Characteristics of Gut Microbiota in Small for Gestational Age Infants with Very Low Birth Weight
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Sample Collection, Microbial DNA Extraction, PCR Amplification, and Sequencing
2.3. Bioinformatics Analysis of Microbiota Composition
2.4. Statistical Analyses
3. Results
3.1. Demographic Characteristics
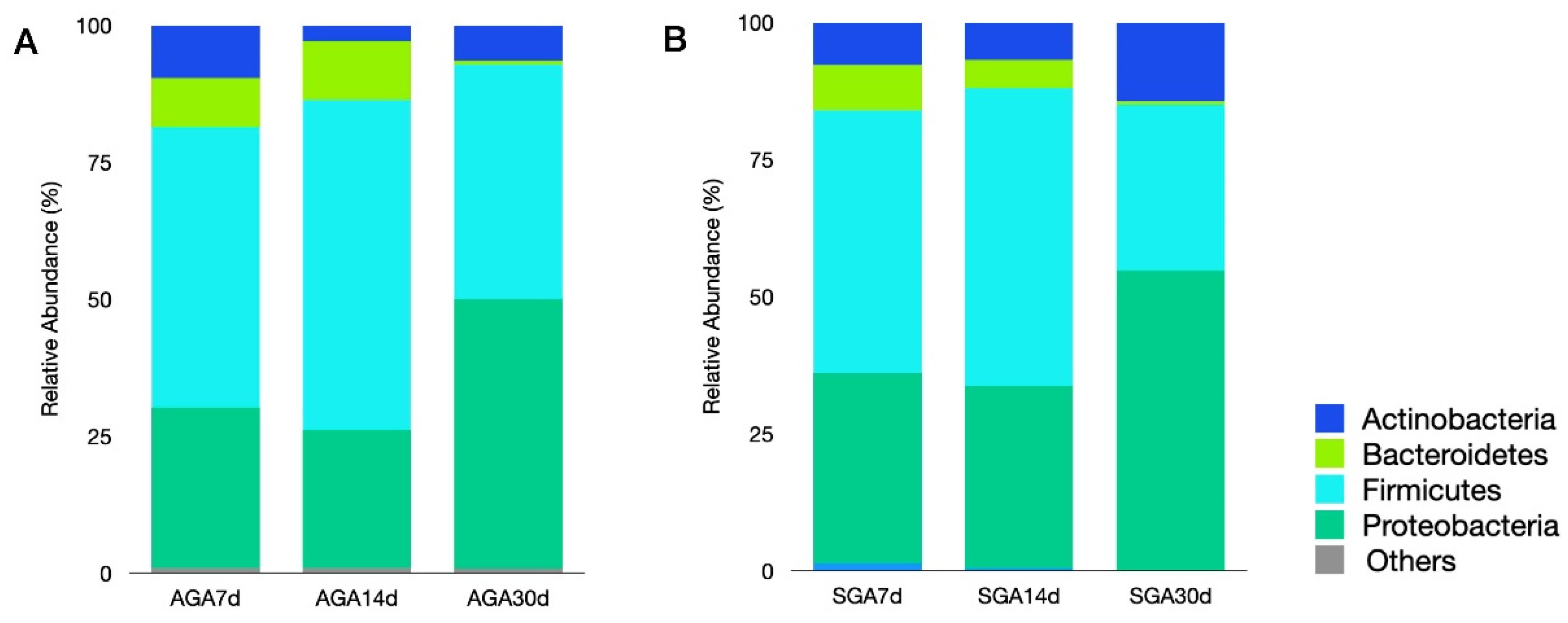
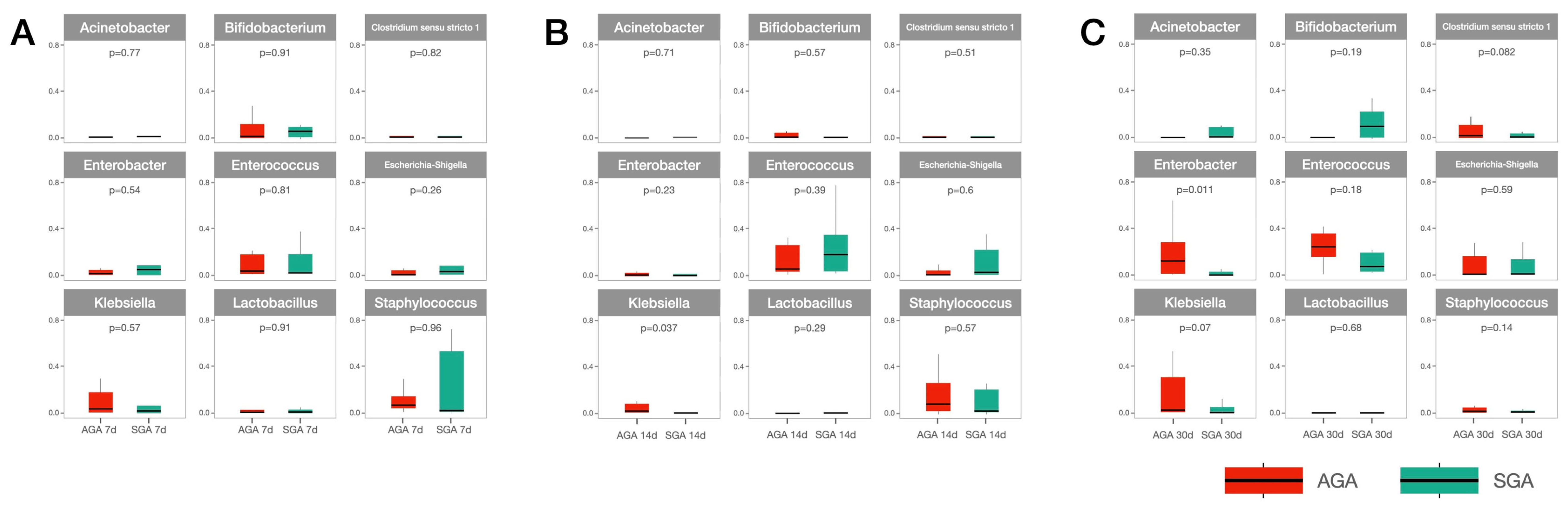
3.2. Analysis of Microbiota Composition
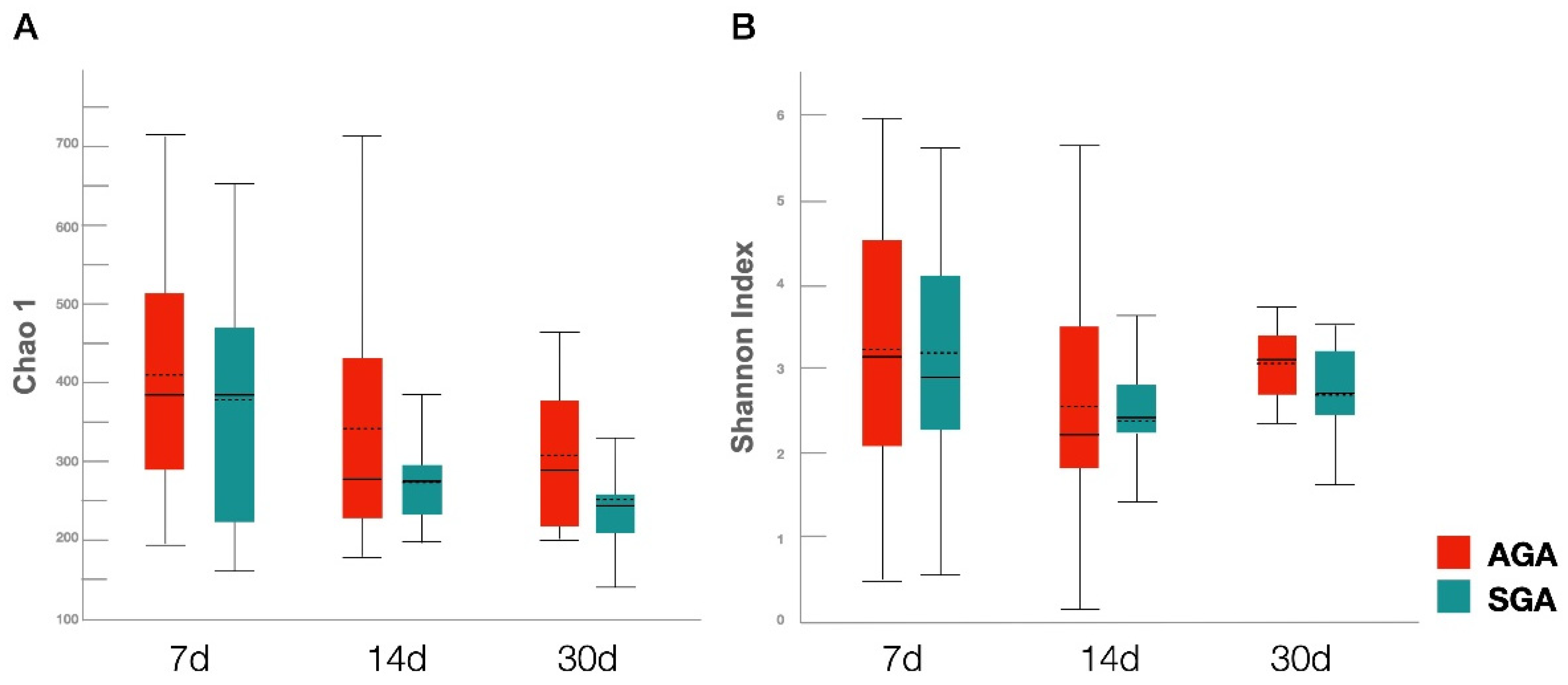
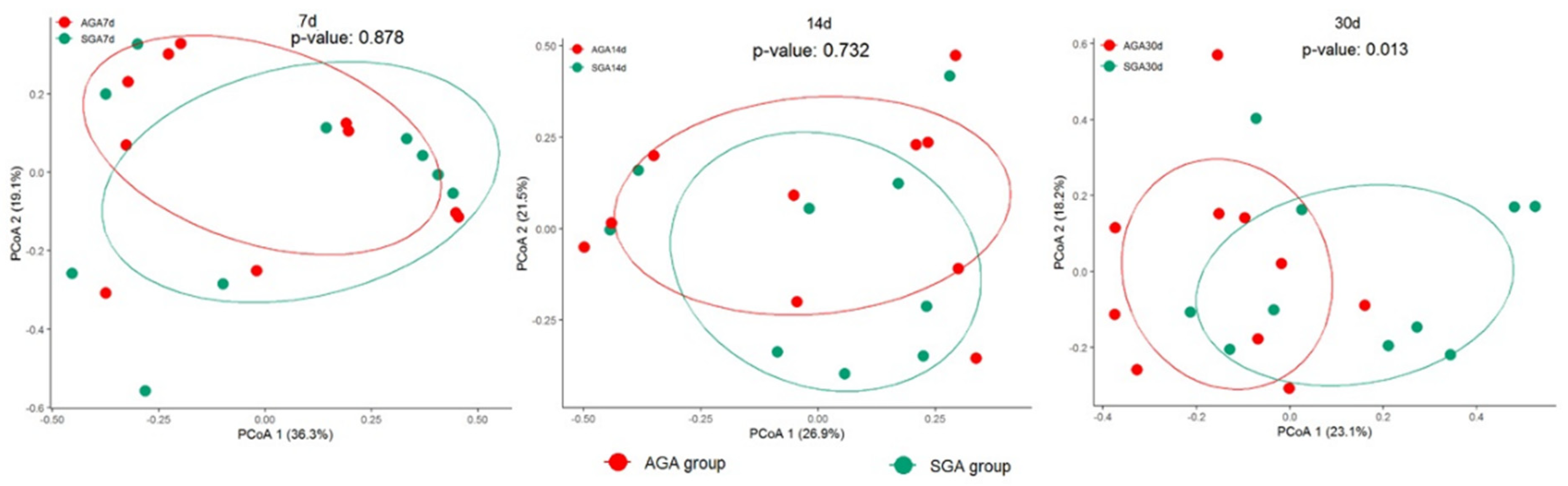
3.3. Diversity Analysis of Gut Microbiota
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Onis, M.; Habicht, J.P. Anthropometric reference data for international use: Recommendations from a World Health Organization Expert Committee. Am. J. Clin. Nutr. 1996, 64, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Shastri, S.; Sharma, P. Intrauterine Growth Restriction: Antenatal and Postnatal Aspects. Clin. Med. Insights. Pediatr. 2016, 10, 67–83. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, H.; Sun, X.; Li, G. Risk factors and complications of small for gestational age. Pak. J. Med. Sci. 2019, 35, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Tsai, L.-Y.; Chen, Y.-L.; Tsou, K.-I.; Mu, S.-C. Taiwan Premature Infant Developmental Collaborative Study, G. The Impact of Small-for-gestational-age on Neonatal Outcome Among Very-low-birth-weight Infants. Pediatr. Neonatol. 2015, 56, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.A.; Foglia, E.E.; Dysart, K.C.; Simmons, R.A.; Aghai, Z.H.; Cook, A.; Greenspan, J.S.; DeMauro, S.B. Adverse effects of small for gestational age differ by gestational week among very preterm infants. Arch. Dis. Child.-Fetal Neonatal Ed. 2018, 104, F192–F198. [Google Scholar] [CrossRef] [PubMed]
