Whole Exome Sequencing Study Identifies Novel Rare Risk Variants for Habitual Coffee Consumption Involved in Olfactory Receptor and Hyperphagia
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethic Statement
2.2. Study Participants from UK Biobank
2.3. UK Biobank Genotyping and Imputation for PRS Calculation
2.4. Exome Sequencing, Genotype Calling, and Data Processing in UK Biobank
2.5. Habitual Coffee Consumption Definition
2.6. Filtering and Annotation of Genetic Variants
2.7. Polygenic Risk Scores Calculation for Habitual Coffee Consumption
2.8. Gene-Based Association Analyses
2.9. Verification for Gene-Based Association Analyses Results
3. Results
3.1. Population Characteristic of Habitual Coffee Consumption
3.2. Annotation of Identified Variants
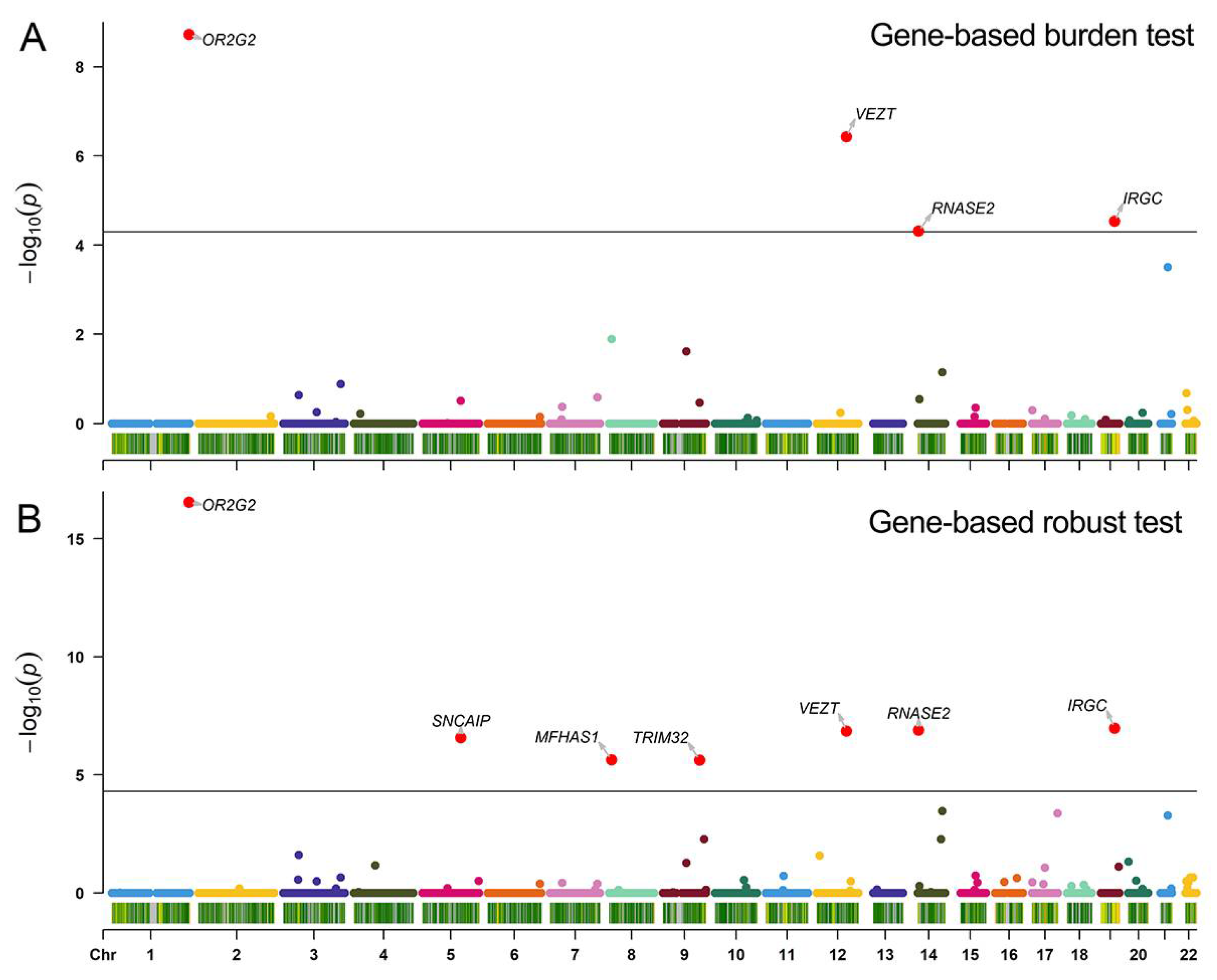
3.3. Gene-Based Burden Test Result
3.4. Verification for Gene-Based Association Analyses Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Socała, K.; Szopa, A. Neuroprotective effects of coffee bioactive compounds: A review. Int. J. Mol. Sci. 2020, 22, 107. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; Byrne, E.M.; Esko, T.; Nalls, M.A.; Ganna, A.; Paynter, N.; Monda, K.L.; Amin, N.; Fischer, K.; Renstrom, F.; et al. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol. Psychiatry 2015, 20, 647–656. [Google Scholar] [PubMed]
- Wierzejska, R. Can coffee consumption lower the risk of alzheimer’s disease and parkinson’s disease? A literature review. Arch. Med. Sci. 2017, 13, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Kolb, H.; Martin, S.; Kempf, K. Coffee and lower risk of type 2 diabetes: Arguments for a causal relationship. Nutrients 2021, 13, 1144. [Google Scholar] [CrossRef]
- Alicandro, G.; Tavani, A.; La Vecchia, C. Coffee and cancer risk: A summary overview. Eur. J. Cancer Prev. 2017, 26, 424–432. [Google Scholar] [CrossRef]
- Rodríguez-Artalejo, F.; López-García, E. Coffee consumption and cardiovascular disease: A condensed review of epidemiological evidence and mechanisms. J. Agric. Food Chem. 2018, 66, 5257–5263. [Google Scholar] [CrossRef]
- Ribeiro, E.M.; Alves, M.; Costa, J.; Ferreira, J.J.; Pinto, F.J.; Caldeira, D. Safety of coffee consumption after myocardial infarction: A systematic review and meta-analysis. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 2146–2158. [Google Scholar] [CrossRef]
- Lee, C.H.; George, O.; Kimbrough, A. Chronic voluntary caffeine intake in male wistar rats reveals individual differences in addiction-like behavior. Pharmacol. Biochem. Behav. 2020, 191, 172880. [Google Scholar] [CrossRef]
- Ágoston, C.; Urbán, R.; Richman, M.J.; Demetrovics, Z. Caffeine use disorder: An item-response theory analysis of proposed dsm-5 criteria. Addict. Behav. 2018, 81, 109–116. [Google Scholar] [CrossRef]
