Blood DNA Methylation Predicts Diabetic Kidney Disease Progression in High Fat Diet-Fed Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Group Classification
2.3. Blood and Urine Assay
2.4. RNA Extraction and Quantitative Real-Time PCR
2.5. Immunoblot
2.6. Reduced Representative Bisulfide Sequencing (RRBS)
2.7. Histology
2.8. Statistical Analysis
3. Results
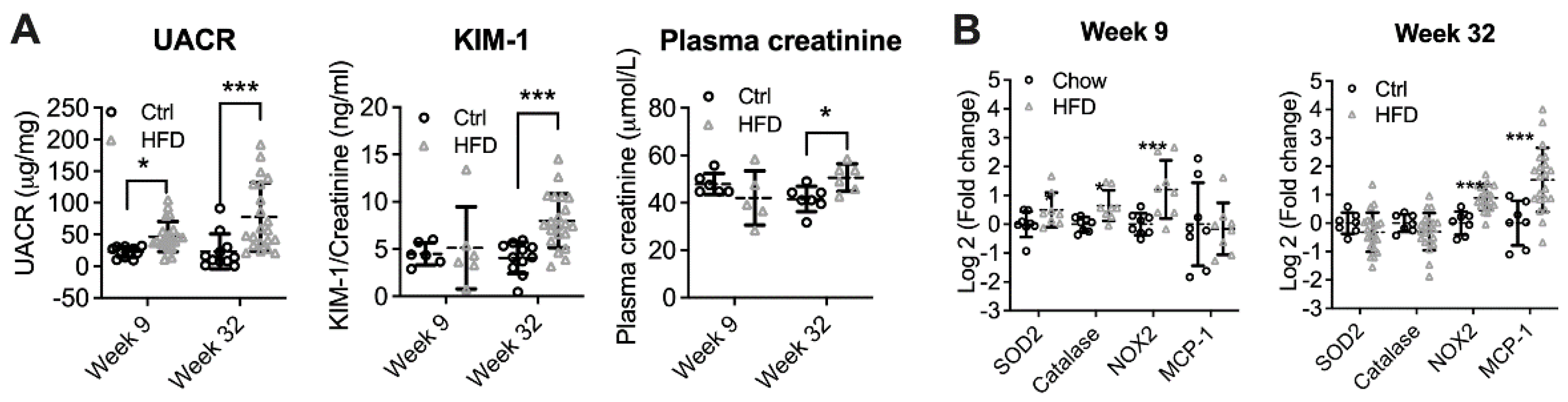
3.1. High-Fat Diet Induced Diabetic Kidney Disease
3.2. DKD Progression Rates Vary among HFD-Fed Mice
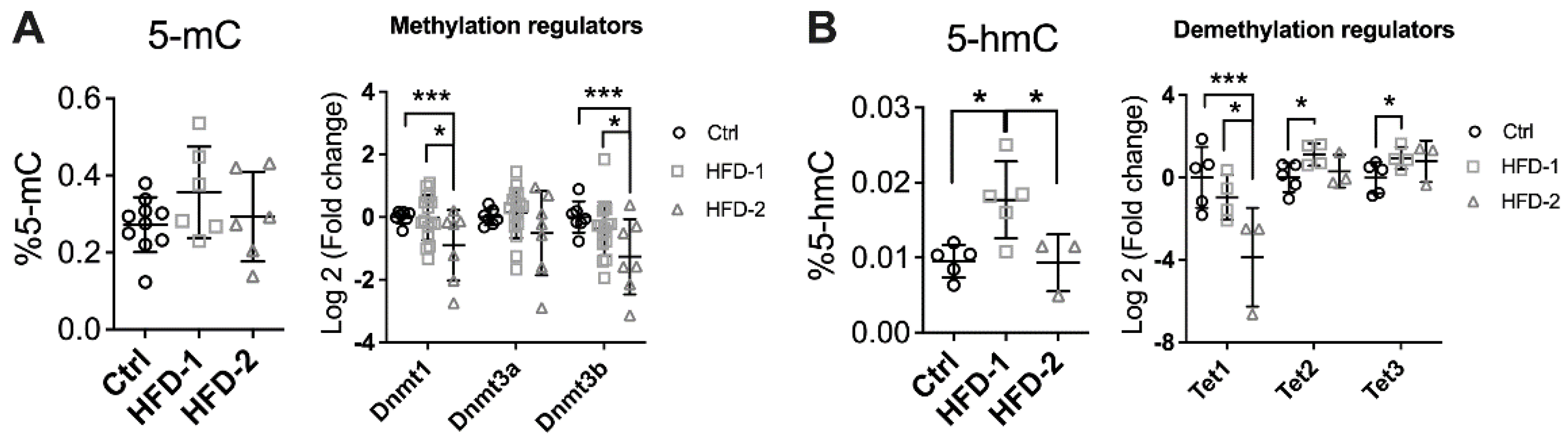
3.3. Renal DNA Methylation Was Impaired in Mice with Advanced DKD
3.4. Different Blood DNA Methylation Profiles between Mice with Advanced and Mild DKD
3.5. Blood DNA Methylation Markers Were Detectable in Early DKD
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. Diabetes. Available online: https://www.who.int/en/news-room/fact-sheets/detail/diabetes (accessed on 25 October 2021).
- Gheith, O.; Farouk, N.; Nampoory, N.; Halim, M.A.; Al-Otaibi, T. Diabetic kidney disease: World wide difference of prevalence and risk factors. J. Nephropharmacol. 2016, 5, 49–56. [Google Scholar] [CrossRef]
- Alicic, R.Z.; Rooney, M.T.; Tuttle, K.R. Diabetic Kidney Disease. Chall. Prog. Possibilities 2017, 12, 2032–2045. [Google Scholar] [CrossRef] [PubMed]
- Glassock, R.J. Is the presence of microalbuminuria a relevant marker of kidney disease? Curr. Hypertens. Rep. 2010, 12, 364–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwyer, J.P.; Parving, H.H.; Hunsicker, L.G.; Ravid, M.; Remuzzi, G.; Lewis, J.B. Renal Dysfunction in the Presence of Normoalbuminuria in Type 2 Diabetes: Results from the DEMAND Study. Cardiorenal Med. 2012, 2, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radcliffe, N.J.; Seah, J.-M.; Clarke, M.; MacIsaac, R.J.; Jerums, G.; Ekinci, E.I. Clinical predictive factors in diabetic kidney disease progression. J. Diabetes Investig. 2017, 8, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Miao, F.; Paterson, A.D.; Lachin, J.M.; Zhang, L.; Schones, D.E.; Wu, X.; Wang, J.; Tompkins, J.D.; Genuth, S. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc. Natl. Acad. Sci. USA 2016, 113, E3002–E3011. [Google Scholar] [CrossRef] [Green Version]