- Karagianni, P.; Kyriakidou, M.; Mitsiakos, G.; Chatzioanidis, H.; Koumbaras, E.; Evangeliou, A.; Nikolaides, N. Neurological Outcome in Preterm Small for Gestational Age Infants Compared to Appropriate for Gestational Age Preterm at the Age of 18 Months: A Prospective Study. J. Child Neurol. 2009, 25, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.H.; Chung, S. Small for gestational age and obesity related comorbidities. Ann. Pediatr. Endocrinol. Metab. 2018, 23, 4–8. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Ye, Y.; Wang, F.; Chen, F.; Zheng, C. The Role of Microbiota in Infant Health: From Early Life to Adulthood. Front. Immunol. 2021, 12, 708472. [Google Scholar] [CrossRef]
- Groer, M.W.; Gregory, K.E.; Louis-Jacques, A.; Thibeau, S.; Walker, W.A. The very low birth weight infant microbiome and childhood health. Birth Defects Res. Part. C Embryo Today Rev. 2015, 105, 252–264. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Chiau, J.-S.C.; Ho, Y.-H.; Chang, J.-H.; Tsai, K.-N.; Liu, C.-Y.; Hsu, C.-H.; Lin, C.-Y.; Ko, M.H.-J.; Lee, H.-C. Impact of Early Empiric Antibiotic Regimens on the Gut Microbiota in Very Low Birth Weight Preterm Infants: An Observational Study. Front. Pediatr. 2021, 9, 651713. [Google Scholar] [CrossRef]
- Sarkar, A.; Yoo, J.Y.; Valeria Ozorio Dutra, S.; Morgan, K.H.; Groer, M. The Association between Early-Life Gut Microbiota and Long-Term Health and Diseases. J. Clin. Med. 2021, 10, 459. [Google Scholar] [CrossRef]
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The Human Microbiome and Child Growth—First 1000 Days and Beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef]
- Hoffman, D.J.; Ponce, M.C.; Taddei, C.; Doak, C.M. Microbiome, growth retardation and metabolism: Are they related? Ann. Hum. Biol. 2017, 44, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.A.; Sohn, K. The Microbiota of the Extremely Preterm Infant. Clin. Perinatol. 2017, 44, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Groer, M.W.; A Luciano, A.; Dishaw, L.J.; Ashmeade, T.L.; Miller, E.; Gilbert, J.A. Development of the preterm infant gut microbiome: A research priority. Microbiome 2014, 2, 38. [Google Scholar] [CrossRef]
