Serum and Urine Metabolites in Healthy Men after Consumption of Acidified Milk and Yogurt
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Test Products
2.3. Study Design
2.4. GC-MS Analysis with Derivatisation
2.4.1. Sample Preparation
2.4.2. Sample Analysis
2.4.3. Data Pre-Treatment
2.5. Nutrivolatilomics Analysis
2.5.1. Sample Preparation
2.5.2. Sample Analysis
2.5.3. Data Pre-Treatment
2.6. Statistical Analyses
3. Results and Discussion
3.1. Overview of the GC-MS Results with Derivatisation
3.2. Overview of Nutrivolatilomic Results
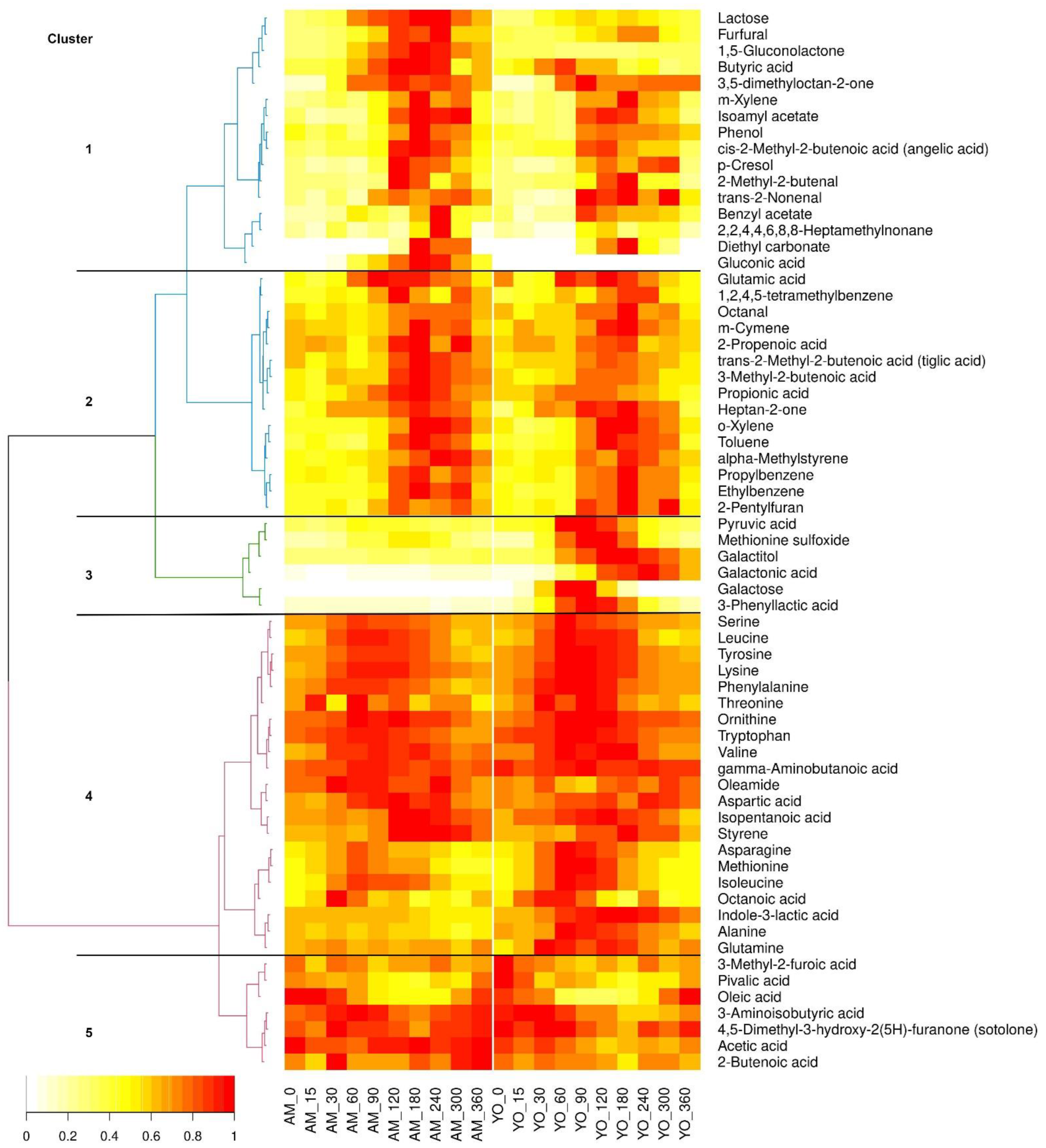
3.3. Kinetic Clustering of Postprandial Metabolites in Serum
3.4. Amino Acids and Their Derivatives
3.5. Carbohydrates and Their Derivatives
3.6. Organic Acids and Oleamide
3.7. Aldehydes
3.8. Esters
3.9. Furans
3.10. Ketons
3.11. Hydrocarbons
3.12. Phenols
3.13. Use of Different Metabolomics Platforms for Biomarker Discovery
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chilton, S.N.; Burton, J.P.; Reid, G. Inclusion of fermented foods in food guides around the world. Nutrients 2015, 7, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Masood, M.I.; Qadir, M.I.; Shirazi, J.H.; Khan, I.U. Beneficial effects of lactic acid bacteria on human beings. Crit. Rev. Microbiol. 2011, 37, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.; Sugrue, I.; Arendt, E.; Hill, C.; Stanton, C.; Ross, R.P. Recent advances in microbial fermentation for dairy and health. F1000Research 2017, 6, 751. [Google Scholar] [CrossRef] [PubMed]
- Walther, B.; Gille, D.; Egger, L. Konsum von Milchprodukten trotz Laktoseintoleranz und Galaktosämie. Schweiz. Z. Für Ernährungsmedizin 2019, 17, 24–27. [Google Scholar]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligne, B.; Ganzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- Savaiano, D.A.; Hutkins, R.W. Yogurt, cultured fermented milk, and health: A systematic review. Nutr. Rev. 2021, 79, 599–614. [Google Scholar] [CrossRef]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health Benefits of Lactic Acid Bacteria (LAB) Fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef]
- Diez-Ozaeta, I.; Astiazaran, O.J. Fermented foods: An update on evidence-based health benefits and future perspectives. Food Res. Int. 2022, 156, 111133. [Google Scholar] [CrossRef]
- Castellone, V.; Bancalari, E.; Rubert, J.; Gatti, M.; Neviani, E.; Bottari, B. Eating Fermented: Health Benefits of LAB-Fermented Foods. Foods 2021, 10, 2639. [Google Scholar] [CrossRef]
- Leeuwendaal, N.K.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. Fermented Foods, Health and the Gut Microbiome. Nutrients 2022, 14, 1527. [Google Scholar] [CrossRef] [PubMed]
- Bonham, M.P.; Linderborg, K.M.; Dordevic, A.; Larsen, A.E.; Nguo, K.; Weir, J.M.; Gran, P.; Luotonen, M.K.; Meikle, P.J.; Cameron-Smith, D.; et al. Lipidomic profiling of chylomicron triacylglycerols in response to high fat meals. Lipids 2013, 48, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Pellis, L.; van Erk, M.J.; van Ommen, B.; Bakker, G.C.M.; Hendriks, H.F.J.; Cnubben, N.H.P.; Kleemann, R.; van Someren, E.P.; Bobeldijk, I.; Rubingh, C.M.; et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics 2012, 8, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Pratico, G.; Scalbert, A.; Vergeres, G.; Kolehmainen, M.; Manach, C.; Brennan, L.; Afman, L.A.; Wishart, D.S.; Andres-Lacueva, C.; et al. A scheme for a flexible classification of dietary and health biomarkers. Genes Nutr. 2017, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Fuchsmann, P.; Tena Stern, M.; Munger, L.H.; Pimentel, G.; Burton, K.J.; Vionnet, N.; Vergeres, G. Nutrivolatilomics of Urinary and Plasma Samples to Identify Candidate Biomarkers after Cheese, Milk, and Soy-Based Drink Intake in Healthy Humans. J. Proteome Res. 2020, 19, 4019–4033. [Google Scholar] [CrossRef]
- Münger, L.; Trimigno, A.; Picone, G.; Freiburghaus, C.; Pimentel, G.; Burton, K.; Vionnet, N.; Capozzi, F.; Pralong, F.; Badertscher, R.; et al. Identification of urinary food intake biomarkers for milk, cheese and soy-based drink by untargeted GC-MS and NMR in healthy humans. J. Proteome Res. 2017, 16, 3321–3335. [Google Scholar] [CrossRef]
- Pimentel, G.; Burton, K.J.; von Ah, U.; Bütikofer, U.; Pralong, F.P.; Vionnet, N.; Portmann, R.; Vergères, G. Metabolic footprinting of fermented milk consumption in serum of healthy men. J. Nutr. 2018, 148, 851–860. [Google Scholar] [CrossRef]
- Kim, J.; Blaser, C.; Portmann, R.; Badertscher, R.; Marmonier, C.; Blot, A.; David, J.; Stoffers, H.; von Ah, U.; Bütikofer, U.; et al. Postprandial Responses on Serum Metabolome to Milk and Yogurt Intake in Young and Older Men. Front. Nutr. 2022, 9, 947. [Google Scholar] [CrossRef]
- Li, K.J.; Brouwer-Brolsma, E.M.; Burton-Pimentel, K.J.; Vergeres, G.; Feskens, E.J.M. A systematic review to identify biomarkers of intake for fermented food products. Genes Nutr. 2021, 16, 5. [Google Scholar] [CrossRef]
- Fuchsmann, P.; Tena Stern, M.; Bischoff, P.; Badertscher, R.; Breme, K.; Walther, B. Development and performance evaluation of a novel dynamic headspace vacuum transfer “In Trap” extraction method for volatile compounds and comparison with headspace solid-phase microextraction and headspace in-tube extraction. J. Chromatogr. A 2019, 1601, 60–70. [Google Scholar] [CrossRef]
