elegans. FASEB J. 2020, 34, 8204–8216. [Google Scholar] [CrossRef] [Green Version]
- Song, B.J.; Akbar, M.; Abdelmegeed, M.A.; Byun, K.; Lee, B.; Yoon, S.K.; Hardwick, J.P. Mitochondrial dysfunction and tissue injury by alcohol, high fat, nonalcoholic substances and pathological conditions through Post-Translational protein modifications. Redox Biol. 2014, 3, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Abdelmegeed, M.A.; Jang, S.; Banerjee, A.; Hardwick, J.P.; Song, B.J. Robust protein nitration contributes to Acetaminophen-Induced mitochondrial dysfunction and acute liver injury. Free Radic. Biol. Med. 2013, 60, 211–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, J.; McFadden, G.E.; Borgerding, A.J.; Norby, F.L.; Ren, B.H.; Ye, G.; Epstein, P.N.; Ren, J. Overexpression of alcohol dehydrogenase exacerbates Ethanol-Induced contractile defect in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 2002, 282, H1216–H1222. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.J.; Lee, H.M.; Kim, J.H.; Park, I.S.; Rho, Y.S. Heavy alcohol drinking downregulates ALDH2 gene expression but heavy smoking up-regulates SOD2 gene expression in head and neck squamous cell carcinoma. World J. Surg. Oncol. 2017, 15, 163. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.M.; Liu, A.J.; Zang, P.; Dong, W.Z.; Ying, L.; Wang, W.; Xu, P.; Song, X.R.; Cai, J.; Zhang, S.Q.; et al. ALDH2 protects against stroke by clearing 4-HNE. Cell Res. 2013, 23, 915–930. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, B.; Zhang, J.; He, D.; Zhang, Q.; Pan, C.; Yuan, Q.; Shi, Y.; Tang, H.; Xu, F.; et al. ALDH2 (Aldehyde Dehydrogenase 2) Protects Against Hypoxia-Induced Pulmonary Hypertension. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 2303–2319. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yu, L.; Byra, E.A.; Hu, N.; Kitagawa, K.; Nakayama, K.I.; Kawamoto, T.; Ren, J. Aldehyde dehydrogenase 2 knockout accentuates Ethanol-Induced cardiac depression: Role of protein phosphatases. J. Mol. Cell. Cardiol. 2010, 49, 322–329. [Google Scholar] [CrossRef] [Green Version]
- Doser, T.A.; Turdi, S.; Thomas, D.P.; Epstein, P.N.; Li, S.Y.; Ren, J. Transgenic overexpression of aldehyde dehydrogenase-2 rescues chronic alcohol intake-induced myocardial hypertrophy and contractile dysfunction. Circulation 2009, 119, 1941–1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wang, C.; Turdi, S.; Richmond, K.L.; Zhang, Y.; Ren, J. ALDH2 protects against high fat Diet-Induced obesity cardiomyopathy and defective autophagy: Role of CaM kinase II, histone H3K9 methyltransferase SUV39H, Sirt1, and PGC-1alpha deacetylation. Int. J. Obes. 2018, 42, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.S.; Chen, Y.H.; Hu, J.T.; Chiu, C.F.; Hung, S.W.; Chang, Y.C.; Chiu, C.C.; Chuang, H.L. Aldehyde Dehydrogenase Mutation Exacerbated High-Fat-Diet-Induced Nonalcoholic Fatty Liver Disease with Gut Microbiota Remodeling in Male Mice. Biology 2021, 10, 737. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Munukutla, S.; Kar, A.; Gardinier, J.; Thandavarayan, R.A.; Palaniyandi, S.S. Type-2 diabetic aldehyde dehydrogenase 2 mutant mice (ALDH 2 * 2) exhibiting heart failure with preserved ejection fraction phenotype can be determined by exercise stress echocardiography. PLoS ONE 2018, 13, e0195796. [Google Scholar] [CrossRef] [Green Version]
- Laudato, J.A.; Tice, A.L.; Dunlap, K.R.; Keith, Z.A.; Steiner, J.L. Dietary Composition Exacerbates Sex Differences in Skeletal Muscle Myopathy following Chronic Alcohol Intake. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Lieber, C.S.; Spritz, N.; DeCarli, L.M. Role of dietary, adipose, and endogenously synthesized fatty acids in the pathogenesis of the alcoholic fatty liver. J. Clin. Investig. 1966, 45, 51–62. [Google Scholar] [CrossRef]
- Dasarathy, J.; McCullough, A.J.; Dasarathy, S. Sarcopenia in Alcoholic Liver Disease: Clinical and Molecular Advances. Alcohol. Clin. Exp. Res. 2017, 41, 1419–1431. [Google Scholar] [CrossRef]
