Long Noncoding RNA, MicroRNA, Zn Transporter Zip14 (Slc39a14) and Inflammation in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Hematoxylin and Eosin Staining (H&E)
2.3. Tissue Preparation and RNA Isolation
2.4. RNA-Sequencing
2.5. Quantitative PCR
2.6. Chromatin Immunoprecipitation (ChIP) Assays
2.7. Preparation and Treatment of Intestinal Organoids (Enteroids)
2.8. Statistics
3. Results
3.1. Influence of WB-Zip14 Ablation on Differential Expression of Specific lncRNA Genes in Proximal Small Intestine
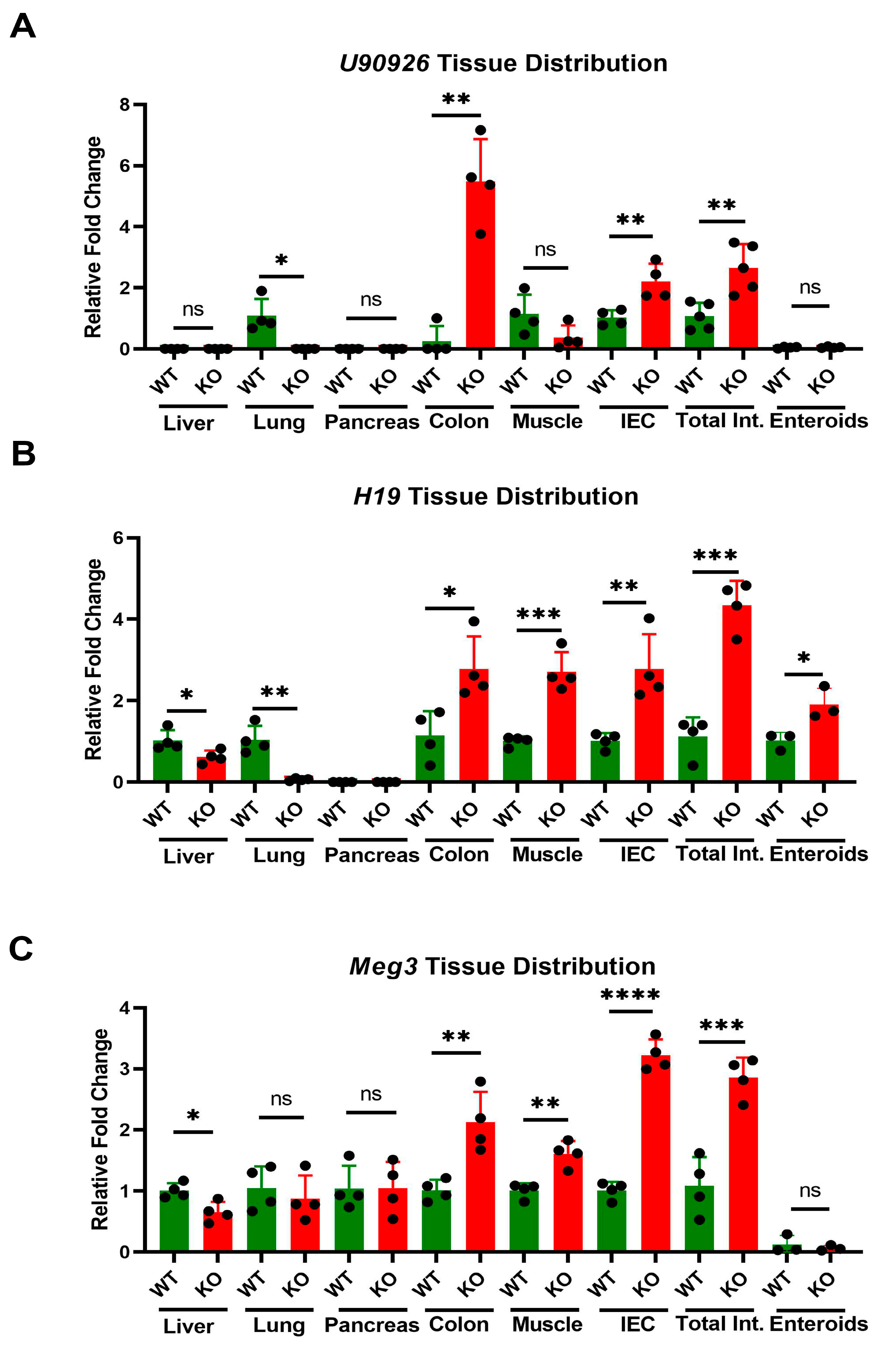
3.2. Tissue Specific Expression of lncRNAs in Mice with WB-Zip14 Deletion
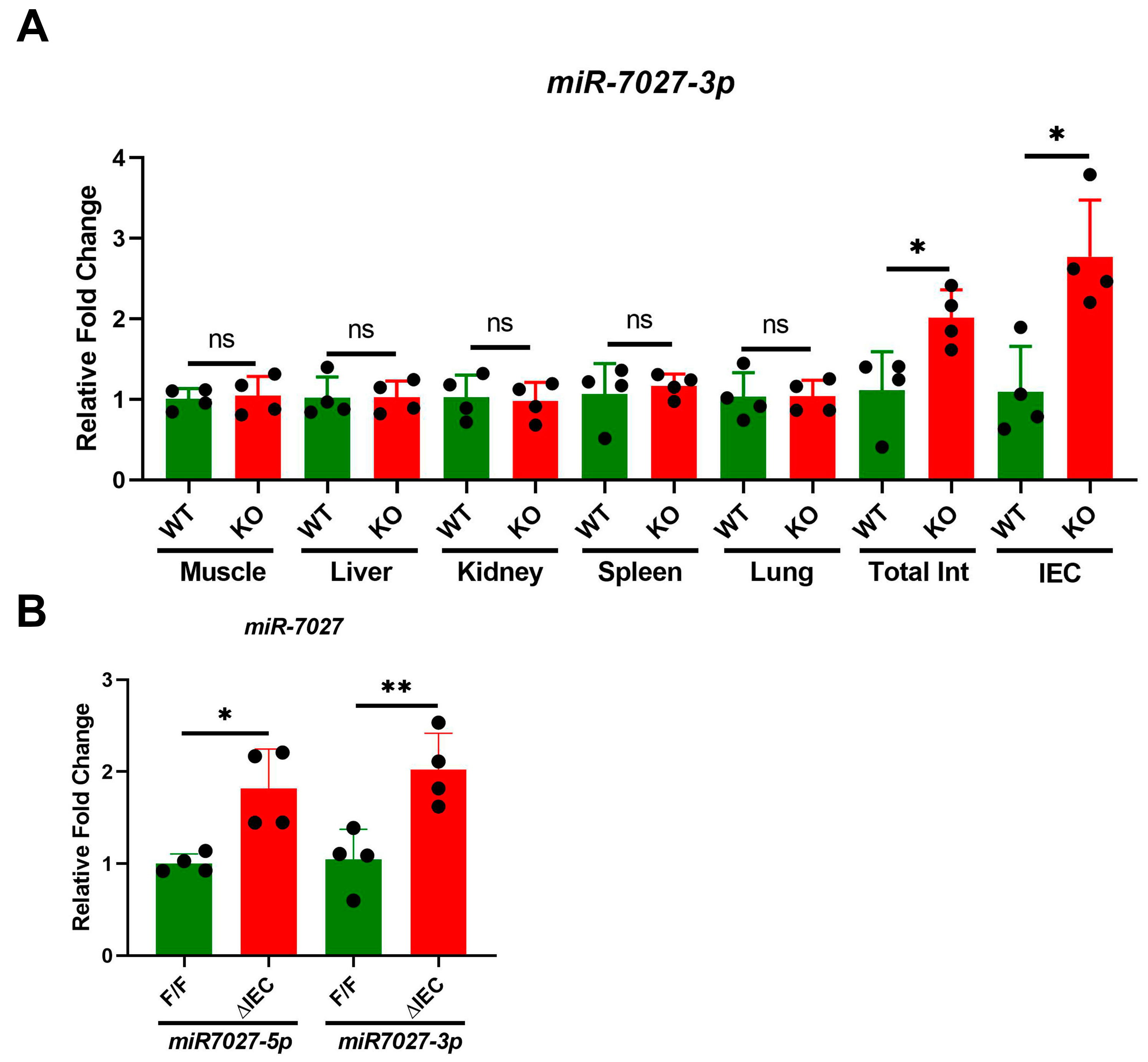
3.3. Tissue Specific Expression of miR-7027
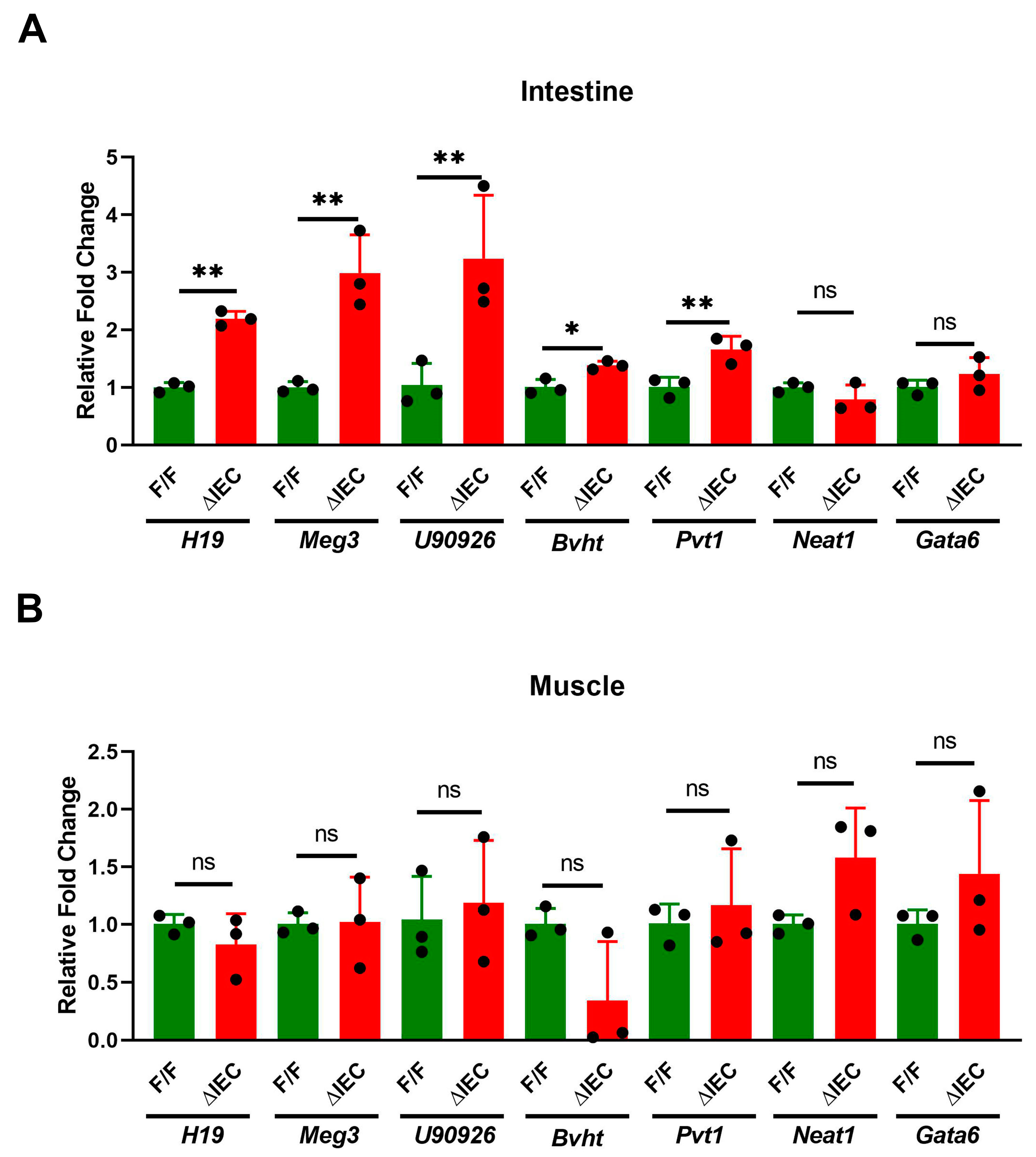
3.4. Tissue Specific Expression of lncRNAs in Mice with Zip14ΔIEC Deletion
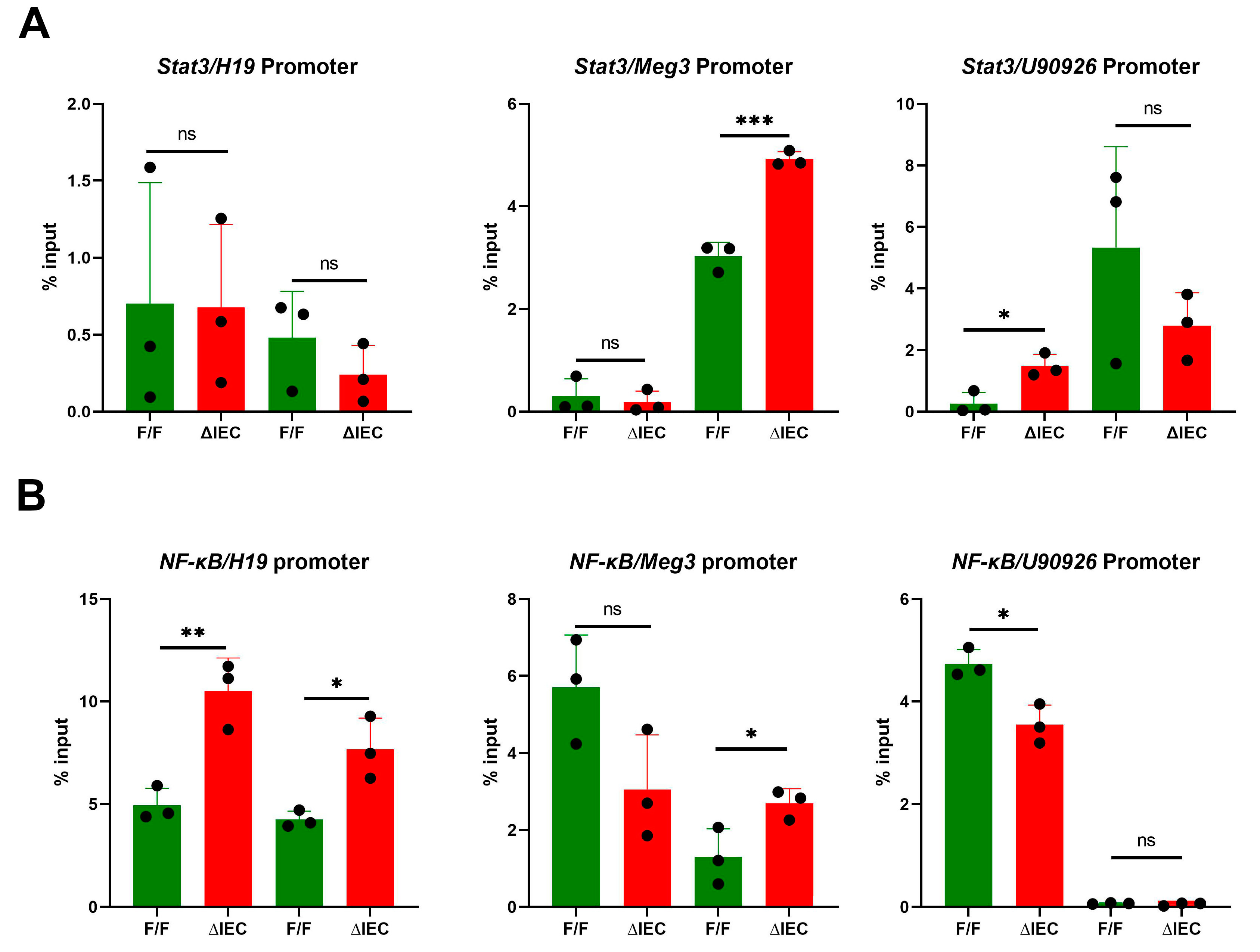
3.5. Promoter Occupancy May Explain Differential Expression of H19, Meg3 and U90926 with Zip14 Ablation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Runtsch, M.C.; Round, J.L.; O’Connell, R.M. MicroRNAs and the regulation of intestinal homeostasis. Front. Genet. 2014, 5, 347. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.C.; Mah, A.T.; Pitman, W.A.; Ding, S.; Lund, P.K.; Sethupathy, P. Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J. Biol. Chem. 2017, 292, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, M.; Vujic Spasić, M.; Altamura, S.; Elmén, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmüller, U.; et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Ross, A.C.; Caballero, B.H.; Cousins, R.J.; Tucker, K.L.; Ziegler, T.R. Modern Nutrition in Health and Disease; Lippincott Williams & Wilkins: Baltimore, MD, USA, 2014; pp. 189–205. [Google Scholar]