- Li, N.; Huang, S.; Jiang, L.; Wang, W.; Li, T.; Zuo, B.; Li, Z.; Wang, J. Differences in the Gut Microbiota Establishment and Metabolome Characteristics Between Low-and Normal-Birth-Weight Piglets During Early-Life. Front. Microbiol. 2018, 9, 1798. [Google Scholar] [CrossRef] [PubMed]
- Wiese, M.; Hui, Y.; Nielsen, D.S.; Williams, A.R.; Lynegaard, J.C.; Weber, N.R.; Amdi, C. Color of Colon Content of Normal and Intrauterine Growth-Restricted Weaned Piglets Is Associated with Specific Microbial Taxa and Physiological Parameters. Animals 2020, 10, 1073. [Google Scholar] [CrossRef]
- Zhang, W.; Ma, C.; Xie, P.; Zhu, Q.; Wang, X.; Yin, Y.; Kong, X. Gut microbiota of newborn piglets with intrauterine growth restriction have lower diversity and different taxonomic abundances. J. Appl. Microbiol. 2019, 127, 354–369. [Google Scholar] [CrossRef]
- Jiang, L.; Feng, C.; Tao, S.; Li, N.; Zuo, B.; Han, D.; Wang, J. Maternal imprinting of the neonatal microbiota colonization in intrauterine growth restricted piglets: A review. J. Anim. Sci. Biotechnol. 2019, 10, 88. [Google Scholar] [CrossRef]
- Fança-Berthon, P.; Hoebler, C.; Mouzet, E.; David, A.; Michel, C. Intrauterine Growth Restriction Not Only Modifies the Cecocolonic Microbiota in Neonatal Rats But Also Affects Its Activity in Young Adult Rats. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 402–413. [Google Scholar] [CrossRef]
- Nutricionist, A.C.T.; Procianoy, R.S.; Roesch, L.F.W.; Corso, A.L.; Dobbler, P.T.; Silveira, R.C. Meconium microbiome and its relation to neonatal growth and head circumference catch-up in preterm infants. PLoS ONE 2020, 15, e0238632. [Google Scholar] [CrossRef]
- Li, H.; He, Z.; Gao, D.; Lv, Y.; Zhou, Q.; Xiao, B.; Huang, W. Characteristics of the Intestinal Microbiota in Very Low Birth Weight Infants with Extrauterine Growth Restriction. Front. Pediatr. 2019, 7, 99. [Google Scholar] [CrossRef]