- Cornelis, M.C. Coffee intake. Prog. Mol. Biol. Transl. Sci. 2012, 108, 293–322. [Google Scholar]
- Luciano, M.; Kirk, K.M.; Heath, A.C.; Martin, N.G. The genetics of tea and coffee drinking and preference for source of caffeine in a large community sample of australian twins. Addiction 2005, 100, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Vink, J.M.; Staphorsius, A.S.; Boomsma, D.I. A genetic analysis of coffee consumption in a sample of dutch twins. Twin Res. Hum. Genet. 2009, 12, 127–131. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Laitala, V.S.; Kaprio, J.; Silventoinen, K. Genetics of coffee consumption and its stability. Addiction 2008, 103, 2054–2061. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.T. Quantile-specific heritability may account for gene-environment interactions involving coffee consumption. Behav. Genet. 2020, 50, 119–126. [Google Scholar] [CrossRef]
- Rasmussen, B.B.; Brix, T.H.; Kyvik, K.O.; Brøsen, K. The interindividual differences in the 3-demthylation of caffeine alias cyp1a2 is determined by both genetic and environmental factors. Pharmacogenetics 2002, 12, 473–478. [Google Scholar] [CrossRef]
- Cornelis, M.C.; El-Sohemy, A.; Campos, H. Genetic polymorphism of the adenosine a2a receptor is associated with habitual caffeine consumption. Am. J. Clin. Nutr. 2007, 86, 240–244. [Google Scholar] [CrossRef]
- Amin, N.; Byrne, E.; Johnson, J.; Chenevix-Trench, G.; Walter, S.; Nolte, I.M.; Vink, J.M.; Rawal, R.; Mangino, M.; Teumer, A.; et al. Genome-wide association analysis of coffee drinking suggests association with cyp1a1/cyp1a2 and nrcam. Mol. Psychiatry 2012, 17, 1116–1129. [Google Scholar] [CrossRef]
- Cornelis, M.C.; Monda, K.L.; Yu, K.; Paynter, N.; Azzato, E.M.; Bennett, S.N.; Berndt, S.I.; Boerwinkle, E.; Chanock, S.; Chatterjee, N.; et al. Genome-wide meta-analysis identifies regions on 7p21 (ahr) and 15q24 (cyp1a2) as determinants of habitual caffeine consumption. PLoS Genet. 2011, 7, e1002033. [Google Scholar] [CrossRef]
- Karabegović, I.; Portilla-Fernandez, E.; Li, Y.; Ma, J.; Maas, S.C.E.; Sun, D.; Hu, E.A.; Kühnel, B.; Zhang, Y. Epigenome-wide association meta-analysis of DNA methylation with coffee and tea consumption. Nat. Commun. 2021, 12, 2830. [Google Scholar] [CrossRef]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of gwas discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of gwas discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Michailidou, K.; Lindström, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemaçon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kraja, A.T.; Smith, J.A. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat. Genet. 2016, 48, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Consortium, I.M.S.G. Low-frequency and rare-coding variation contributes to multiple sclerosis risk. Cell 2018, 175, 1679–1687.e1677. [Google Scholar]