- Kang, Y.; Park, H.; Choe, B.-H.; Kang, B. The role and function of mucins and its relationship to inflammatory bowel disease. Front. Med. 2022, 9, 848344. [Google Scholar] [CrossRef]
- Yusuf, A.; Abubakar, M.; Malami, I.; Ibrahim, K.; Abubakar, B.; Bello, M.; Qusty, N.; Elazab, S.; Imam, M.; Alexiou, A.; et al. Zinc metalloproteins in epigenetics and their crosstalk. Life 2021, 11, 186. [Google Scholar] [CrossRef]
- Ryu, M.-S.; Langkamp-Henken, B.; Chang, S.-M.; Shankar, M.N.; Cousins, R.J. Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc. Natl. Acad. Sci. USA 2011, 108, 20970–20975. [Google Scholar] [CrossRef]
- Hlavna, M.; Raudenska, M.; Hudcova, K.; Gumulec, J.; Sztalmachova, M.; Tanhäuserova, V.; Babula, P.; Adam, V.; Eckschlager, T.; Kizek, R.; et al. MicroRNAs and zinc metabolism-related gene expression in prostate cancer cell lines treated with zinc(II) ions. Int. J. Oncol. 2012, 41, 2237–2244. [Google Scholar] [CrossRef]
- Qiu, Y.; Gao, Y.; Yu, D.; Zhong, L.; Cai, W.; Ji, J.; Geng, F.; Tang, G.; Zhang, H.; Cao, J.; et al. Genome-wide analysis reveals zinc transporter ZIP9 regulated by DNA methylation promotes radiation-induced skin fibrosis via the TGF-Β signaling pathway. J. Investig. Dermatol. 2020, 140, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bai, J.; Si, W.; Yuan, S.; Li, Y.; Chen, X. SLC39A7, regulated by miR-139-5p, induces cell proliferation, migration and inhibits apoptosis in gastric cancer via Akt/mTOR signaling pathway. Biosci. Rep. 2020, 40, BSR20200041. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Pan, R.; Weaver, J.; Jia, M.; Yang, X.; Yang, T.; Liang, J.; Liu, K.J. MicroRNA-30a regulates acute cerebral ischemia-induced blood-brain barrier damage through ZnT4/zinc pathway. J. Cereb. Blood Flow Metab. 