- Hu, J.; Benny, P.; Wang, M.; Ma, Y.; Lambertini, L.; Peter, I.; Xu, Y.; Lee, M.-J. Intrauterine Growth Restriction Is Associated with Unique Features of the Reproductive Microbiome. Reprod. Sci. 2020, 28, 828–837. [Google Scholar] [CrossRef]
- Hsieh, W.-S.; Wu, H.-C.; Jeng, S.-F.; Liao, H.-F.; Su, Y.-N.; Lin, S.-J.; Hsieh, C.-J.; Chen, P.-C. Nationwide singleton birth weight percentiles by gestational age in Taiwan, 1998–2002. Acta. Paediatr. Taiwan 2006, 47, 25–33. [Google Scholar] [PubMed]
- Yeung, C.-Y.; Chiang Chiau, J.S.; Cheng, M.L.; Chan, W.T.; Chang, S.W.; Chang, Y.H.; Jiang, C.B.; Lee, H.C. Modulations of probiotics on gut microbiota in a 5-fluorouracil-induced mouse model of mucositis. J. Gastroenterol. Hepatol. 2019, 35, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Di Simone, N.; Santamaria Ortiz, A.; Specchia, M.; Tersigni, C.; Villa, P.; Gasbarrini, A.; Scambia, G.; D’Ippolito, S. Recent Insights on the Maternal Microbiota: Impact on Pregnancy Outcomes. Front. Immunol. 2020, 11, 528202. [Google Scholar] [CrossRef] [PubMed]
- Nogacka, A.; Salazar, N.; Suárez, M.; Milani, C.; Arboleya, S.; Solís, G.; Fernández, N.; Alaez, L.; Hernández-Barranco, A.M.; de Los Reyes-Gavilan, C.; et al. Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. Microbiome 2017, 5, 93. [Google Scholar] [CrossRef]
- Zou, Z.-H.; Liu, D.; Li, H.-D.; Zhu, D.-P.; He, Y.; Hou, T.; Yu, J.-L. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 9. [Google Scholar] [CrossRef]
- Aloisio, I.; Quagliariello, A.; De Fanti, S.; Luiselli, D.; De Filippo, C.; Albanese, D.; Corvaglia, L.T.; Faldella, G.; Di Gioia, D. Evaluation of the effects of intrapartum antibiotic prophylaxis on newborn intestinal microbiota using a sequencing approach targeted to multi hypervariable 16S rDNA regions. Appl. Microbiol. Biotechnol. 2016, 100, 5537–5546. [Google Scholar] [CrossRef]