- Park, J.; Lucas, A.M.; Zhang, X. Exome-wide evaluation of rare coding variants using electronic health records identifies new gene-phenotype associations. Nat. Med. 2021, 27, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Zhou, S.; Wu, H.; Forgetta, V.; Greenwood, C.M.T.; Richards, J.B. Individuals with common diseases but with a low polygenic risk score could be prioritized for rare variant screening. Genet. Med. 2021, 23, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Esplin, E.D.; Oei, L.; Snyder, M.P. Personalized sequencing and the future of medicine: Discovery, diagnosis and defeat of disease. Pharmacogenomics 2014, 15, 1771–1790. [Google Scholar] [CrossRef]
- Cheng, S.; Cheng, B.; Liu, L. Exome-wide screening identifies novel rare risk variants for major depression disorder. Mol. Psychiatry 2022, 27, 3069–3074. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Szustakowski, J.D.; Balasubramanian, S.; Kvikstad, E. Advancing human genetics research and drug discovery through exome sequencing of the uk biobank. Nat. Genet. 2021, 53, 942–948. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Cirulli, E.T.; White, S. Genome-wide rare variant analysis for thousands of phenotypes in over 70,000 exomes from two cohorts. Nat. Commun. 2020, 11, 542. [Google Scholar] [CrossRef]
- Sun, Y.V.; Sung, Y.J.; Tintle, N.; Ziegler, A. Identification of genetic association of multiple rare variants using collapsing methods. Genet. Epidemiol. 2011, 35 (Suppl. S1), S101–S106. [Google Scholar] [CrossRef]
- Lee, S.; Wu, M.C.; Lin, X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012, 13, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Pan, W. Comparison of statistical tests for disease association with rare variants. Genet. Epidemiol. 2011, 35, 606–619. [Google Scholar] [CrossRef]
- Zhong, V.W.; Kuang, A.; Danning, R.D.; Kraft, P.; van Dam, R.M.; Chasman, D.I.; Cornelis, M.C. A genome-wide association study of bitter and sweet beverage consumption. Hum. Mol. Genet. 2019, 28, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Euesden, J.; Lewis, C.M.; O’Reilly, P.F. Prsice: Polygenic risk score software. Bioinformatics 2015, 31, 1466–1468. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Fuchsberger, C.; Kim, S.; Scott, L. An efficient resampling method for calibrating single and gene-based rare variant association analysis in case-control studies. Biostatistics 2016, 17, 1–15. [Google Scholar] [CrossRef]
- Lee, S.; Emond, M.J.; Bamshad, M.J.; Barnes, K.C.; Rieder, M.J.; Nickerson, D.A.; Christiani, D.C.; Wurfel, M.M.; Lin, X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am. J. Hum. Genet. 2012, 91, 224–237. [Google Scholar] [CrossRef]