- Burton, K.J.; Rosikiewicz, M.; Pimentel, G.; Butikofer, U.; von Ah, U.; Voirol, M.J.; Croxatto, A.; Aeby, S.; Drai, J.; McTernan, P.G.; et al. Probiotic yogurt and acidified milk similarly reduce postprandial inflammation and both alter the gut microbiota of healthy, young men. Br. J. Nutr. 2017, 17, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef] [PubMed]
- Edmands, W.M.B.; Ferrari, P.; Scalbert, A. Normalization to Specific Gravity Prior to Analysis Improves Information Recovery from High Resolution Mass Spectrometry Metabolomic Profiles of Human Urine. Anal. Chem. 2014, 86, 10925–10931. [Google Scholar] [CrossRef]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dormann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef]
- Deporte, R.; Amiand, M.; Moreau, A.; Charbonnel, C.; Campion, L. High-performance liquid chromatographic assay with UV detection for measurement of dihydrouracil/uracil ratio in plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2006, 834, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 12 May 2022).
- Ekstrøm, C.T. MESS: Miscellaneous Esoteric Statistical Scripts. R package version 0.5.7. Available online: https://cran.r-project.org/web/packages/MESS/index.html (accessed on 18 June 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F.; et al. Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies. Mol. Nutr. Food Res. 2018, 63, e1800384. [Google Scholar] [CrossRef]
- Bütikofer, U.; Eberhard, P.; Sieber, R.; Fuchs, D. Über Veränderungen der freien Aminosäuren während der Lagerung von Joghurt. Schweiz. Milchwirtsch. Forsch. 1995, 24, 3–6. [Google Scholar]
- Bos, C.; Metges, C.C.; Gaudichon, C.; Petze, K.J.; Pueyo, M.E.; Morens, C.; Everwand, J.; Benamouzig, R.; Tome, D. Postprandial kinetics of dietary amino acids are the main determinant of their metabolism after soy or milk protein ingestion in humans. J. Nutr. 2003, 133, 1308–1315. [Google Scholar] [CrossRef]
- Trimigno, A.; Münger, L.; Picone, G.; Freiburghaus, C.; Pimentel, G.; Vionnet, N.; Pralong, F.; Capozzi, F.; Badertscher, R.; Vergères, G. GC-MS Based Metabolomics and NMR Spectroscopy Investigation of Food Intake Biomarkers for Milk and Cheese in Serum of Healthy Humans. Metabolites 2018, 8, 26. [Google Scholar] [CrossRef]
- Sun, D.; Li, H.; Song, D.; Zhang, L.; Zhao, X.; Xu, X. Genome, transcriptome and fermentation analyses of Lactobacillus plantarum LY-78 provide new insights into the mechanism of phenyllactate biosynthesis in lactic acid bacteria. Biochem. Biophys. Res. Commun. 2019, 519, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Achilli, C.; Ciana, A.; Minetti, G. The discovery of methionine sulfoxide reductase enzymes: An historical account and future perspectives. BioFactors 2015, 41, 135–152. [Google Scholar] [CrossRef] [PubMed]
- Baxter, J.H.; Lai, C.S.; Phillips, R.R.; Dowlati, L.; Chio, J.J.; Luebbers, S.T.; Dimler, S.R.; Johns, P.W. Direct determination of methionine sulfoxide in milk proteins by enzyme hydrolysis/high-performance liquid chromatography. J. Chromatogr. A 2007, 1157, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Stanstrup, J.; Rasmussen, J.E.; Ritz, C.; Holmer-Jensen, J.; Hermansen, K.; Dragsted, L.O. Intakes of whey protein hydrolysate and whole whey proteins are discriminated by LC–MS metabolomics. Metabolomics 2014, 10, 719–736. [Google Scholar] [CrossRef]
- Smart, T.G.; Stephenson, F.A. A half century of gamma-aminobutyric acid. Brain Neurosci. Adv. 2019, 3, 2398212819858249. [Google Scholar] [CrossRef]
- Cui, Y.; Miao, K.; Niyaphorn, S.; Qu, X. Production of Gamma-Aminobutyric Acid from Lactic Acid Bacteria: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 995. [Google Scholar] [CrossRef]
- Leslie, N.D. Insights into the pathogenesis of galactosemia. Annu. Rev. Nutr. 2003, 23, 59–80. [Google Scholar] [CrossRef]
- Vionnet, N.; Münger, L.H.; Freiburghaus, C.; Burton, K.J.; Pimentel, G.; Pralong, F.P.; Badertscher, R.; Vergères, G. Assessment of lactase activity in humans by measurement of galactitol and galactonate in serum and urine after milk intake. Am. J. Clin. Nutr. 2019, 109, 470–477. [Google Scholar] [CrossRef]
- Fernandez, A.; Ogawa, J.; Penaud, S.; Boudebbouze, S.; Ehrlich, D.; van de Guchte, M.; Maguin, E. Rerouting of pyruvate metabolism during acid adaptation in Lactobacillus bulgaricus. Proteomics 2008, 8, 3154–3163. [Google Scholar] [CrossRef]
- Settachaimongkon, S.; van Valenberg, H.J.; Winata, V.; Wang, X.; Nout, M.J.; van Hooijdonk, T.C.; Zwietering, M.H.; Smid, E.J. Effect of sublethal preculturing on the survival of probiotics and metabolite formation in set-yoghurt. Food Microbiol. 2015, 49, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Trimigno, A.; Boge Lyndgaard, C.; Atladottir, G.A.; Aru, V.; Balling Engelsen, S.; Harder Clemmensen, L.K. An NMR Metabolomics Approach to Investigate Factors Affecting the Yoghurt Fermentation Process and Quality. Metabolites 2020, 10, 293. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Leroy, F. Functional role of yeasts, lactic acid bacteria and acetic acid bacteria in cocoa fermentation processes. FEMS Microbiol. Rev. 2020, 44, 432–453. [Google Scholar] [CrossRef] [PubMed]