2021, 41, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, G.; Borra, V.M.; Steenackers, E.; Yorgan, T.A.; Hermans, C.; Boudin, E.; Waterval, J.J.; Jansen, I.D.C.; Aydemir, T.B.; Kamerling, N.; et al. Conditional mouse models support the role of SLC39A14 (ZIP14) in hyperostosis cranialis interna and in bone homeostasis. PLoS Genet. 2018, 14, e1007321-e21. [Google Scholar] [CrossRef]
- Tuschl, K.; Meyer, E.; Valdivia, L.E.; Zhao, N.; Dadswell, C.; Abdul-Sada, A.; Hung, C.Y.; Simpson, M.; Chong, W.K.; Jacques, T.S.; et al. Mutations in SLC39A14 disrupt manganese homeostasis and cause childhood-onset parkinsonism-dystonia. Nat. Commun. 2016, 7, 11601. [Google Scholar] [CrossRef]
- Kim, J.; Aydemir, T.B.; Jimenez-Rondan, F.R.; Ruggiero, C.H.; Kim, M.-H.; Cousins, R.J. Deletion of metal transporter Zip14 (slc39a14) produces skeletal muscle wasting, endotoxemia, mef2c activation and induction of miR-675 and Hspb7. Sci. Rep. 2020, 10, 4050. [Google Scholar] [CrossRef]
- Aydemir, T.B.; Chang, S.-M.; Guthrie, G.J.; Maki, A.B.; Ryu, M.-S.; Karabiyik, A.; Cousins, R.J. Zinc transporter ZIP14 functions in hepatic zinc, iron and glucose homeostasis during the innate immune response (endotoxemia). PLoS ONE 2012, 7, e48679. [Google Scholar] [CrossRef]
- Aydemir, T.B.; Thorn, T.L.; Ruggiero, C.H.; Pompilus, M.; Febo, M.; Cousins, R.J. Intestine-specific deletion of metal transporter Zip14 (slc39a14) causes brain manganese overload and locomotor defects of manganism. Am. J. Physiol. Liver Physiol. 2020, 318, G673–G681. [Google Scholar] [CrossRef]