- Zhao, Z.; Bi, W.; Zhou, W.; VandeHaar, P.; Fritsche, L.G.; Lee, S. Uk biobank whole-exome sequence binary phenome analysis with robust region-based rare-variant test. Am. J. Hum. Genet. 2020, 106, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Bis, J.C.; Jian, X.; Kunkle, B.W.; Chen, Y. Whole exome sequencing study identifies novel rare and common alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol. Psychiatry 2020, 25, 1859–1875. [Google Scholar] [CrossRef]
- Glezer, I.; Malnic, B. Olfactory receptor function. Handb. Clin. Neurol. 2019, 164, 67–78. [Google Scholar]
- Hiroi, M.; Tanimura, T.; Marion-Poll, F. Hedonic taste in drosophila revealed by olfactory receptors expressed in taste neurons. PLoS ONE 2008, 3, e2610. [Google Scholar] [CrossRef]
- Malnic, B.; Godfrey, P.A.; Buck, L.B. The human olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2584–2589. [Google Scholar] [CrossRef]
- Meng, G.; Inazawa, J.; Ishida, R.; Tokura, K.; Nakahara, K.; Aoki, K.; Kasai, M. Structural analysis of the gene encoding rp58, a sequence-specific transrepressor associated with heterochromatin. Gene 2000, 242, 59–64. [Google Scholar] [CrossRef]
- Boland, E.; Clayton-Smith, J.; Woo, V.G.; McKee, S.; Manson, F.D.; Medne, L.; Zackai, E.; Swanson, E.A.; Fitzpatrick, D.; Millen, K.J.; et al. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase akt3 in postnatal microcephaly and agenesis of the corpus callosum. Am. J. Hum. Genet. 2007, 81, 292–303. [Google Scholar] [CrossRef]
- Bramswig, N.C.; Lüdecke, H.J.; Hamdan, F.F.; Altmüller, J.; Beleggia, F.; Elcioglu, N.H.; Freyer, C.; Gerkes, E.H.; Demirkol, Y.K.; Knupp, K.G.; et al. Heterozygous hnrnpu variants cause early onset epilepsy and severe intellectual disability. Hum. Genet. 2017, 136, 821–834. [Google Scholar] [CrossRef]
- Liu, J.; Li, T.; Thomas, J.M.; Pei, Z.; Jiang, H.; Engelender, S.; Ross, C.A.; Smith, W.W. Synphilin-1 attenuates mutant lrrk2-induced neurodegeneration in parkinson’s disease models. Hum. Mol. Genet. 2016, 25, 672–680. [Google Scholar] [CrossRef]
- Shishido, T.; Nagano, Y.; Araki, M.; Kurashige, T.; Obayashi, H.; Nakamura, T.; Takahashi, T.; Matsumoto, M.; Maruyama, H. Synphilin-1 has neuroprotective effects on mpp(+)-induced parkinson’s disease model cells by inhibiting ros production and apoptosis. Neurosci. Lett. 2019, 690, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X.; Ma, R. Ampk signaling mediates synphilin-1-induced hyperphagia and obesity in drosophila. J. Cell Sci. 2021, 134, jcs247742. [Google Scholar] [CrossRef]
- Li, X.; Tamashiro, K.L.; Liu, Z.; Bello, N.T.; Wang, X.; Aja, S.; Bi, S.; Ladenheim, E.E.; Ross, C.A.; Moran, T.H.; et al. A novel obesity model: Synphilin-1-induced hyperphagia and obesity in mice. Int. J. Obes. 2012, 36, 1215–1221. [Google Scholar] [CrossRef]