- Ostertag, L.M.; Philo, M.; Colquhoun, I.J.; Tapp, H.S.; Saha, S.; Duthie, G.G.; Kemsley, E.K.; de Roos, B.; Kroon, P.A.; Le Gall, G. Acute Consumption of Flavan-3-ol-Enriched Dark Chocolate Affects Human Endogenous Metabolism. J. Proteome Res. 2017, 16, 2516–2526. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, J.A.; Madrid-Gambin, F.; Garcia-Aloy, M.; Andres-Lacueva, C.; Logue, C.; Gallagher, A.M.; Mack, C.; Kulling, S.E.; Gao, Q.; Pratico, G.; et al. Biomarkers of intake for coffee, tea, and sweetened beverages. Genes Nutr. 2018, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Van Dorsten, F.A.; Daykin, C.A.; Mulder, T.P.; Van Duynhoven, J.P. Metabonomics approach to determine metabolic differences between green tea and black tea consumption. J. Agric. Food Chem. 2006, 54, 6929–6938. [Google Scholar] [CrossRef] [PubMed]
- Rabinowitz, J.D.; Enerbäck, S. Lactate: The ugly duckling of energy metabolism. Nat. Metab. 2020, 2, 566–571. [Google Scholar] [CrossRef]
- Collomb, M.; Bühler, T. Analyse de la composition en acides gras de la graisse de lait, I. Optimisation et validation d’une méthode générale à haute résolution. Trav. Chim. Aliment. D’hygiène 2000, 91, 306–332. [Google Scholar]
- Mansson, H.L. Fatty acids in bovine milk fat. Food Nutr. Res. 2008, 52, 1821. [Google Scholar] [CrossRef]
- Gu, Y.; Li, X.; Chen, H.; Guan, K.; Qi, X.; Yang, L.; Ma, Y. Evaluation of FAAs and FFAs in yogurts fermented with different starter cultures during storage. J. Food Comp. Anal. 2021, 96, 103666. [Google Scholar] [CrossRef]
- Vénica, C.I.; Perotti, M.C.; Bergamini, C.V. Organic acids profiles in lactose-hydrolyzed yogurt with different matrix composition. Dairy Sci. Tech. 2014, 94, 561–580. [Google Scholar] [CrossRef]
- Regula, A. Free fatty acid profiles of fermented beverages made from ewe’s milk. Le Lait 2007, 87, 71–77. [Google Scholar] [CrossRef]
- Beshkova, D.; Simova, E.; Frengova, G.; Simov, Z. Production of flavour compounds by yogurt starter cultures. J. Ind. Microbiol. Biotech. 1998, 20, 180–186. [Google Scholar] [CrossRef]
- Jackson, K.G.; Wolstencroft, E.J.; Bateman, P.A.; Yaqoob, P.; Williams, C.M. Acute effects of meal fatty acids on postprandial NEFA, glucose and apo E response: Implications for insulin sensitivity and lipoprotein regulation? Br. J. Nutr. 2005, 93, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Spégel, P.; Danielsson, A.P.H.; Bacos, K.; Nagorny, C.L.F.; Moritz, T.; Mulder, H.; Filipsson, K. Metabolomic analysis of a human oral glucose tolerance test reveals fatty acids as reliable indicators of regulated metabolism. Metabolomics 2009, 6, 56–66. [Google Scholar] [CrossRef]
- Collomb, M.; Sollberger, H.; Bütikofer, U.; Sieber, R.; Stoll, W.; Schaeren, W. Impact of a basal diet of hay and fodder beet supplemented with rapeseed, linseed and sunflowerseed on the fatty acid composition of milk fat. Int. Dairy J. 2004, 14, 549–559. [Google Scholar] [CrossRef]
- Hertli, S.; Zimmermann, P. Molecular interactions between the intestinal microbiota and the host. Mol. Microbiol. 2022, 117, 1297–1307. [Google Scholar] [CrossRef]
- Shafaei, A.; Vamathevan, V.; Pandohee, J.; Lawler, N.G.; Broadhurst, D.; Boyce, M.C. Sensitive and quantitative determination of short-chain fatty acids in human serum using liquid chromatography mass spectrometry. Anal. Bioanal. Chem. 2021, 413, 6333–6342. [Google Scholar] [CrossRef]
- Bondia-Pons, I.; Maukonen, J.; Mattila, I.; Rissanen, A.; Saarela, M.; Kaprio, J.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Hyotylainen, T.; et al. Metabolome and fecal microbiota in monozygotic twin pairs discordant for weight: A Big Mac challenge. FASEB J. 2014, 28, 4169–4179. [Google Scholar] [CrossRef]
- Lepine, G.; Tremblay-Franco, M.; Bouder, S.; Dimina, L.; Fouillet, H.; Mariotti, F.; Polakof, S. Investigating the Postprandial Metabolome after Challenge Tests to Assess Metabolic Flexibility and Dysregulations Associated with Cardiometabolic Diseases. Nutrients 2022, 14, 472. [Google Scholar] [CrossRef]
- Iversen, K.N.; Dicksved, J.; Zoki, C.; Fristedt, R.; Pelve, E.A.; Langton, M.; Landberg, R. The Effects of High Fiber Rye, Compared to Refined Wheat, on Gut Microbiota Composition, Plasma Short Chain Fatty Acids, and Implications for Weight Loss and Metabolic Risk Factors (the RyeWeight Study). Nutrients 2022, 14, 1669. [Google Scholar] [CrossRef] [PubMed]
- Ba, Z.; Lee, Y.; Meng, H.; Kris-Etherton, P.M.; Rogers, C.J.; Lewis, Z.T.; Mills, D.A.; Furumoto, E.J.; Rolon, M.L.; Fleming, J.A.; et al. Matrix Effects on the Delivery Efficacy of Bifidobacterium animalis subsp. lactis BB-12 on Fecal Microbiota, Gut Transit Time, and Short-Chain Fatty Acids in Healthy Young Adults. mSphere 2021, 6, e0008421. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, T.; Koyama, H.; Okada, M.; Ushida, K. Stimulation of butyrate production by gluconic acid in batch culture of pig cecal digesta and identification of butyrate-producing bacteria. J. Nutr. 2002, 132, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Kameue, C.; Tsukahara, T.; Yamada, K.; Koyama, H.; Iwasaki, Y.; Nakayama, K.; Ushida, K. Dietary sodium gluconate protects rats from large bowel cancer by stimulating butyrate production. J. Nutr. 2004, 134, 940–944. [Google Scholar] [CrossRef]
- Anonymous. Pivalic Acid. Available online: https://foodb.ca/compounds/FDB111687 (accessed on 20 April 2022).