- Chu, A.Y.; Tin, A.; Schlosser, P.; Ko, Y.-A.; Qiu, C.; Yao, C.; Joehanes, R.; Grams, M.E.; Liang, L.; Gluck, C.A.; et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat. Commun. 2017, 8, 1286. [Google Scholar] [CrossRef]
- Wing, M.R.; Devaney, J.M.; Joffe, M.M.; Xie, D.; Feldman, H.I.; Dominic, E.A.; Guzman, N.J.; Ramezani, A.; Susztak, K.; Herman, J.G.; et al. DNA methylation profile associated with rapid decline in kidney function: Findings from the CRIC study. Nephrol. Dial. Transplant. 2014, 29, 864–872. [Google Scholar] [CrossRef] [Green Version]
- Smyth, L.J.; Patterson, C.C.; Swan, E.J.; Maxwell, A.P.; McKnight, A.J. DNA methylation associated with diabetic kidney disease in Blood-Derived DNA. Front. Cell Dev. Biol. 2020, 8, 561907. [Google Scholar] [CrossRef]
- Volkmar, M.; Dedeurwaerder, S.; Cunha, D.A.; Ndlovu, M.N.; Defrance, M.; Deplus, R.; Calonne, E.; Volkmar, U.; Igoillo-Esteve, M.; Naamane, N.; et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012, 31, 1405–1426. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Bae, J.H.; Park, K.S.; Sung, J.; Kwak, S.H. DNA Methylation Changes Associated With Type 2 Diabetes and Diabetic Kidney Disease in an East Asian Population. J. Clin. Endocrinol. Metab. 2021, 106, e3837–e3851. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Mak, C.H.; Chen, H.; Zaky, A.A.; Wong, M.G.; Pollock, C.A.; Saad, S. SIRT1 Attenuates Kidney Disorders in Male Offspring Due to Maternal High-Fat Diet. Nutrients 2019, 11, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.T.; Chen, H.; Zaky, A.; Pollock, C.; Saad, S. SIRT1 overexpression attenuates offspring metabolic and liver disorders as a result of maternal high-fat feeding. J. Physiol. 2019, 597, 467–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glastras, S.J.; Chen, H.; Tsang, M.; Teh, R.; McGrath, R.T.; Zaky, A.; Chen, J.; Wong, M.G.; Pollock, C.A.; Saad, S. The renal consequences of maternal obesity in offspring are overwhelmed by postnatal high fat diet. PLoS ONE 2017, 12, e0172644. [Google Scholar] [CrossRef] [Green Version]