- Arboleya, S.; Sánchez, B.; Milani, C.; Duranti, S.; Solís, G.; Fernández, N.; de Los Reyes-Gavilan, C.G.; Ventura, M.; Margolles, A.; Gueimonde, M. Intestinal Microbiota Development in Preterm Neonates and Effect of Perinatal Antibiotics. J. Pediatr. 2015, 166, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Sánchez, B.; Solís, G.; Fernández, N.; Suárez, M.; Hernández-Barranco, A.M.; Milani, C.; Margolles, A.; de los Reyes-Gavilán, C.G.; Ventura, M.; et al. Impact of Prematurity and Perinatal Antibiotics on the Developing Intestinal Microbiota: A Functional Inference Study. Int. J. Mol. Sci. 2016, 17, 649. [Google Scholar] [CrossRef]
- Klopp, J.; Ferretti, P.; Meyer, C.U.; Hilbert, K.; Haiß, A.; Marißen, J.; Henneke, P.; Hudalla, H.; Pirr, S.; Viemann, D.; et al. Meconium Microbiome of Very Preterm Infants across Germany. mSphere 2022, 7, e0080821. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The Placenta Harbors a Unique Microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef]
- Barrett, E.; Kerr, C.; Murphy, K.; O’Sullivan, O.; Ryan, C.A.; Dempsey, E.M.; Murphy, B.P.; O’Toole, P.W.; Cotter, P.D.; Fitzgerald, G.F.; et al. The individual-specific and diverse nature of the preterm infant microbiota. Arch. Dis. Child.-Fetal Neonatal Ed. 2013, 98, F334–F340. [Google Scholar] [CrossRef]
- Conge, G.; Gouache, P.; Joyeux, Y.; Goichot, J.; Fournier, J. Influence of different types of experimental obesity on resistance of the mouse to infection by Salmonella typhimurium and Klebsiella pneumoniae. Ann. Nutr. Metab. 1988, 32, 113–120. [Google Scholar] [CrossRef]
- Keskitalo, A.; Munukka, E.; Toivonen, R.; Hollmén, M.; Kainulainen, H.; Huovinen, P.; Jalkanen, S.; Pekkala, S. Enterobacter cloacae administration induces hepatic damage and subcutaneous fat accumulation in high-fat diet fed mice. PLoS ONE 2018, 13, e0198262. [Google Scholar] [CrossRef] [PubMed]
- Ardissone, A.N.; De La Cruz, D.M.; Davis-Richardson, A.G.; Rechcigl, K.T.; Li, N.; Drew, J.C.; Murgas-Torrazza, R.; Sharma, R.; Hudak, M.L.; Triplett, E.W.; et al. Meconium Microbiome Analysis Identifies Bacteria Correlated with Premature Birth. PLoS ONE 2014, 9, e90784. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-J.; Yeung, C.