- Parada, F.; Duque, C.; Fujimoto, Y. Free and bound volatile composition and characterization of some glucoconjugates as aroma precursors in melón de olor fruit pulp (Sicana odorifera). J. Agric. Food Chem. 2000, 48, 6200–6204. [Google Scholar] [CrossRef]
- Jahan, I.; Tona, M.R.; Sharmin, S.; Sayeed, M.A.; Tania, F.Z.; Paul, A.; Chy, M.N.U.; Rakib, A.; Emran, T.B.; Simal-Gandara, J. GC-MS Phytochemical Profiling, Pharmacological Properties, and In Silico Studies of Chukrasia velutina Leaves: A Novel Source for Bioactive Agents. Molecules 2020, 25, 3536. [Google Scholar] [CrossRef]
- Anonymous. Angelic Acid. Available online: https://hmdb.ca/metabolites/HMDB0029608 (accessed on 21 April 2022).
- Anonymous. Senecioic Acid. Available online: https://foodb.ca/compounds/FDB000736 (accessed on 21 April 2022).
- Anonymous. Tiglic Acid. Available online: https://foodb.ca/compounds/FDB000774 (accessed on 21 April 2022).
- Curoni, P.M.G.; Bosset, J.O. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 2002, 12, 959–984. [Google Scholar] [CrossRef]
- Orhan, H.; van Holland, B.; Krab, B.; Moeken, J.; Vermeulen, N.P.; Hollander, P.; Meerman, J.H. Evaluation of a multi-parameter biomarker set for oxidative damage in man: Increased urinary excretion of lipid, protein and DNA oxidation products after one hour of exercise. Free Radic. Res. 2004, 38, 1269–1279. [Google Scholar] [CrossRef]
- Dennis, E.G.; Keyzers, R.A.; Kalua, C.M.; Maffei, S.M.; Nicholson, E.L.; Boss, P.K. Grape contribution to wine aroma: Production of hexyl acetate, octyl acetate, and benzyl acetate during yeast fermentation is dependent upon precursors in the must. J. Agric. Food Chem. 2012, 60, 2638–2646. [Google Scholar] [CrossRef]
- Anonymous. 2-Pentylfuran. Available online: https://hmdb.ca/metabolites/HMDB0013824 (accessed on 26 April 2022).
- Frank, N.; Dubois, M.; Huertas Perez, J.F. Detection of Furan and five Alkylfurans, including 2-Pentylfuran, in various Food Matrices. J. Chromatogr. A 2020, 1622, 461119. [Google Scholar] [CrossRef]
- Anonymous. Furfural. Available online: https://hmdb.ca/metabolites/HMDB0032914 (accessed on 26 June 2022).
- USDA. Dr. Duke’s Phytochemical and Ethnobotanical Databases. Available online: https://phytochem.nal.usda.gov/phytochem/search (accessed on 2 June 2022).
- Chen, Y.C.; Wu, H.F. Revolving hollow fiber-liquid phase microextraction coupled to GC/MS using electron ionization for quantification of five aromatic hydrocarbon isomers. J. Sep. Sci. 2009, 32, 3013–3019. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.L.; Sparling, M.; Pelletier, L.; Dabeka, R. Styrene in foods and dietary exposure estimates. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess 2018, 35, 2045–2051. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Pan, H.; Yuan, Y.; Zhou, X.; Chen, L.; Wang, X.; Fang, F.; Hu, L.; Xie, Y.; Shen, C. Sera Metabolomics Characterization of Patients at Different Stages in Wuhan Identifies Critical Biomarkers of COVID-19. Front. Cell Infect. Microbiol. 2022, 12, 882661. [Google Scholar] [CrossRef]
- Bernalte, M.J.; Hernandez, M.T. Physical, Chemical, Flavor and Sensory Characteristics of Two Sweet Cherry Varieties Grown In ‘Valle Del Jerte’ (Spain). J. Food Qual. 1999, 22, 403–416. [Google Scholar] [CrossRef]
- Lopez-Blanco, R.; Gilbert-Lopez, B.; Rojas-Jimenez, R.; Robles-Molina, J.; Ramos-Martos, N.; Garcia-Reyes, J.F.; Molina-Diaz, A. Evaluation of processing factors for selected organic contaminants during virgin olive oil production: Distribution of BTEXS during olives processing. Food Chem. 2016, 199, 273–279. [Google Scholar] [CrossRef]
- Grob, K. Mineral oil hydrocarbons in food: A review. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess 2018, 35, 1845–1860. [Google Scholar] [CrossRef]
- Sanchis, Y.; Yusa, V.; Coscolla, C. Analytical strategies for organic food packaging contaminants. J. Chromatogr. A 2017, 1490, 22–46. [Google Scholar] [CrossRef]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef]
- Duranton, F.; Cohen, G.; De Smet, R.; Rodriguez, M.; Jankowski, J.; Vanholder, R.; Argiles, A.; European Uremic Toxin Work, G. Normal and pathologic concentrations of uremic toxins. J. Am. Soc. Nephrol. 2012, 23, 1258–1270. [Google Scholar] [CrossRef]
- Li, K.J.; Burton-Pimentel, K.J.; Brouwer-Brolsma, E.M.; Feskens, E.J.M.; Blaser, C.; Badertscher, R.; Portmann, R.; Vergères, G. Evaluating the Robustness of Biomarkers of Dairy Food Intake in a Free-Living Population Using Single- and Multi-Marker Approaches. Metabolites 2021, 11, 395. [Google Scholar] [CrossRef]
- D’Angelo, S.; Gormley, I.C.; McNamara, A.E.; Brennan, L. multiMarker: Software for modelling and prediction of continuous food intake using multiple biomarkers measurements. BMC Bioinform. 2021, 22, 469. [Google Scholar] [CrossRef] [PubMed]
 milk and
milk and  yogurt; A.U. arbitrary units (● outlier).
yogurt; A.U. arbitrary units (● outlier).