- Zeisberg, M.; Neilson, E.G. Mechanisms of Tubulointerstitial Fibrosis. J. Am. Soc. Nephrol. 2010, 21, 1819–1834. [Google Scholar] [CrossRef] [Green Version]
- Han, W.K.; Bailly, V.; Abichandani, R.; Thadhani, R.; Bonventre, J.V. Kidney Injury Molecule-1 (KIM-1): A novel biomarker for human renal proximal tubule injury. Kidney Int. 2002, 62, 237–244. [Google Scholar] [CrossRef] [Green Version]
- Sheanon, N.M.; Mottl, A.K.; Dagostino, R.; Suerken, C.; Afkarian, M.; Dabelea, D.; Imperatore, G.; Marcovina, S.M.; Pettitt, D.J.; Saydah, S.; et al. Urinary NGAL and KIM-1 Are Significantly Elevated in Young Adults (YA) with Type 1 (T1D) and Type 2 (T2D) Diabetes. Diabetes 2018, 67, 525-P. [Google Scholar] [CrossRef]
- Khan, F.A.; Fatima, S.S.; Khan, G.M.; Shahid, S. Evaluation of kidney injury molecule-1 as a disease progression biomarker in diabetic nephropathy. Pak. J. Med. Sci 2019, 35, 992–996. [Google Scholar] [CrossRef] [Green Version]
- Feldreich, T.; Nowak, C.; Fall, T.; Carlsson, A.C.; Carrero, J.J.; Ripsweden, J.; Qureshi, A.R.; Heimbürger, O.; Barany, P.; Stenvinkel, P.; et al. Circulating proteins as predictors of cardiovascular mortality in end-stage renal disease. J. Nephrol. 2019, 32, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Carlsson, A.C.; Nowak, C.; Lind, L.; Östgren, C.J.; Nyström, F.H.; Sundström, J.; Carrero, J.J.; Riserus, U.; Ingelsson, E.; Fall, T.; et al. Growth differentiation factor 15 (GDF-15) is a potential biomarker of both diabetic kidney disease and future cardiovascular events in cohorts of individuals with type 2 diabetes: A proteomics approach. Upsala J. Med. Sci. 2020, 125, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Saad, S.; Shi, Y.; Wang, R.; Chou, A.S.Y.; Gill, A.; Yao, Y.; Jarolimek, W.; Pollock, C.A. Lysyl oxidase inhibitors attenuate cyclosporin A-induced nephropathy in mouse. Sci. Rep. 2021, 11, 12437. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 27 November 2021).