- Hillje, A.L.; Beckmann, E.; Pavlou, M.A.; Jaeger, C.; Pacheco, M.P.; Sauter, T.; Schwamborn, J.C.; Lewejohann, L. The neural stem cell fate determinant trim32 regulates complex behavioral traits. Front. Cell. Neurosci. 2015, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Hillje, A.L.; Pavlou, M.A.; Beckmann, E.; Worlitzer, M.M.; Bahnassawy, L.; Lewejohann, L.; Palm, T.; Schwamborn, J.C. Trim32-dependent transcription in adult neural progenitor cells regulates neuronal differentiation. Cell Death Dis. 2013, 4, e976. [Google Scholar] [CrossRef]
- Lionel, A.C.; Tammimies, K.; Vaags, A.K.; Rosenfeld, J.A.; Ahn, J.W.; Merico, D.; Noor, A.; Runke, C.K.; Pillalamarri, V.K.; Carter, M.T.; et al. Disruption of the astn2/trim32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, adhd and other neurodevelopmental phenotypes. Hum. Mol. Genet. 2014, 23, 2752–2768. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Xiong, L.L.; Lu, B.T.; Zhang, H.X.; Zhang, P.; Wang, T.H. Suppression of trim32 enhances motor function repair after traumatic brain injury associated with antiapoptosis. Cell Transplant. 2017, 26, 1276–1285. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, W.; Yang, H.; Zhou, Z.; Zhu, X.; Sun, C.; Liu, Y.; Yu, Z.; Chen, Y.; Wang, Y. Upregulated expression of trim32 is involved in schwann cell differentiation, migration and neurite outgrowth after sciatic nerve crush. Neurochem. Res. 2017, 42, 1084–1095. [Google Scholar] [CrossRef]
- Srinivasan, B.; Samaddar, S.; Mylavarapu, S.V.S.; Clement, J.P.; Banerjee, S. Homeostatic scaling is driven by a translation-dependent degradation axis that recruits mirisc remodeling. PLoS Biol. 2021, 19, e3001432. [Google Scholar] [CrossRef]
- Kudryashova, E.; Wu, J.; Havton, L.A.; Spencer, M.J. Deficiency of the e3 ubiquitin ligase trim32 in mice leads to a myopathy with a neurogenic component. Hum. Mol. Genet. 2009, 18, 1353–1367. [Google Scholar] [CrossRef]
- Derkach, A.; Zhang, H.; Chatterjee, N. Power analysis for genetic association test (pageant) provides insights to challenges for rare variant association studies. Bioinformatics 2018, 34, 1506–1513. [Google Scholar] [CrossRef]
- Lipchock, S.V.; Spielman, A.I.; Mennella, J.A.; Mansfield, C.J.; Hwang, L.D.; Douglas, J.E.; Reed, D.R. Caffeine Bitterness is Related to Daily Caffeine Intake and Bitter Receptor mRNA Abundance in Human Taste Tissue. Perception 2017, 46, 245–256. [Google Scholar] [CrossRef]
- Wei, W.; Cheng, B.; He, D.; Zhao, Y.; Qin, X.; Cai, Q.; Zhang, N.; Chu, X.; Shi, S.; Zhang, F. Identification of Human Brain Proteins for Bitter-Sweet Taste Perception: A Joint Proteome-Wide and Transcriptome-Wide Association Study. Nutrients 2022, 14, 2177. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.S.; Hwang, L.D.; Zhong, V.W.; An, J.; Gharahkhani, P.; Breslin, P.A.S.; Wright, M.J.; Lawlor, D.A.; Whitfield, J.; MacGregor, S.; et al. Understanding the role of bitter taste perception in coffee, tea and alcohol consumption through Mendelian randomization. Sci. Rep. 2018, 8, 16414. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; van Dam, R.M. Genetic determinants of liking and intake of coffee and other bitter foods and beverages. Sci. Rep. 2021, 11, 23845. [Google Scholar] [CrossRef] [PubMed]