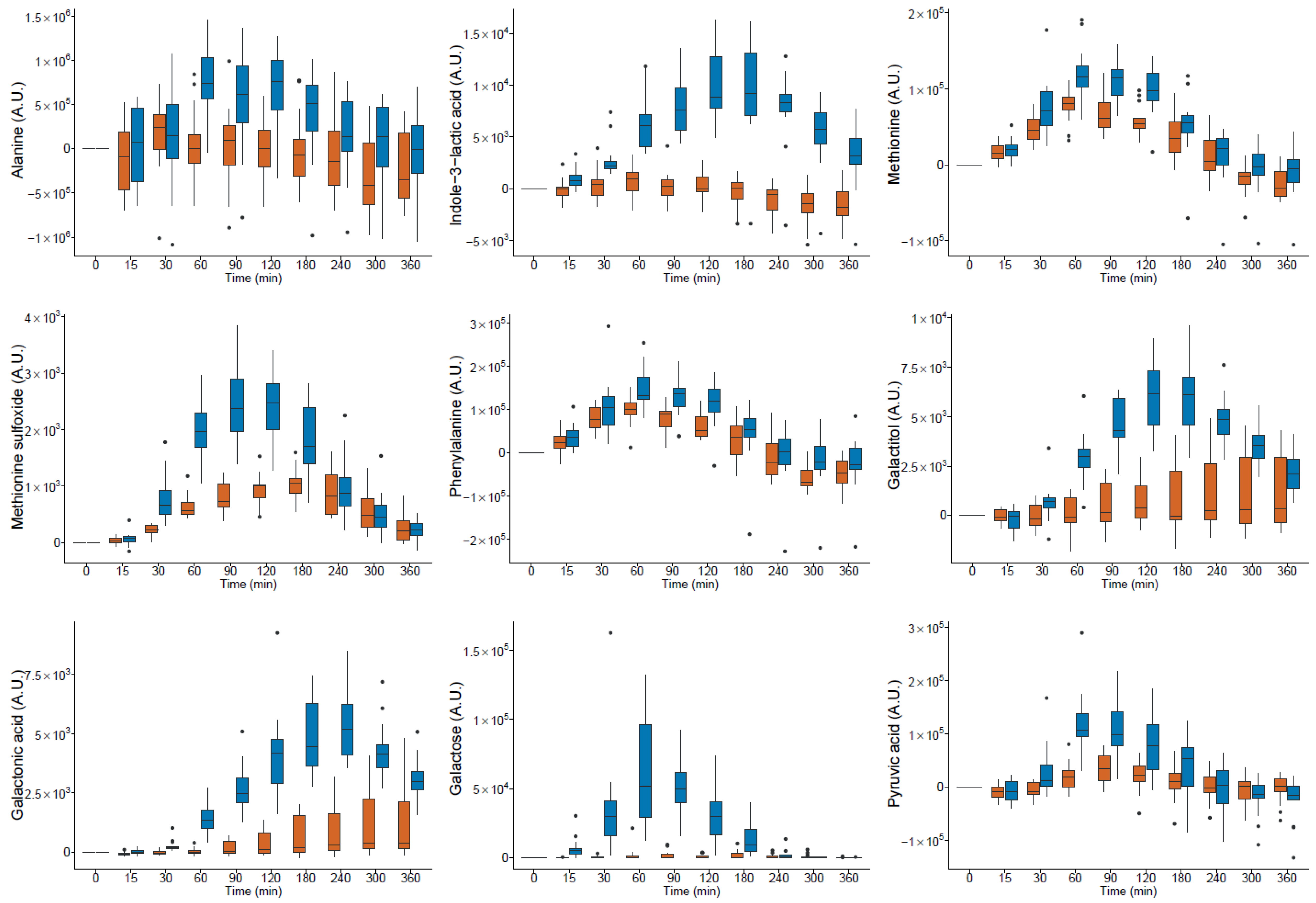
 milk and
milk and  yogurt; A.U. arbitrary units (● outlier).