- Krueger, F. Trim Galore!: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 2 January 2019).
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Han, Y.; Zhou, L.; Chen, E.; Lu, B.; Liu, Y.; Pan, X.; Cowley, A.W., Jr.; Liang, M.; Wu, Q.; et al. A comprehensive evaluation of alignment software for reduced representation bisulfite sequencing data. Bioinformatics 2018, 34, 2715–2723. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Weinhold, L.; Wahl, S.; Pechlivanis, S.; Hoffmann, P.; Schmid, M. A statistical model for the analysis of beta values in DNA methylation studies. BMC Bioinform. 2016, 17, 480. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Chen, H.; Mak, C.; Zaky, A.; Pollock, C.; Saad, S. SRT1720 attenuates obesity and insulin resistance but not liver damage in the offspring due to maternal and postnatal high-fat diet consumption. Am. J. Physiol.-Endocrinol. Metab. 2018, 315, E196–E203. [Google Scholar] [CrossRef]
- Nowak, N.; Skupien, J.; Niewczas, M.A.; Yamanouchi, M.; Major, M.; Croall, S.; Smiles, A.; Warram, J.H.; Bonventre, J.V.; Krolewski, A.S. Increased plasma kidney injury molecule-1 suggests early progressive renal decline in non-proteinuric patients with type 1 diabetes. Kidney Int. 2016, 89, 459–467. [Google Scholar] [CrossRef] [Green Version]
- Gluck, C.; Qiu, C.; Han, S.Y.; Palmer, M.; Park, J.; Ko, Y.-A.; Guan, Y.; Sheng, X.; Hanson, R.L.; Huang, J.; et al. Kidney cytosine methylation changes improve renal function decline estimation in patients with diabetic kidney disease. Nat. Commun. 2019, 10, 2461. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanner, N.; Vornweg, J.; Combes, A.; Wilson, S.; Plappert, J.; Rafflenbeul, G.; Puelles, V.G.; Rahman, R.U.; Liwinski, T.; Lindner, S.; et al. DNA Methyltransferase 1 Controls Nephron Progenitor Cell Renewal and Differentiation. J. Am. Soc. Nephrol. JASN 2019, 30, 63–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Hu, M.; Lyu, X.; Li, C.; Thannickal, V.J.; Sanders, Y.Y. DNA methylation regulated gene expression in organ fibrosis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Osborn, J.W.; Tyshynsky, R.; Vulchanova, L. Function of Renal Nerves in Kidney Physiology and Pathophysiology. Annu. Rev. Physiol. 2021, 83, 429–450. [Google Scholar] [CrossRef]
- Kim, J.; Padanilam, B.J. Renal nerves drive interstitial fibrogenesis in obstructive nephropathy. J. Am. Soc. Nephrol. 2013, 24, 229–242. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Kirabo, A.; Wu, J.; Saleh, M.A.; Zhu, L.; Wang, F.; Takahashi, T.; Loperena, R.; Foss, J.D.; Mernaugh, R.L. Renal denervation prevents immune cell activation and renal inflammation in angiotensin II–induced hypertension. Circ. Res. 2015, 117, 547–557. [Google Scholar] [CrossRef]