-Y. Recent advance in infant nutrition: Human milk oligosaccharides. Pediatr. Neonatol. 2021, 62, 347–353. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Chen, J.-H.; Chang, J.-H.; Lin, H.-C.; Lin, C.-Y.; Peng, C.-C. Multiple strains probiotics appear to be the most effective probiotics in the prevention of necrotizing enterocolitis and mortality: An updated meta-analysis. PLoS ONE 2017, 12, e0171579. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Su, W.; Ying, Z.; Chen, Y.; Zhang, L.; Lu, Z.; Wang, T. Effects of dietary Bacillus amyloliquefaciens supplementation on growth performance, intestinal morphology, inflammatory response, and microbiota of intra-uterine growth retarded weanling piglets. J. Anim. Sci. Biotechnol. 2018, 9, 22. [Google Scholar] [CrossRef]
- Cong, X.; Xu, W.; Janton, S.; Henderson, W.A.; Matson, A.; McGrath, J.M.; Maas, K.; Graf, J. Gut Microbiome Developmental Patterns in Early Life of Preterm Infants: Impacts of Feeding and Gender. PLoS ONE 2016, 11, e0152751. [Google Scholar] [CrossRef]
- Stewart, C.J.; Embleton, N.D.; Clements, E.; Luna, P.N.; Smith, D.P.; Fofanova, T.Y.; Nelson, A.; Taylor, G.; Orr, C.H.; Petrosino, J.F.; et al. Cesarean or Vaginal Birth Does Not Impact the Longitudinal Development of the Gut Microbiome in a Cohort of Exclusively Preterm Infants. Front. Microbiol. 2017, 8, 1008. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Zwittink, R.D.; Renes, I.B.; A van Lingen, R.; van Zoeren-Grobben, D.; Konstanti, P.; Norbruis, O.F.; Martin, R.; Jebbink, L.J.M.G.; Knol, J.; Belzer, C. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 475–483. [Google Scholar] [CrossRef]
- Esaiassen, E.; Hjerde, E.; Cavanagh, J.P.; Pedersen, T.; Andresen, J.H.; Rettedal, S.I.; Støen, R.; Nakstad, B.; Willassen, N.P.; Klingenberg, C. Effects of Probiotic Supplementation on the Gut Microbiota and Antibiotic Resistome Development in Preterm Infants. Front. Pediatr. 2018, 6, 347. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Li, D.; Yin, Y.; Wang, X.; Li, P.; Dangott, L.J.; Hu, W.; Wu, G. Intrauterine Growth Restriction Affects the Proteomes of the Small Intestine, Liver, and Skeletal Muscle in Newborn Pigs. J. Nutr. 2008, 138, 60–66. [Google Scholar] [CrossRef]