yogurt; A.U. arbitrary units (● outlier).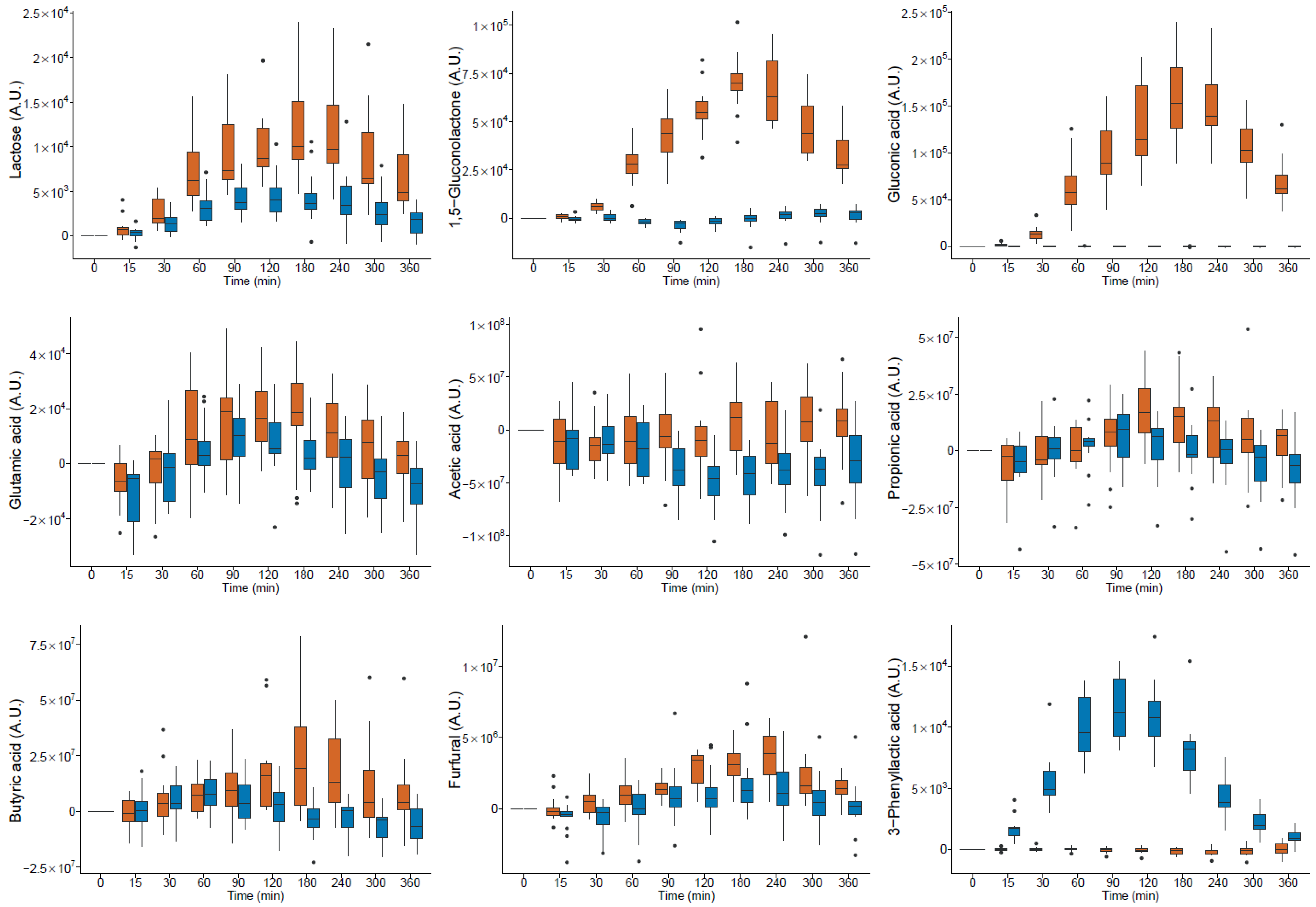
| Compound | HMDB Database Entry | RT | RI | Quantifier Ion | Qualifier Ion | Level ID b | Postprandial Response (iAUC) in Serum after Milk Intake c | Postprandial Response (iAUC) in Serum after Yogurt Intake d | Product with Higher Post-Prandial Response (iAUC) in Serum f | Product with Higher Content in 6 h Urine Pool g | Product with Higher Content h | Postprandial Response (iAUC) in Serum after HFM Intake i | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (min) | ( ) | (m/z) a | (m/z) a | Median | SD | Median | SD | |||||||||
| Amino acids and derivatives | ||||||||||||||||
| Alanine | HMDB0000161 | 14.87 | 1094 | 116 | 190 | 1 | −4.58 × 107 | 1.20 × 108 | 1.42 × 108 | 1.24 × 108 | * | YO * | YO * | YO * | pos * | |
| Asparagine | HMDB0000168 | 27.46 | 1658 | 116 | 231 | 1 | 2.92 × 106 | 4.48 × 106 | * | 6.09 × 106 | 4.41 × 106 | * | YO | YO * | YO * | pos * |
| Aspartic acid | HMDB0000191 | 25.13 | 1508 | 232 | 218 | 1 | 2.52 × 106 | 3.08 × 106 | * | 2.35 × 106 | 2.14 × 106 | * | AM | YO | YO* | pos |
| Glutamic acid | HMDB0000148 | 26.74 | 1607 | 246 | 128 | 1 | 4.00 × 106 | 4.51 × 106 | * | 9.36 × 105 | 3.36 × 106 | AM * | ND | YO | pos | |
| Glutamine | HMDB0000641 | 28.80 | 1764 | 156 | 245 | 2 | 6.02 × 106 | 1.75 × 107 | 1.88 × 107 | 3.05 × 107 | * | YO | ND | ND | pos | |
| Isoleucine | HMDB0000172 | 20.27 | 1286 | 158 | 218 | 1 | 8.20 × 107 | 2.00 × 107 | * | 7.76 × 107 | 3.02 × 107 | * | AM | YO | YO* | pos * |
| Leucine | HMDB0000687 | 19.69 | 1265 | 158 | 232 | 1 | 1.20 × 108 | 3.98 × 107 | * | 1.15 × 108 | 5.41 × 107 | * | AM | YO | YO* | pos * |
| Lysine | HMDB0000182 | 30.64 | 1914 | 174 | 317 | 1 | 3.51 × 107 | 1.98 × 107 | * | 4.46 × 107 | 1.58 × 107 | * | YO | YO* | YO* | pos * |
| Methionine | HMDB0000696 | 25.21 | 1513 | 176 | 128 | 1 | 7.16 × 106 | 5.51 × 106 | * | 1.81 × 107 | 8.09 × 106 | * | YO * | YO | YO* | pos * |
| Phenylalanine | HMDB0000159 | 26.97 | 1623 | 218 | 192 | 1 | 6.44 × 106 | 8.97 × 106 | * | 1.93 × 107 | 1.65 × 107 | * | YO * | YO | YO* | pos * |
| Serine | HMDB0062263 | 21.88 | 1351 | 204 | 218 | 1 | 2.42 × 107 | 1.88 × 107 | * | 2.72 × 107 | 3.16 × 107 | * | YO | YO* | YO* | pos * |
| Threonine | HMDB0000167 | 20.31 | 1288 | 117 | 130 | 1 | 1.38 × 106 | 4.84 × 106 | 2.60 × 106 | 2.14 × 106 | * | YO | YO | YO* | pos | |
| Tryptophan | HMDB0000929 | 33.52 | 2215 | 202 | 203 | 2 | 3.65 × 106 | 2.79 × 107 | 2.03 × 107 | 3.76 × 107 | YO | YO* | YO* | pos | ||
| Tyrosine | HMDB0000158 | 30.82 | 1932 | 218 | 280 | 1 | 4.35 × 107 | 2.31 × 107 | * | 6.12 × 107 | 2.27 × 107 | * | YO | YO* | YO* | pos |
| Valine | HMDB0000883 | 18.10 | 1209 | 144 | 218 | 1 | 1.17 × 108 | 5.87 × 107 | * | 1.21 × 108 | 7.53 × 107 | * | YO | AM | YO* | pos * |
| 3-Aminoisobutyric acid (BAIBA) | HMDB0003911 | 24.17 | 1455 | 248 | 304 | 1 | −3.62 × 104 | 2.07 × 105 | −1.14 × 105 | 2.00 × 105 | * | -YO | AM | ND | pos | |
| 3-Phenyllactic acid | HMDB0000779 | 26.33 | 1579 | 193 | 220 | 1 | −6.67 × 103 | 9.32 × 104 | 2.18 × 106 | 2.46 × 105 | * | YO * | YO* | YO* | neg | |
| gamma-Amino-butanoic acid (GABA) | HMDB0000112 | 25.39 | 1524 | 174 | 304 | 1 | 7.76 × 104 | 5.71 × 105 | 6.27 × 104 | 1.20 × 106 | AM | YO | YO* | pos | ||
| Indole-3-lactic acid | HMDB0000671 | 33.14 | 2172 | 202 | 203 | 1 | −1.62 × 105 | 4.69 × 105 | 2.38 × 106 | 9.47 × 105 | * | YO * | YO* | YO* | neg * | |
| Methionine sulfoxide | HMDB0002005 | 28.95 | 1775 | 128 | 174 | 1 | 2.22 × 105 | 6.61 × 104 | * | 4.38 × 105 | 8.02 × 104 | * | YO * | ND | ND | pos |
| Ornithine | HMDB0000214 | 29.34 | 1806 | 142 | 174 | 1 | 2.32 × 107 | 1.41 × 107 | * | 1.90 × 107 | 1.21 × 107 | * | AM | ND | ND | pos * |
| Lipid compounds | ||||||||||||||||
| Oleamide | HMDB0002117 | 35.28 | 2415 | 144 | 338 | 1 | 2.03 × 106 | 4.79 × 106 | 2.57 × 106 | 5.70 × 106 | YO | ND | ND | neg | ||