- Zeineldin, M.; Neufeld, K. Isolation of epithelial cells from mouse gastrointestinal tract for western blot or RNA analysis. Bio-Protoc. 2012, 2, e292. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing (R version 4.2.2); R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Jimenez, F.R.; Ruggiero, C.H.; Cousins, R.J. Intestinal Zip14 ablation dysregulates tight junction protein expression, inflammatory genes and lncrnas in intestinal epithelial cells and enteroids. FASEB J. 2022, 36. [Google Scholar] [CrossRef]
- Guthrie, G.J.; Aydemir, T.B.; Troche, C.; Martin, A.B.; Chang, S.-M.; Cousins, R.J. Influence of Zip14 (slc39a14) on intestinal zinc processing and barrier function. Am. J. Physiol. Liver Physiol. 2015, 308, G171–G178. [Google Scholar] [CrossRef] [PubMed]
- Aydemir, T.B.; Cousins, R.J. The multiple faces of the metal transporter ZIP14 (slc39a14). J. Nutr. 2018, 148, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Pfeifer, K.; Dutta, A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Landecker, H. Food as exposure: Nutritional epigenetics and the new metabolism. Biosocieties 2011, 6, 167–194. [Google Scholar] [CrossRef] [PubMed]
- Olechnowicz, J.; Tinkov, A.; Skalny, A.; Suliburska, J. Zinc status is associated with inflammation, oxidative stress, lipid, and glucose metabolism. J. Physiol. Sci. 2018, 68, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ahmad, A.; Azmi, A.; Li, Y.; Prasad, A.; Sarkar, F.H. Chapter 2—The biological significance of zinc in inflammation and aging. In Inflammation, Advancing Age and Nutrition; Rahman, I., Bagchi, D., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 15–27. [Google Scholar]
- Wan, Y.; Zhang, B. The impact of zinc and zinc homeostasis on the intestinal mucosal barrier and intestinal diseases. Biomolecules 2022, 12, 900. [Google Scholar] [CrossRef]