- Santos, T.G.; Fernandes, S.D.; de Oliveira Araujo, S.B.; Felicioni, F.; de Merici Domingues, E.P.T.; Caldeira-Brant, A.L.; Ferreira, S.V.; de Paula Naves, L.; de Souza, S.P.; Campos, P.; et al. Intrauterine growth restriction and its impact on intestinal morphophysiology throughout postnatal development in pigs. Sci. Rep. 2022, 12, 11810. [Google Scholar] [CrossRef] [PubMed]
- Ishimwe, J.A. Maternal microbiome in preeclampsia pathophysiology and implications on offspring health. Physiol. Rep. 2021, 9, e14875. [Google Scholar] [CrossRef]
- Gregory, K.E.; Samuel, B.S.; Houghteling, P.; Shan, G.; Ausubel, F.M.; Sadreyev, R.I.; Walker, W.A. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 2016, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-C.; Sun, Q.; Ji, Y.-C.; Fu, L.-Z.; Wang, Z.-L.; He, Y.; Li, L.-Q. Differences in the Gut Microbiota Composition and Metabolites Associated with Feeding Intolerance in VLBW Infants with a Gestational Age of ≤ 30 Weeks: A Pilot Study. Front. Cell. Infect. Microbiol. 2022, 12, 726322. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ao, D.; Cai, X.; Huang, P.; Cai, N.; Lin, S.; Wu, B. Early gut microbiota in very low and extremely low birth weight preterm infants with feeding intolerance: A prospective case-control study. J. Microbiol. 2022, 60, 1021–1031. [Google Scholar] [CrossRef]
- Hu, X.; Chang, Y.; Wang, Z.; Bao, W.; Li, Z. Altered gut microbiota is associated with feeding intolerance in preterm infants. Turk. J. Pediatr. 2021, 63, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Camblor, P.M.; Solís, G.; Suárez, M.; Fernández, N.; de Los Reyes-Gavilan, C.D.L.; Gueimonde, M. Intestinal Microbiota and Weight-Gain in Preterm Neonates. Front. Microbiol. 2017, 8, 183. [Google Scholar] [CrossRef] [PubMed]
| SGA Group a (n = 10) | AGA Group a (n = 10) | p Value | |