| Oleic acid | HMDB0000207 | 33.50 | 2213 | 339 | 117 | 1 | −3.91 × 107 | 3.35 × 107 | * | −2.44 × 107 | 3.00 × 107 | * | -AM | YO | YO * | neg * |
| Carbohydrates and derivatives | ||||||||||||||||
| Lactose | HMDB0041627 | 37.90 | 2664 | 480 | 451 | 1 | 2.65 × 106 | 1.38 × 106 | * | 9.57 × 105 | 6.75 × 105 | * | AM * | AM * | AM *,e | neg |
| Galactose | HMDB0000143 | 30.51 | 1906 | 319 | 205 | 1 | 7.67 × 104 | 6.13 × 105 | * | 6.12 × 106 | 2.38 × 106 | * | YO * | YO * | YO * | ND |
| Galactitol | HMDB0000107 | 30.70 | 1924 | 217 | 307 | 1 | 1.82 × 104 | 5.13 × 105 | 1.49 × 106 | 3.22 × 105 | * | YO * | YO * | ND | pos | |
| Galactonic acid | HMDB0000565 | 31.28 | 1978 | 292 | 333 | 1 | 6.15 × 104 | 2.81 × 105 | * | 1.22 × 106 | 3.62 × 105 | * | YO * | YO * | YO * | neg * |
| Pyruvic acid | HMDB0000243 | 13.33 | 1038 | 174 | 115 | 1 | 3.21 × 106 | 7.33 × 106 | 1.22 × 107 | 1.26 × 107 | * | YO * | YO * | ND | pos | |
| GDL and derivatives | ||||||||||||||||
| 1,5-Glucono-lactone | HMDB0000150 | 30.11 | 1870 | 229 | 189 | 1 | 1.70 × 107 | 2.90 × 106 | * | 2.26 × 105 | 1.03 × 106 | AM * | AM * | ND | neg * | |
| Gluconic acid | HMDB0000625 | 31.33 | 1990 | 292 | 333 | 1 | 3.87 × 107 | 6.99 × 106 | * | −7.88 × 103 | 1.13 × 105 | AM * | AM * | AM * | neg * | |
| Compound | HMDB Database Entry | RT | RI | Quantifier Ion | Postprandial Response (iAUC) in Serum after Milk Intake | Postprandial Response (iAUC) in Serum after Yogurt Intake | Product with Higher Post-Prandial Response (iAUC) in Serum | Product with Higher Content in 6 h Urine Pool | Postprandial Response (iAUC) in Serum after HFM Intake | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (min) | ( ) | (m/z) | Median | SD | Median | SD | |||||||
| Carboxylic acids | |||||||||||||
| Acetic acid | HMDB0000042 | 29.10 | 1476 | 60 | −1.9 × 107 | 8.5 × 109 | −1.2 × 1010 | 7.3 × 109 | * | -YO * | YO | neg* | |
| Propionic acid | HMDB0000237 | 32.64 | 1560 | 74 | 2.8 × 109 | 4.2 × 109 | * | 2.6 × 108 | 4.2 × 109 | AM * | YO | pos * | |
| 2,2-Dimethyl-propionic acid (pivalic acid) | HMDB0041992 | 34.16 | 1596 | 69 | −2.3 × 107 | 8.4 × 107 | −6.2 × 107 | 8.2 × 107 | * | -YO | ND | pos | |
| Butyric acid | HMDB0000039 | 36.38 | 1652 | 73 | 3.4 × 109 | 5.3 × 109 | * | 1.2 × 108 | 2.6 × 109 | AM * | AM | pos | |
| 2-Propenoic acid | HMDB0031647 | 36.63 | 1658 | 72 | 4.0 × 108 | 1.2 × 109 | 3.0 × 108 | 4.8 × 108 | AM | AM | neg | ||
| Isopentanoic acid | HMDB0000718 | 37.98 | 1692 | 87 | 4.5 × 109 | 8.2 × 109 | 3.0 × 109 | 6.3 × 109 | AM | YO | pos | ||
| 2-Butenoic acid | HMDB0010720 | 42.02 | 1800 | 86 | 2.3 × 108 | 1.1 × 109 | −2.8 × 108 | 7.8 × 108 | * | AM | ND | neg | |
| cis-2-Methyl-2-butenoic acid (angelic acid) | HMDB0029608 | 42.26 | 1806 | 100 | 1.0 × 108 | 1.6 × 108 | * | 1.1 × 108 | 8.7 × 107 | * | YO | AM | pos * |
| 3-Methyl-2-butenoic acid (senecioic acid) | HMDB0000509 | 42.80 | 1821 | 100 | 4.1 × 108 | 7.6 × 108 | 3.2 × 108 | 7.6 × 108 | AM | YO | pos * | ||
| trans-2-Methyl-2-but-enoic acid (tiglic acid) | HMDB0001470 | 44.54 | 1869 | 100 | 8.9 × 108 | 1.7 × 109 | 6.1 × 108 | 1.2 × 109 | AM | YO | pos | ||
| Octanoic acid | HMDB0000482 | 51.83 | 2083 | 115 | 1.1 × 109 | 1.1 × 109 | * | 9.6 × 108 | 2.0 × 109 | AM | AM | neg | |
| 3-Methyl-2-furoic acid | NA | 66.70 | 2569 | 126 | −3.2 × 108 | 1.9 × 109 | −1.5 × 109 | 1.8 × 109 | * | AM | YO | pos * | |
| Aldehydes | |||||||||||||
| 2-Methyl-2-butenal (tiglic aldehyde) | HMDB0031512 | 12.74 | 1124 | 84 | 6.7 × 108 | 6.7 × 108 | 7.0 × 108 | 4.7 × 108 | * | YO | YO | pos | |
| Octanal | HMDB0001140 | 21.88 | 1316 | 41 | 3.0 × 107 | 6.5 × 107 | * | 5.4 × 107 | 5.9 × 107 | * | YO | YO | pos |
| trans-2-Nonenal | HMDB0255708 | 32.82 | 1564 | 81 | 1.5 × 108 | 1.6 × 108 | * | 1.9 × 108 | 1.7 × 108 | * | YO | YO | pos * |
| Esters | |||||||||||||
| Diethyl carbonate | HMDB0059844 | 13.27 | 1135 | 91 | 1.4 × 107 | 1.0 × 107 | * | 1.5 × 107 | 1.5 × 107 | YO | ND | pos * | |