- Rankin, C.R.; Theodorou, E.; Law, I.K.M.; Rowe, L.; Kokkotou, E.; Pekow, J.; Wang, J.; Martin, M.G.; Pothoulakis, C.; Padua, D.M. Identification of novel mRNAs and lncRNAs associated with mouse experimental colitis and human inflammatory bowel disease. Am. J. Physiol. Liver Physiol. 2018, 315, G722–G733. [Google Scholar] [CrossRef]
- Liu, H.; Li, T.; Zhong, S.; Yu, M.; Huang, W. Intestinal epithelial cells related lncRNA and mRNA expression profiles in dextran sulphate sodium-induced colitis. J. Cell. Mol. Med. 2021, 25, 1060–1073. [Google Scholar] [CrossRef]
- Lin, L.; Zhou, G.; Chen, P.; Wang, Y.; Han, J.; Chen, M.; He, Y.; Zhang, S. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020, 11, 456. [Google Scholar] [CrossRef]
- Fong, L.Y.; Taccioli, C.; Jing, R.; Smalley, K.J.; Alder, H.; Jiang, Y.; Fadda, P.; Farber, J.L.; Croce, C.M. MicroRNA dysregulation and esophageal cancer development depend on the extent of zinc dietary deficiency. Oncotarget 2016, 7, 10723–10738. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, J.; Kim, D.; Lee, C.H.; Lee, M.S.; Chun, C.-H.; Jin, E.-J. MicroRNA-488 regulates zinc transporter SLC39A8/ZIP8 during pathogenesis of osteoarthritis. J. Biomed. Sci. 2013, 20, 31. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Aydemir, F.; Nam, H.; Knutson, M.D.; Cousins, R.J. Cousins. Zip14 (slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13612–13617. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, R.; Engers, J.; Abdulqadir, R. Talk about micromanaging! Role of microRNAs in intestinal barrier function. Am. J. Physiol. Liver Physiol. 2020, 319, G170–G174. [Google Scholar] [CrossRef]
- Ohsaka, F.; Sonoyama, K. Murine intestinal organoids resemble intestinal epithelium in their microRNA profiles. Biosci. Biotechnol. Biochem. 2018, 82, 1560–1567. [Google Scholar] [CrossRef]