|---|---|---|---|
| Maternal age a | 35 (18–39) | 32 (25–45) | 0.690 |
| Prenatal steroid use | 10 | 9 | 0.343 |
| Prenatal antibiotics use | 3 | 5 | 0.388 |
| Preeclampsia | 7 | 1 | 0.005 |
| Cesarean delivery | 7 | 9 | 0.290 |
| GA (week) a | 29.5 (27–32) | 29.0 (27–32) | 0.235 |
| BW (g) a | 1119 (734–1361) | 1270 (1120–1465) | 0.034 |
| Male | 3 | 6 | 0.196 |
| APGAR score at 5 min a | 9 (7–10) | 9 (7–10) | 1.000 |
| Reach full feeding (d) a | 21.0 (9.0–44) | 17.5 (9–21) | 0.709 |
| RDS needs surfactant | 0 | 3 | 0.081 |
| CLD | 3 | 3 | 1.000 |
| ROP needs treatment | 0 | 1 | 0.343 |
| Total hospitalization days a | 48.5 (36–77) | 51.5 (36–98) | 0.715 |
| GA at discharge a | 38.0 (36–43) | 37.0 (34–44) | 0.488 |
| Weight at discharge a | 2380 (2260–2582) | 2408 (2294–4184) | 0.109 |
| Weight less than 10th percentile at discharge | 9 | 0 | 0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.-Y.; Chiang Chiau, J.-S.; Chang, J.-H.; Hsu, C.-H.; Lin, C.-Y.; Ko, M.H.-J.; Lee, H.-C. Characteristics of Gut Microbiota in Small for Gestational Age Infants with Very Low Birth Weight. Nutrients 2022, 14, 5158. https://doi.org/10.3390/nu14235158
Chang H-Y, Chiang Chiau J-S, Chang J-H, Hsu C-H, Lin C-Y, Ko MH-J, Lee H-C. Characteristics of Gut Microbiota in Small for Gestational Age Infants with Very Low Birth Weight. Nutrients. 2022; 14(23):5158. https://doi.org/10.3390/nu14235158
Chicago/Turabian StyleChang, Hung-Yang, Jen-Shiu Chiang Chiau, Jui-Hsing Chang, Chyong-Hsin Hsu, Chia-Ying Lin, Mary Hsin-Ju Ko, and Hung-Chang Lee. 2022. "Characteristics of Gut Microbiota in Small for Gestational Age Infants with Very Low Birth Weight" Nutrients 14, no. 23: 5158. https://doi.org/10.3390/nu14235158
APA StyleChang, H.-Y., Chiang Chiau, J.-S., Chang, J.-H., Hsu, C.-H., Lin, C.-Y., Ko, M. H.-J., & Lee, H.-C. (2022). Characteristics of Gut Microbiota in Small for Gestational Age Infants with Very Low Birth Weight. Nutrients, 14(23), 5158. https://doi.org/10.3390/nu14235158