- Bradford, B.U.; O’Connell, T.M.; Han, J.; Kosyk, O.; Shymonyak, S.; Ross, P.K.; Winnike, J.; Kono, H.; Rusyn, I. Metabolomic profiling of a modified alcohol liquid diet model for liver injury in the mouse uncovers new markers of disease. Toxicol. Appl. Pharmacol. 2008, 232, 236–243. [Google Scholar] [CrossRef] [Green Version]
- Bertola, A.; Mathews, S.; Ki, S.H.; Wang, H.; Gao, B. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat. Protoc. 2013, 8, 627–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanji, A.A.; Sadrzadeh, S.M.; Yang, E.K.; Fogt, F.; Meydani, M.; Dannenberg, A.J. Dietary saturated fatty acids: A novel treatment for alcoholic liver disease. Gastroenterology 1995, 109, 547–554. [Google Scholar] [CrossRef]
- Zhong, W.; Li, Q.; Xie, G.; Sun, X.; Tan, X.; Sun, X.; Jia, W.; Zhou, Z. Dietary fat sources differentially modulate intestinal barrier and hepatic inflammation in alcohol-induced liver injury in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G919–G932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, M.; Considine, R.V.; Leone, T.C.; Kelly, D.P.; Crabb, D.W. Role of adiponectin in the protective action of dietary saturated fat against alcoholic fatty liver in mice. Hepatology 2005, 42, 568–577. [Google Scholar] [CrossRef]
- Warner, D.R.; Liu, H.; Dastidar, S.G.; Warner, J.B.; Prodhan, M.A.I.; Yin, X.; Zhang, X.; Feldstein, A.E.; Gao, B.; Prough, R.A.; et al. Ethanol and unsaturated dietary fat induce unique patterns of hepatic omega-6 and omega-3 PUFA oxylipins in a mouse model of alcoholic liver disease. PLoS ONE 2018, 13, e0204119. [Google Scholar] [CrossRef] [Green Version]
- Kirpich, I.A.; Petrosino, J.; Ajami, N.; Feng, W.; Wang, Y.; Liu, Y.; Beier, J.I.; Barve, S.S.; Yin, X.; Wei, X.; et al. Saturated and Unsaturated Dietary Fats Differentially Modulate Ethanol-Induced Changes in Gut Microbiome and Metabolome in a Mouse Model of Alcoholic Liver Disease. Am. J. Pathol. 2016, 186, 765–776. [Google Scholar] [CrossRef] [Green Version]
| Diet Name | Carbohydrates | Fat | Protein | Alcohol |
|---|---|---|---|---|
| EtOH-free HF Diet | 47 | 35 | 18 | 0 |
| HF + EtOH Diet | 15 | 35 | 18 | 32 |
| LF + EtOH Diet | 38 | 12 | 18 | 32 |
| mRNA | 5′—————3′ | Forward/Reverse | Amplicon Size (Base Pairs) |
|---|---|---|---|
| GCLC | AACACAGACCCAACCCAGAG TGGCACATTGATGACAACCT | Forward Reverse | 143 |
| GCLM | TGTGTGATGCCACCAGATTT GCTTCAATGTCAGGGATGCT | Forward Reverse | 137 |
| HMOX1 | TTACCTTCCCGAACATCGAC TCCTCTGTCAGCATCACCTG | Forward Reverse | 172 |
| Keap1 | GGCAGGACCAGTTGAACAGT ATCACTGTCCGGGTCATAGC | Forward Reverse | 188 |
| Nrf2 | AGCAAGTTTGGCAGGAGCTA TTCTTTTTCCAGCGAGGAGA | Forward Reverse | 164 |
| PRDX6 | AAACACCCACGGAAAAGTTG GTTTCTTGTCAGGGCCAAAA | Forward Reverse | 153 |
| POR | GTCTTCTGCATGGCCACATA TGGCGTTGAAGTGCTCATAG | Forward Reverse | 151 |
| B2M 1 | TGGTCTTTCTGGTGCTTGTC GGGTGGAACTGTGTTACGTAG | Forward Reverse | 112 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismaeel, A.; Laudato, J.A.; Fletcher, E.; Papoutsi, E.; Tice, A.; Hwa, L.S.; Miserlis, D.; Jamurtas, A.Z.; Steiner, J.; Koutakis, P. High-Fat Diet Augments the Effect of Alcohol on Skeletal Muscle Mitochondrial Dysfunction in Mice. Nutrients 2022, 14, 1016. https://doi.org/10.3390/nu14051016
Ismaeel A, Laudato JA, Fletcher E, Papoutsi E, Tice A, Hwa LS, Miserlis D, Jamurtas AZ, Steiner J, Koutakis P. High-Fat Diet Augments the Effect of Alcohol on Skeletal Muscle Mitochondrial Dysfunction in Mice. Nutrients. 2022; 14(5):1016. https://doi.org/10.3390/nu14051016
Chicago/Turabian StyleIsmaeel, Ahmed, Joseph A. Laudato, Emma Fletcher, Evlampia Papoutsi, Abigail Tice, Lara S. Hwa, Dimitrios Miserlis, Athanasios Z. Jamurtas, Jennifer Steiner, and Panagiotis Koutakis. 2022. "High-Fat Diet Augments the Effect of Alcohol on Skeletal Muscle Mitochondrial Dysfunction in Mice" Nutrients 14, no. 5: 1016. https://doi.org/10.3390/nu14051016
APA StyleIsmaeel, A., Laudato, J. A., Fletcher, E., Papoutsi, E., Tice, A., Hwa, L. S., Miserlis, D., Jamurtas, A. Z., Steiner, J., & Koutakis, P. (2022). High-Fat Diet Augments the Effect of Alcohol on Skeletal Muscle Mitochondrial Dysfunction in Mice. Nutrients, 14(5), 1016. https://doi.org/10.3390/nu14051016