| n = 20,566 | Mean ± SD | Range |
|---|---|---|
| Age, years | 56.55 ± 7.94 | 40–70 |
| Coffee intake, cups/day | 2.14 ± 2.10 | 0–10 |
| Coffee intake, PRS | 0.01 ± 0.01 | −0.02–0.04 |
| Smoking, frequency/day | 5.47 ± 9.64 | 0–80 |
| Alcohol use, frequency/week | 8.76 ± 9.44 | 0–235 |
| Energy | 8861.30 ± 3066.13 | 1009.84–41,830.10 |
| BMI | 27.04 ± 4.56 | 15.2–63.4 |
| TDI | −1.62 ± 2.68 | −6.26–9.64 |
| Gene | No. of Marker Test | PSKAT Bonferroni adjust | PSKAT Robust Bonferroni adjust |
|---|---|---|---|
| OR2G2 | 5 | 1.88 × 10−9 | 2.91 × 10−17 |
| VEZT | 3 | 3.72 × 10−7 | 1.41 × 10−7 |
| IRGC | 6 | 2.92 × 10−5 | 1.07 × 10−7 |
| RNASE2 | 2 | 4.85 × 10−5 | 1.29 × 10−7 |
| SNCAIP | 6 | / | 2.72 × 10−7 |
| MFHAS1 | 2 | / | 2.32 × 10−6 |
| TRIM32 | 5 | / | 2.42 × 10−6 |
| SNP | Gene | Chromosome | REF | ALT | GWAS P |
|---|---|---|---|---|---|
| rs12737801 | OR2G2 | 1 | C | G | 0.002 |
| rs1151687 | OR2G2 | 1 | G | C | 0.002 |
| rs201317857 | VEZT | 12 | C | A | 0.020 |
| rs34439296 | IRGC | 19 | C | T | 0.008 |
| rs346049 | IRGC | 19 | C | T | 0.011 |
| rs55712196 | SNCAIP | 5 | G | C | 0.028 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, B.; Pan, C.; Cheng, S.; Meng, P.; Liu, L.; Wei, W.; Yang, X.; Jia, Y.; Wen, Y.; Zhang, F. Whole Exome Sequencing Study Identifies Novel Rare Risk Variants for Habitual Coffee Consumption Involved in Olfactory Receptor and Hyperphagia. Nutrients 2022, 14, 4330. https://doi.org/10.3390/nu14204330
Cheng B, Pan C, Cheng S, Meng P, Liu L, Wei W, Yang X, Jia Y, Wen Y, Zhang F. Whole Exome Sequencing Study Identifies Novel Rare Risk Variants for Habitual Coffee Consumption Involved in Olfactory Receptor and Hyperphagia. Nutrients. 2022; 14(20):4330. https://doi.org/10.3390/nu14204330
Chicago/Turabian StyleCheng, Bolun, Chuyu Pan, Shiqiang Cheng, Peilin Meng, Li Liu, Wenming Wei, Xuena Yang, Yumeng Jia, Yan Wen, and Feng Zhang. 2022. "Whole Exome Sequencing Study Identifies Novel Rare Risk Variants for Habitual Coffee Consumption Involved in Olfactory Receptor and Hyperphagia" Nutrients 14, no. 20: 4330. https://doi.org/10.3390/nu14204330
APA StyleCheng, B., Pan, C., Cheng, S., Meng, P., Liu, L., Wei, W., Yang, X., Jia, Y., Wen, Y., & Zhang, F. (2022). Whole Exome Sequencing Study Identifies Novel Rare Risk Variants for Habitual Coffee Consumption Involved in Olfactory Receptor and Hyperphagia. Nutrients, 14(20), 4330. https://doi.org/10.3390/nu14204330