- Luippold, G.; Beilharz, M.; Mühlbauer, B. Chronic renal denervation prevents glomerular hyperfiltration in diabetic rats. Nephrol. Dial. Transplant. 2004, 19, 342–347. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.; Fomison-Nurse, I.C.; Harrison, J.C.; Walker, R.J.; Davis, G.; Sammut, I.A. Chronic bilateral renal denervation attenuates renal injury in a transgenic rat model of diabetic nephropathy. Am. J. Physiol.-Ren. Physiol. 2014, 307, F251–F262. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Jiang, F.; Zhang, Z.; Wang, X.; Li, H.; Kuang, Y.; Yang, G. Catheter-Based Renal Denervation Attenuates Kidney Interstitial Fibrosis in a Canine Model of High-Fat Diet-Induced Hypertension. Kidney Blood Press. Res. 2019, 44, 628–642. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, C.J.; Liu, Y. Wnt Signaling in Kidney Development and Disease. Prog. Mol. Biol. Transl. Sci. 2018, 153, 181–207. [Google Scholar] [CrossRef]
- He, W.; Dai, C.; Li, Y.; Zeng, G.; Monga, S.P.; Liu, Y. Wnt/β-Catenin Signaling Promotes Renal Interstitial Fibrosis. J. Am. Soc. Nephrol. 2009, 20, 765–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.; He, X.; Cheng, R.; Zhang, B.; Zhang, R.R.; Chen, Y.; Takahashi, Y.; Murray, A.R.; Lee, K.; Gao, G.; et al. Implication of dysregulation of the canonical wingless-type MMTV integration site (WNT) pathway in diabetic nephropathy. Diabetologia 2012, 55, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, T.; Shiizaki, K.; Miura, Y.; Matsui, M.; Kosaki, K.; Mori, S.; Yamagata, K.; Maeda, S.; Kishi, T.; Usui, N.; et al. Increased fibroblast growth factor-21 in chronic kidney disease is a trade-off between survival benefit and blood pressure dysregulation. Sci. Rep. 2019, 9, 19247. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, H.; Matsushita, A.; Futaya, M.; Teraoka, A.; Akiyama, K.-i.; Usui, N.; Nagano, N.; Nitta, K.; Tsuchiya, K. Fibroblast growth factor 23 is upregulated in the kidney in a chronic kidney disease rat model. PLoS ONE 2018, 13, e0191706. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Chen, M.; Chen, M.; Ye, J.; Ying, Y.; Wu, Q.; Dou, H.; Bai, L.; Mao, F.; Ni, W.; et al. Fibroblast Growth Factor 22 Inhibits ER Stress-Induced Apoptosis and Improves Recovery of Spinal Cord Injury. Front. Pharmacol. 2020, 11, 18. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Fan, X.; Zhao, M.; Wu, M.; Li, H.; Ji, B.; Zhu, X.; Li, L.; Ding, H.; Sun, M.; et al. DNA Methylation-Reprogrammed Ang II (Angiotensin II) Type 1 Receptor-Early Growth Response Gene 1-Protein Kinase C ε Axis Underlies Vascular Hypercontractility in Antenatal Hypoxic Offspring. Hypertension 2021, 77, 491–506. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, L.T.; Larkin, B.P.; Wang, R.; Faiz, A.; Pollock, C.A.; Saad, S. Blood DNA Methylation Predicts Diabetic Kidney Disease Progression in High Fat Diet-Fed Mice. Nutrients 2022, 14, 785. https://doi.org/10.3390/nu14040785
Nguyen LT, Larkin BP, Wang R, Faiz A, Pollock CA, Saad S. Blood DNA Methylation Predicts Diabetic Kidney Disease Progression in High Fat Diet-Fed Mice. Nutrients. 2022; 14(4):785. https://doi.org/10.3390/nu14040785
Chicago/Turabian StyleNguyen, Long T., Benjamin P. Larkin, Rosy Wang, Alen Faiz, Carol A. Pollock, and Sonia Saad. 2022. "Blood DNA Methylation Predicts Diabetic Kidney Disease Progression in High Fat Diet-Fed Mice" Nutrients 14, no. 4: 785. https://doi.org/10.3390/nu14040785
APA StyleNguyen, L. T., Larkin, B. P., Wang, R., Faiz, A., Pollock, C. A., & Saad, S. (2022). Blood DNA Methylation Predicts Diabetic Kidney Disease Progression in High Fat Diet-Fed Mice. Nutrients, 14(4), 785. https://doi.org/10.3390/nu14040785