- Sun, L.; Xia, R.; Jiang, J.; Wen, T.; Huang, Z.; Qian, R.; Zhang, M.-D.; Zhou, M.; Peng, C. MicroRNA-96 is required to prevent allodynia by repressing voltage-gated sodium channels in spinal cord. Prog. Neurobiol. 2021, 202, 102024. [Google Scholar] [CrossRef]
- Geng, H.; Bu, H.-F.; Liu, F.; Wu, L.; Pfeifer, K.; Chou, P.M.; Wang, X.; Sun, J.; Lu, L.; Pandey, A.; et al. In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology 2018, 155, 144–155. [Google Scholar] [CrossRef]
- Zou, T.; Jaladanki, S.K.; Liu, L.; Xiao, L.; Chung, H.K.; Wang, J.-Y.; Xu, Y.; Gorospe, M.; Wang, J.-Y. H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein hur. Mol. Cell. Biol. 2016, 36, 1332–1341. [Google Scholar] [CrossRef]
- Du, X.; Tian, D.; Wei, J.; Yan, C.; Hu, P.; Wu, X.; Yang, W. MEG3 alleviated LPS-induced intestinal injury in sepsis by modulating miR-129-5p and surfactant protein d. Mediat. Inflamm. 2020, 2020, 8232734. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, Y.; Huang, X.; Guo, X.; Liu, Y.; Zhong, J.; Yuan, J. STAT3-induced upregulation of lncRNA MEG3 regulates the growth of cardiac hypertrophy through miR-361-5p/HDAC9 axis. Sci. Rep. 2019, 9, 460. [Google Scholar] [CrossRef]
- Do, K.N.; Fink, L.; Jensen, T.; Gautier, L.; Parlesak, A. TLR2 controls intestinal carcinogen detoxication by CYP1A1. PLoS ONE 2012, 7, e32309. [Google Scholar] [CrossRef] [PubMed]
- Dann, S.M.; Manthey, C.F.; Le, C.; Miyamoto, Y.; Gima, L.; Abrahim, A.; Cao, A.T.; Hanson, E.M.; Kolls, J.K.; Raz, E.; et al. IL-17A promotes protective IgA responses and expression of other potential effectors against the lumen-dwelling enteric parasite giardia. Exp. Parasitol. 2015, 156, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Ai, L.; Qian, J.; Fang, J.-Y.; Xu, J. Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 2015, 5, 11763. [Google Scholar] [CrossRef] [PubMed]