| Isoamyl acetate | HMDB0031528 | 13.85 | 1147 | 43 | 5.0 × 107 | 5.1 × 107 | * | 4.3 × 107 | 6.8 × 107 | * | AM | ND | pos * |
| Benzyl acetate | HMDB0031310 | 40.62 | 1762 | 108 | 7.5 × 107 | 9.4 × 107 | * | 6.7 × 107 | 1.6 × 108 | AM | AM * | pos * | |
| Furans | |||||||||||||
| 2-Pentylfuran | HMDB0013824 | 19.01 | 1255 | 81 | 2.0 × 108 | 3.0 × 108 | * | 3.5 × 108 | 2.0 × 108 | * | YO | YO | pos * |
| 2-Furancarboxal-dehyde (furfural) | HMDB0032914 | 30.16 | 1500 | 96 | 7.1 × 108 | 2.8 × 108 | * | 3.3 × 108 | 4.6 × 108 | AM * | AM * | pos * | |
| 4,5-Dimethyl-3-hydroxy-2(5H)-furanone (sotolone) | HMDB0031306 | 56.66 | 2238 | 128 | 1.1 × 108 | 2.1 × 109 | −8.8 × 108 | 4.8 × 109 | AM | AM | pos | ||
| Ketones | |||||||||||||
| Heptan-2-one | HMDB0003671 | 16.90 | 1211 | 114 | 8.2 × 106 | 5.8 × 106 | * | 1.1 × 107 | 5.4 × 106 | * | YO | ND | pos * |
| 3,5-dimethyloctan-2-one | NA | 24.51 | 1374 | 72 | 4.2 × 107 | 2.3 × 107 | * | 4.6 × 107 | 2.1 × 107 | * | YO | ND | neg |
| Hydrocarbons | |||||||||||||
| Toluene | HMDB0034168 | 10.15 | 1067 | 91 | 4.6 × 109 | 4.0 × 109 | * | 4.9 × 109 | 3.4 × 109 | * | YO | YO | pos * |
| Ethylbenzene | HMDB0059905 | 13.93 | 1149 | 91 | 2.7 × 108 | 3.2 × 108 | * | 3.1 × 108 | 1.8 × 108 | * | YO | YO | pos * |
| m-Xylene | HMDB0059810 | 14.62 | 1163 | 91 | 8.1 × 108 | 1.2 × 109 | * | 8.7 × 108 | 4.2 × 108 | * | YO | YO | pos * |
| o-Xylene | HMDB0059851 | 16.90 | 1211 | 91 | 3.6 × 108 | 3.3 × 108 | * | 4.1 × 108 | 2.8 × 108 | * | YO | YO | pos * |
| Propylbenzene | HMDB0059877 | 17.94 | 1233 | 91 | 1.2 × 108 | 1.3 × 108 | * | 1.6 × 108 | 7.8 × 107 | * | YO | AM | pos * |
| 2,2,4,4,6,8,8-Heptamethylnon-ane (isocetane) | NA | 20.08 | 1278 | 57 | 1.9 × 108 | 1.9 × 108 | * | 9.0 × 107 | 2.0 × 108 | * | AM | ND | pos * |
| Styrene | HMDB0034240 | 20.50 | 1287 | 104 | 8.7 × 108 | 8.7 × 108 | * | 7.5 × 108 | 9.0 × 108 | * | AM | AM | pos * |
| m-Cymene | HMDB0037051 | 20.86 | 1294 | 119 | 3.4 × 108 | 3.0 × 108 | * | 3.1 × 108 | 3.0 × 108 | * | AM | YO | pos * |
| alpha-Methyl-styrene | HMDB0059899 | 23.87 | 1360 | 103 | 1.4 × 108 | 9.6 × 107 | * | 1.0 × 108 | 1.4 × 108 | * | AM | YO | pos * |
| 1,2,4,5-tetra-methyl-benzene (durene) | HMDB0244147 | 28.53 | 1463 | 119 | 1.9 × 108 | 3.0 × 108 | * | 2.5 × 108 | 3.4 × 108 | * | YO | YO | pos * |
| Phenols | |||||||||||||
| Phenol | HMDB0000228 | 50.57 | 2044 | 94 | 4.5 × 109 | 4.6 × 109 | * | 3.8 × 109 | 1.1 × 1010 | * | AM | AM | pos |
| p-Cresol | HMDB0001858 | 53.37 | 2131 | 77 | 7.3 × 108 | 2.0 × 109 | * | 5.2 × 108 | 1.5 × 109 | * | AM | AM | pos * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bütikofer, U.; Badertscher, R.; Blaser-Freiburghaus, C.; Fuchsmann, P.; Tena Stern, M.; Kuert, P.A.; Pimentel, G.; Burton-Pimentel, K.J.; Vionnet, N.; Vergères, G. Serum and Urine Metabolites in Healthy Men after Consumption of Acidified Milk and Yogurt. Nutrients 2022, 14, 4794. https://doi.org/10.3390/nu14224794
Bütikofer U, Badertscher R, Blaser-Freiburghaus C, Fuchsmann P, Tena Stern M, Kuert PA, Pimentel G, Burton-Pimentel KJ, Vionnet N, Vergères G. Serum and Urine Metabolites in Healthy Men after Consumption of Acidified Milk and Yogurt. Nutrients. 2022; 14(22):4794. https://doi.org/10.3390/nu14224794
Chicago/Turabian StyleBütikofer, Ueli, René Badertscher, Carola Blaser-Freiburghaus, Pascal Fuchsmann, Mireille Tena Stern, Philipp A. Kuert, Grégory Pimentel, Kathryn Jane Burton-Pimentel, Nathalie Vionnet, and Guy Vergères. 2022. "Serum and Urine Metabolites in Healthy Men after Consumption of Acidified Milk and Yogurt" Nutrients 14, no. 22: 4794. https://doi.org/10.3390/nu14224794
APA StyleBütikofer, U., Badertscher, R., Blaser-Freiburghaus, C., Fuchsmann, P., Tena Stern, M., Kuert, P. A., Pimentel, G., Burton-Pimentel, K. J., Vionnet, N., & Vergères, G. (2022). Serum and Urine Metabolites in Healthy Men after Consumption of Acidified Milk and Yogurt. Nutrients, 14(22), 4794. https://doi.org/10.3390/nu14224794