| Symbol | log2FoldChange | LOG(FDR) | padj | p-Value | Name |
|---|---|---|---|---|---|
| U90926 | 5.826542 | 3.95 | 0.000111 | 8.0 × 106 | cDNA sequence U90926 |
| H19 | 2.107413 | 1.51 | 0.030793 | 7.5 × 103 | H19, imprinted maternally expressed transcript |
| Bvht | 1.132362 | 2.15 | 0.007004 | 1.2 × 103 | Braveheart long non-coding RNA |
| Pvt1 | 1.052021 | 1.66 | 0.021957 | 4.8 × 103 | Plasmacytoma_variant_translocation_1 |
| Meg3 | 1.044219 | 1.60 | 0.025132 | 5.8 × 103 | Maternally expressed 3 |
| Neat1 | −0.009449 | 0.68 | 0.968169 | 9.9 × 101 | Nuclear paraspeckle assembly transcript 1 (non-protein coding) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jimenez-Rondan, F.R.; Ruggiero, C.H.; Cousins, R.J. Long Noncoding RNA, MicroRNA, Zn Transporter Zip14 (Slc39a14) and Inflammation in Mice. Nutrients 2022, 14, 5114. https://doi.org/10.3390/nu14235114
Jimenez-Rondan FR, Ruggiero CH, Cousins RJ. Long Noncoding RNA, MicroRNA, Zn Transporter Zip14 (Slc39a14) and Inflammation in Mice. Nutrients. 2022; 14(23):5114. https://doi.org/10.3390/nu14235114
Chicago/Turabian StyleJimenez-Rondan, Felix R., Courtney H. Ruggiero, and Robert J. Cousins. 2022. "Long Noncoding RNA, MicroRNA, Zn Transporter Zip14 (Slc39a14) and Inflammation in Mice" Nutrients 14, no. 23: 5114. https://doi.org/10.3390/nu14235114
APA StyleJimenez-Rondan, F. R., Ruggiero, C. H., & Cousins, R. J. (2022). Long Noncoding RNA, MicroRNA, Zn Transporter Zip14 (Slc39a14) and Inflammation in Mice. Nutrients, 14(23), 5114. https://doi.org/10.3390/nu14235114






